Abstract
Objective
To develop a single nucleotide polymorphism- and informatics-based non-invasive prenatal test that detects sex chromosome aneuploidies early in pregnancy.
Methods
Fifteen aneuploid samples, including thirteen 45,X, two 47,XXY, and one 47,XYY, along with 185 euploid controls, were analyzed. Cell-free DNA was isolated from maternal plasma, amplified in a single multiplex PCR assay that targeted 19,488 polymorphic loci covering chromosomes 13, 18, 21, X, and Y, and sequenced. Sequencing results were analyzed using a Bayesian-based maximum likelihood statistical method to determine copy number of interrogated chromosomes, calculating sample-specific accuracies.
Results
Of the samples that passed a stringent quality control metric (93%), the algorithm correctly identified copy number at all five chromosomes in all 187 samples, for 934/935 correct calls as early as 9.4 weeks of gestation. We detected 45,X with 91.7% sensitivity (CI: 61.5-99.8%) and 100% specificity (CI: 97.9-100%), and 47,XXY and 47,XYY. The average calculated accuracy was 99.78%.
Conclusion
This method non-invasively detected 45,X, 47,XXY, and 47,XYY fetuses from cfDNA isolated from maternal plasma with high calculated accuracies, and thus offers a non-invasive method with the potential to function as a routine screen allowing for early prenatal detection of rarely diagnosed yet commonly occurring sex aneuploidies.
Introduction
During the last two decades, noninvasive prenatal screening tests based on biochemical analysis of maternal serum and/or ultrasonography have been increasingly adopted into clinical practice to identify women at increased risk of carrying a fetus with a chromosomal aneuploidy. Until recently, screening focused primarily on autosomal trisomies: detection of trisomy 21 (Down syndrome), which is seen in approximately one in 600 live births, and to a lesser extent trisomy 18 (Edwards syndrome) and trisomy 13 (Patau syndrome). However, the combined at-birth prevalence of sex chromosome aneuploidies (aneuploidies involving the X and/or Y chromosomes, including Turner syndrome [45,X], Klinefelter syndrome [47,XXY], 47,XYY, and 47,XXX) is higher than that of the autosomal aneuploidies, occurring in approximately one in 400 live births.1 Neither Turner syndrome nor the sex chromosome trisomies are consistently detected using the current non-invasive screening approaches.2,3 Thus, diagnosis of sex chromosome abnormalities generally occurs during pregnancy when invasive diagnostic testing is completed, often for other reasons, or after the child is born and presents with signs and symptoms, which may not occur until the teen years or when there are eventual reproductive issues.4 Although there are challenges which accompany the diagnosis, there are now several articles that demonstrate the benefits of early detection and treatment, as well as neurodevelopmental progression in Turner and Klinefelter syndromes.5,6 A non-invasive prenatal test that expands clinical coverage to include the X and Y chromosomes thus has the potential to offer significant value to parents and physicians in the form of early detection and the choice of possible biological treatments.5,6
Recent advances based on genetic analysis of cell-free DNA (cfDNA, a mixture of maternal and fetal cfDNA) isolated from maternal plasma provide the opportunity to non-invasively detect fetal aneuploidies, including sex chromosome aneuploidies, early in pregnancy.7-9 Current methods detect trisomy 21 and trisomy 18 with good accuracy; however, trisomy 13 and sex chromosome aneuploidy detection rates and sensitivities are decreased;10-22 for example, reported Monosomy X sensitivities for commercially available methods have reached 94.4%, less than the up to >99% reported for trisomies 18 and 21.13,18,23-26 This is thought to be partially due to highly variable amplification of these chromosomes.27,28 Recent reports indicated that GC bias correction techniques may correct for these issues. However, this has only proven effective for trisomy 13 detection,15 whereas accurate detection of X chromosome copy number continues to prove problematic. This is exacerbated at early gestational ages (GAs) when the fetal cfDNA fraction in maternal plasma is more likely to be low, and the signal-to-noise ratio is smaller.
We developed a novel non-invasive prenatal aneuploidy test that interrogates single-nucleotide polymorphisms (SNPs).29-32 The method involves massively multiplexed polymerase chain reaction (PCR) amplification of cfDNA isolated from maternal plasma, targeting 19,488 SNPs, followed by high-throughput sequencing. This SNP-based method employs a novel algorithm called Next-generation Aneuploidy Test Using SNPs (NATUS), which incorporates parental genotypic data, known inheritance patterns, and complex data models to correctly interpret abnormally distributed data; this is especially relevant for the X chromosome, resulting in consistently accurate copy number determination across all interrogated chromosomes.32
This report focuses on the use of a SNP-based non-invasive method to identify sex chromosome aneuploidies, which have proven difficult to accurately detect.
Methods
Subjects and Sample Collection
The cohort of 201 pregnancies analyzed here included thirteen 45,X, two 47,XXY, one 47,XYY, and 185 euploid samples. Pregnant couples were enrolled at selected prenatal care centers under an Institutional Review Board- or NHS Research Ethics Committee-approved protocol.33 Women were at least 18 years of age, had singleton pregnancies, and signed an informed consent. All samples with known mosaicism, autosomal trisomy, or triploidy were excluded from analysis, and all available samples with known sex chromosome aneuploidy were included. Copy number on all samples was verified through standard invasive diagnostic testing or genetic testing of cord blood, buccal sample, saliva, or products of conception.
Sample Preparation and Measurement
20-40mL of blood were collected from each woman into Cell-free DNA BCT™ tubes (Streck). Plasma (between 7 and 20mL) was isolated from each sample via a double centrifugation protocol of 2000 g for 20 min, followed by 3220 g for 30 min, with supernatant transfer following the first spin. Cell-free DNA was isolated from 7-10mL plasma using the QIAGEN QIAamp Circulating Nucleic Acid kit and eluted in 50uL TE buffer. Maternal genomic DNA was isolated from the buffy coat obtained following the first centrifugation, and paternal genomic DNA was prepared from either a blood or buccal swab sample.
Samples were pre-amplified for 15 cycles using polymerase chain reaction (PCR) and 19,488 target-specific assays. Then, an aliquot was transferred to a second nested 15-cycle PCR reaction. Finally, samples were prepared for sequencing by adding barcoded tags in a third round of 12-cycle PCR reaction. Thus, 19,488 targets were amplified in a single reaction; targets included single nucleotide polymorphisms (SNPs) from chromosomes 13, 18, 21, X, and Y. SNPs targeted from the Y chromosome are in the homologous non-recombining regions of the X and Y chromosomes where the loci are common to the two chromosomes. Amplicons were sequenced using an Illumina HiSeq 2000 sequencer. Parental samples were measured using the same multiplex PCR protocol, but sequenced at a lower depth-of-read (approximately 20%) than that for cfDNA analysis.
NATUS Methodology and Data Analysis
Sequence alignment to the genome was performed using a proprietary algorithm adapted from the Novoalign (Novocraft, Selangor, Malaysia) commercial software package. A chromosome copy number classification algorithm was implemented in MATLAB (MathWorks, Natick, MA, USA) leveraging a proprietary statistical algorithm termed Next-generation Aneuploid Test Using SNPs (NATUS). The NATUS algorithm is an advanced version of the Parental Support™ algorithm.29,31,32 Briefly, NATUS considers parental genotypes and crossover frequency data34,35 to calculate in silico the expected allele distributions for 19,488 SNPs and billions of possible fetal genotypes based on recombination sites in the parent chromosomes. It compares these predicted distributions to actual allelic distributions as measured from the cfDNA sample, employing a Bayesian-based Maximum Likelihood approach to determine the relative likelihood of each hypothesis and fetal fraction given the observed data: it calculates the likelihood of each copy number hypothesis (monosomy, disomy, or trisomy, for which there are numerous sub-hypotheses based on recombination sites), sums the likelihoods of each copy number sub-hypothesis, and calls the hypothesis with the maximum likelihood as the copy number and fetal fraction. The maximum likelihood represents the sample-specific calculated accuracy. NATUS takes into account numerous QC metrics. These include identifying sub-optimal lab or sequencing results, estimating the amount of total starting DNA, determining the fetal fraction, and calculating the extent to which the measured cfDNA data fits expected distributions. Here, a fetal copy number call was not made with <3.5% fetal fraction, <1,500 genome equivalents of input DNA, or >0.2% contamination. All nine hypotheses (46,XX; 46,XY; trisomy 13; trisomy 18; trisomy 21; 45,X; 47,XXX; 47,XXY; and 47,XYY) were considered by the algorithm as possible fetal genotypes for each sample in this cohort. It is important to note that although trisomy 13, trisomy 18, and trisomy 21 samples were excluded from this analysis, the algorithm called copy number at all five chromosomes (13, 18, 21, X, and Y; Table S1). Combining the maximum likelihood ratio with a priori risk can generate a risk score that is analogous to that generated by serum screening. Paternal genomic samples, when available, were included in the NATUS analysis; in the absence of a paternal sample, the NATUS algorithm considers population allele frequencies.34 To determine test performance in the absence of paternal samples, a parallel analysis was performed on all samples excluding paternal genotypic information. Non-paternity will be detected by the NATUS algorithm; however, non-paternity was not identified in this sample cohort.
Results
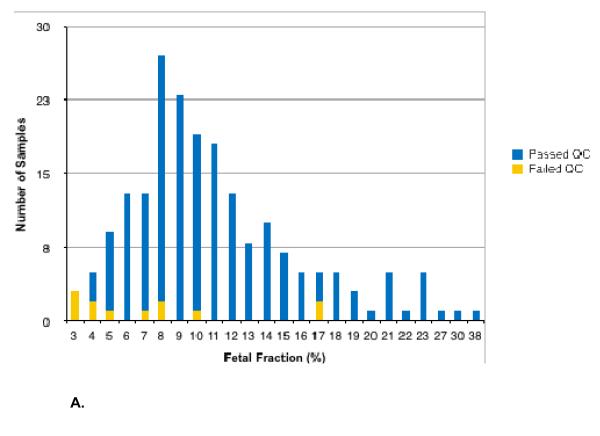
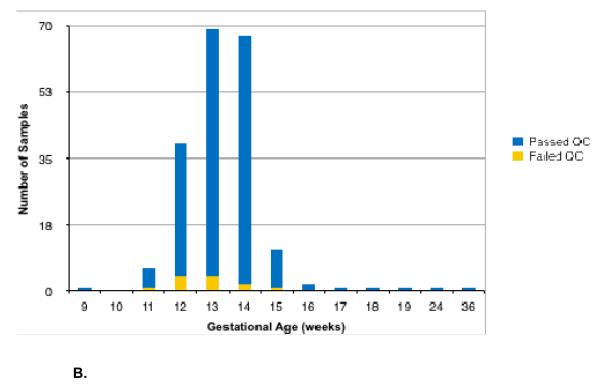
The mean GA was 13.2 and 15.3 weeks for euploid and aneuploid samples, respectively (overall range: 9.4-36.4 weeks). The mean measured fetal fraction was 10.9% and 12.1% for euploid and aneuploid samples, respectively (overall range: 2.9-37.7%). Of the 201 analyzed samples, fourteen (7.0%) did not pass quality control (QC). Excluding the sex chromosome trisomies (i.e. only considering the following hypotheses: 46,XX; 46,XY; trisomy 13; trisomy 18; trisomy 21; and 45,X) reduces this rate to 6.0% (12/201). Figure 1 shows the distribution of samples that passed QC as a function of fetal fraction (Figure 1A) or gestational age (Figure 1B). The method accurately called copy number at all five chromosomes in 186 of the 187 samples that passed QC, for 934/935 correct calls as early as 9.4 weeks of gestation (Table S1) and as low as 4% fetal fraction (Figure 1). The sensitivity for Monosomy X detection was 91.7% (CI: 61.5-99.8%). The three sex chromosome trisomy cases were called correctly; due to the small sample size, sensitivities are not reported. The average calculated accuracy across all five chromosomes for samples that passed QC was 99.78%. When paternal genotypic samples are excluded from analysis, one additional sample (0.5%) did not pass QC, and the average calculated accuracy was 99.76%.
Figure 1.
Histogram of samples stratified by fetal fraction (Figure 1A) and gestational age (Figure 1B).
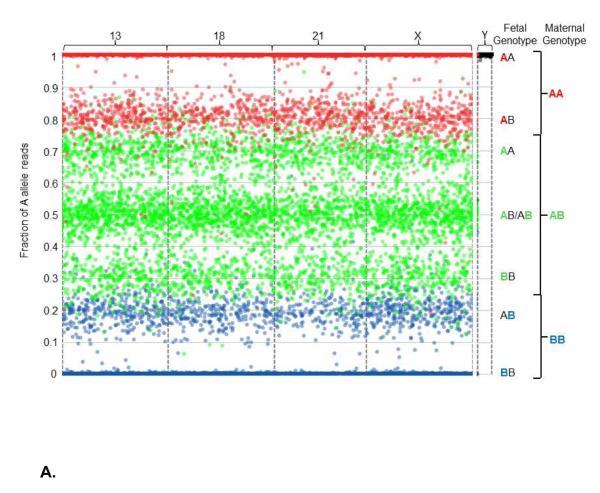
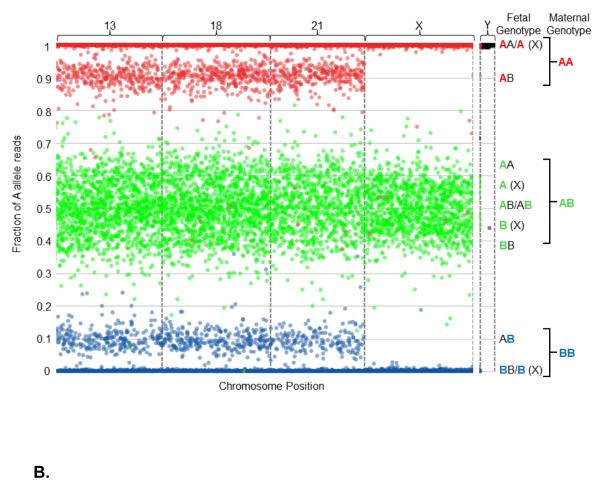
The data can be visualized graphically (Figure 2). Note that this is not how the algorithm makes copy number calls, but is one method for visualizing the data that result in specific ploidy calls. Specifically, Figure 2 includes graphical representations of the sequencing data obtained from one euploid sample (Figure 2A), one 45,X sample (Figure 2B), and one 47,XXY sample (Figure 2C). These are described in detail in the Figure 2 legend.
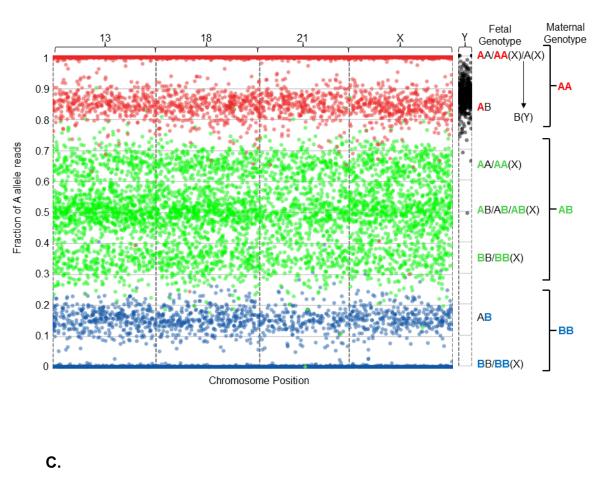
Figure 2.
Graphical representation of sequencing data obtained from one euploid (Figure 2A), one Monosomy X (Figure 2B), and one 47,XXY (Klinefelter syndrome, Figure 2C) sample. All interrogated SNPs are assumed to be dimorphic and are designated as A and B for simplicity. Briefly, for each graph the number of A allele reads as a fraction of the total reads is plotted (y-axis) against the position of each of the several thousand interrogated SNPs on the chromosomes-of-interest (x-axis). The x-axis represents the linear position of each SNP along the chromosome, and each spot corresponds to a single SNP. As plasma cfDNA is a mixture of fetal and maternal cfDNA, the vertical position of each spot represents the sum of the contribution of both fetal and maternal allele reads, and is a function of the fetal fraction. To more readily visualize the maternal and fetal contributions, the spots are colored according to maternal genotype: SNPs for which the mother is homozygous for the A allele (AA) are colored red, SNPs for which the mother is homozygous for the B allele (BB) are colored blue, and SNPs for which the mother is heterozygous (AB) are colored green. Since the majority of plasma cfDNA is maternal in origin, the spots mainly distribute according to maternal genotype. The contribution of fetal allele reads results in segregation into distinct clusters. Because the loci targeted on the Y chromosome are homologous to loci on the X chromosome, but differ by one nucleotide, probes hybridize to both chromosomes. However, the targeted alleles have chromosomally distinct, non-dimorphic identities, so are not color-coded; all alleles from the X chromosome are assigned as A alleles, and all alleles from the Y chromosome are assigned as B alleles. Interrogated chromosomes are indicated above the plot. Fetal and maternal genotypes at individual SNPs are indicated to the right of the plot, with the maternal contribution to the fetal genotype color-coded. A. Euploidy, 38% fetal cfDNA fraction. The presence of three green clusters in the center of the plot (around 0.31, 0.50, and 0.69), as well as the presence of two red (around 1 and 0.81) and two blue (around 0 and 0.19) clusters, indicate the presence of two fetal chromosomes. Thus, this fetus has two copies of chromosomes 13, 18, 21, and X. For the Y chromosome, the A alleles are tightly associated with the upper limit of the plot, indicating the absence of a Y chromosome. Together, this indicates a 46,XX fetal chromosomal complement. B. Monosomy X, 18% fetal cfDNA fraction. The center trio of green clusters (centered around 0.41, 0.50, and 0.59) condense towards the center of the plot, and the red and blue peripheral clusters (centered around 0.91 and 0.09, respectively) regress towards the limits of the plot, due to the decreased contribution of fetal alleles to the overall number of A allele reads. Although copy number is difficult to identify by eye using this method at lower fetal fractions, the NATUS algorithm is able to make highly accurate copy number calls as low as 4% fetal fraction. Here, the pattern indicates two copies of chromosomes 13, 18, and 21. However, for the X chromosome, the center trio of clusters is replaced with a duo of clusters and the peripheral red and blue clusters (around 0.91 and 0.09, respectively) are absent, indicating the presence of a single fetal X chromosome. Additionally, the Y chromosome cluster remains tightly associated with the upper limit of the plot, indicating the lack of a Y chromosome. Together, this indicates a 45,X fetal chromosomal complement. C. Klinefelter syndrome, 30% fetal cfDNA fraction. For the autosomal chromosomes the typical “two-chromosome” cluster distribution is apparent. However, this pattern also appears for the X chromosome, indicating the presence of two copies. In the absence of reads from the Y chromosome, the fetal chromosomal complement would be identified as 46,XX. However, the presence of B alleles from the Y chromosome shifts the cluster downward, indicating the presence of a single Y chromosome (based on the distribution of the reads). Together, this indicates a fetal chromosomal complement of 47,XXY. The method is similarly able to call 47,XYY using SNPs from the homologous non-recombining regions of the X and Y chromosomes.
Discussion
We describe here accurate detection of fetal chromosomal copy number at chromosomes 13, 18, 21, X, and Y using a non-invasive SNP-based approach coupled with NATUS analysis. We detected 45,X, 47,XXY, and 47,XYY, for 934 of 935 accurate copy number calls in 187 samples and with one false negative.
Serendipitous detection of sex chromosome disorders was not uncommon when prenatal genetic diagnosis routinely involved testing by invasive procedures. This is currently much less common now that standard care generally involves prior prenatal screening, characteristically maternal serum analyte and fetal ultrasound (including NT) analysis.36 Nevertheless, these disorders are remain underdiagnosed. NIPT thus offers parents who desire the information an opportunity for prenatal detection and, as a result, early treatment that can be beneficial when initiated in the first year of life.5,6,37-41
While existing NIPT methods demonstrate high sensitivity and specificity when detecting autosomal trisomies, none accomplishes similar levels of accuracy with sex chromosome aneuploidy detection. This is partially attributed to variable amplification that results from different guanosine-cytosine (GC) levels in chromosomes 13 and X as compared to chromosomes 21 and 18,10,27,28 and is particularly problematic for methods that require a reference chromosome. The NATUS method uses SNP measurements to determine chromosome copy number, and does not require a reference chromosome. This obviates amplification bias issues that can reduce detection rates of sex chromosome aneuploidies. Specifically, relative amplification of the two alleles at one SNP locus, whose DNA sequence differs by only one base pair, is inherently more consistent than the relative amplification of unrelated chromosomal regions, which typically have little-to-no nucleotide correlation. Another SNP-based method identifies copy number by comparing fetal to maternal SNP ratios between target and reference chromosomes.42 Although the use of SNPs may mitigate the chromosome-to-chromosome amplification variability, the need for a reference chromosome partly negates this advantage. Additionally, the study only interrogated chromosome 21, so how this method performs on other chromosomes was not examined.
Recently, GC correction was shown to improve trisomy 13 detection.15 That GC correction has not been reported to improve X chromosome quantification suggests that measurement may be complicated by biological phenomena unique to the X chromosome. One possible explanation for anomalous behavior is X inactivation, which may affect how this chromosome is represented in the cfDNA fraction. It’s been postulated that cfDNA is enriched for nucleosome-bound fragments,18 and specific histone variants are differentially enriched on or excluded from the inactivated X chromosome;43-45 this could result in differential degradation and, consequently, representation of this chromosome in the cfDNA fraction. This or other biological factors, such as maternal Monosomy X mosaicism, that affect a chromosome’s cfDNA background signal would be expected to complicate measurement. Accurate detection of chromosomes with anomalous behaviors will thus rely on the ability to model and correct for such behavior.
By utilizing genotypic rather than purely quantitative information, the NATUS algorithm can employ data modeling techniques that measure and interpret a large amount of data. In addition to measuring SNPs, which contain qualitative (genotypic) and quantitative (read count) information, the NATUS algorithm also measures maternal and, when available, paternal genotypic data. Incorporating parental data allows the model to correct for non-standard parental genomes, for example in cases of low-level maternal Monosomy X mosaicism. Specifically, the hypotheses are created in silico given the parental data and recombination data34,35 using complex models of the expected data distributions. Bayesian statistics are utilized to compare possible fetal copy number states to the actual sequencing data, and a Maximum Likelihood approach determines the hypothesis which best fits the actual data. Thus, the NATUS algorithm takes into account cases with complex biology. Ultimately, this allows the algorithm to precisely identify chromosome copy number when the chromosomes do not behave in a straightforward manner, even with poor quality or lower fetal cfDNA fractions.
Empirically, differentiating monosomy from disomy is significantly more straightforward than differentiating disomy from trisomy. This is because the expected allele distributions for a disomic chromosome are less well-differentiated from a trisomic than from a monosomic chromosome. This may contribute to the slight increase in redraw rate when considering sex chromosome trisomies. In this data set, the NATUS algorithm returned no result for 12/201 (6.0%) of the samples due to low fetal fraction or poor DNA quality. An additional two samples (1.0%), which returned a result for the autosomal trisomies and Monosomy X, did not return a result for sex chromosome trisomies. When paternal genotypic information is available, it contributes additional information to the fetal genotype probabilities, increasing the overall power of the test to distinguish from among the modeled fetal ploidy states, especially at low fetal fraction. When the cohort is analyzed without the benefit of the paternal information, the redraw rate when considering only autosomal trisomies and Monosomy X is 13/201 (6.5%), and including sex chromosome trisomies increases the redraw rate to 15/201 (7.5%). When only considering the 113 cases with father sample and including this paternal data in the analysis, the redraw rate when considering the autosomal trisomies and Monosomy X is 5/113 (4.4%), with no additional redraws when including the sex chromosome trisomies. Thus, inclusion of paternal genotypic information may improve detection of aneuploidies, like sex chromosome trisomies, that may be affected by complicated biological phenomena.
The use of sophisticated data modeling and various metrics, such as fetal fraction and DNA quality, also allows the NATUS algorithm to identify those samples with insufficient data to make a high confidence determination, and to report only highly accurate results. This is particularly relevant at early GAs, when fetal fraction is lower and samples are more likely to have an insufficient fetal cfDNA fraction to make an accurate call. Indeed, sensitivity is unaffected using this method at fetal fractions below 8%. This is in contrast to another existing method, where trisomy 21 detection has been reported at 75% below 8% fetal fraction.14 It is thus possible that this method may be less-affected by fetal fractions in this questionable range.
Because fetal cfDNA is thought to originate from the placenta,46 all cfDNA-based methods are subject to the limits of detection as defined by mosaicism. Thus, some discrepant results, including the false negative in this cohort, could be explained by unidentified placental mosaicism. However, the false negative case returned a redraw result when re-analyzed using the current production protocol, which employed slightly altered QC metrics.
Conclusion
With early detection of sex chromosomal variations, biological and neurodevelopmental treatments may allow families to improve their child’s neurologic function, mitigate disease-related phenotypes, and maximize the chance that an affected child will live a normal life. The method described here uses highly multiplex PCR amplification and sequencing of cfDNA to measure fetal and maternal genotypic data at 19,488 SNP loci. The NATUS algorithm augments this inherently noisy data with high-quality parental genotypic data and uses sophisticated data models to interpret data from the X chromosome, whose complex biology presents a unique challenge. This method has the potential to function as an accurate and safe clinical option for early sex chromosome aneuploidy detection.
Supplementary Material
Table S1: List of 189 samples with NATUS-generated copy number result when analyzed for autosomal aneuploidies and Monosomy X. The sample indicated in gray did not return a result when paternal samples were excluded from analysis; samples indicated in pink did not return a result for the sex chromosome trisomies.
Bulleted Statements.
What’s already known
Current routine prenatal screening methods are not designed to detect sex chromosome aneuploidies. Although increased nuchal translucency may indicate Turner syndrome, maternal serum markers are not consistent indicators and are not used to screen for either Turner or the sex chromosome trisomies. Thus, diagnoses are often missed. Current non-invasive prenatal aneuploidy testing methods either do not detect sex chromosome aneuploidy or do not detect them with accuracies comparable to trisomy 21 and 18.
What this study adds
We accurately detect 45,X (Turner) with 91.7% sensitivity (CI: 61.5-99.8%) and 100% specificity (CI: 97.9-100%), as well as 47,XXY and 47,XYY, using a novel non-invasive and informatics-based prenatal method for detecting fetal aneuploidy. The method identified copy number at chromosomes 13, 18, 21, X, and Y with high accuracy across all five chromosomes.
Acknowledgements
Samples were provided by Prof. Kypros Nicolaides (King’s College, London, UK), Goodman & Partridge Obstetrics and Gynecology (Gilbert, AZ), Desert West Obstetrics and Gynecology (Glendale, AZ), Wellspan Maternal Fetal Medicine (York, PA), Dr. Lech Dudarewicz (Polish Mother’s Memorial Hospital, Poland), DNA Diagnostics Center (Fairfield, OH), Gennet s.r.o. (Prague, Czech Republic), and Stem Express (Placerville, CA).
Supported in part by a grant from the National Institute of Health, National Institute of Child Health and Human Development (4R44HD062114-02).
Footnotes
Conflicts of interest: MB, AR, SS, BZ, MH, MPH, MW, JS, ZD, and MR are employees of Natera.
References
- 1.Nielsen J, Wohlert M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum Genet. 1991 May;87:81–3. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- 2.Ranke MB, Saenger P. Turner’s syndrome. Lancet. 2001;358:309–314. doi: 10.1016/S0140-6736(01)05487-3. [DOI] [PubMed] [Google Scholar]
- 3.Kagan KO, Avgidou K, Molina FS, Gajewska K, Nicolaides KH. Relation between increased fetal nuchal translucency thickness and chromosomal defects. Obstet Gynecol. 2006;107:6–10. doi: 10.1097/01.AOG.0000191301.63871.c6. [DOI] [PubMed] [Google Scholar]
- 4.Saenger P. Turner’s Syndrome. New Engl J Med. 1996;335:1749–1754. doi: 10.1056/NEJM199612053352307. [DOI] [PubMed] [Google Scholar]
- 5.Samango-Sprouse CA, Sadeghin T, Mitchell FL, et al. Positive effects of short course androgen therapy on the neurodevelopmental outcome in boys with 47,XXY syndrome at 36 and 72 months of age. Am J Med Genet Part A. doi: 10.1002/ajmg.a.35769. In Press. [DOI] [PubMed] [Google Scholar]
- 6.Samango-Sprouse CA, Gropman AL, Sadeghin T, Kingery M, Lutz-Armstrong M, Rogol AD. Effects of short-course androgen therapy on the neurodevelopmental profile of infants and children with 49,XXXXY syndrome. Acta Paediatr. 2011;100:861–865. doi: 10.1111/j.1651-2227.2011.02252.x. [DOI] [PubMed] [Google Scholar]
- 7.Simpson JL, Elias S. Fetal cells in maternal blood: overview and historical perspective. Ann N Y Acad Sci. 1994;7:1–8. doi: 10.1111/j.1749-6632.1994.tb55743.x. [DOI] [PubMed] [Google Scholar]
- 8.Kazakov VI, Bozhkov VM, Linde VA, Repina MA, Mikhailov VM. Extracellular DNA in the blood of pregnant women. Tsitologia. 1995;37:232–236. [PubMed] [Google Scholar]
- 9.Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 10.Fan HC, Blumenfeld YJ, Chitkara U, et al. Non-invasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105:266–271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu RW, Chan KC, Gao Y, et al. Non-invasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrich M, Deciu C, Zwiefelhofer T, et al. Non-invasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204:205.e1–205.e11. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 13.Liao GJ, Lun FM, Zheng YW, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem. 2011;57:92–101. doi: 10.1373/clinchem.2010.154336. [DOI] [PubMed] [Google Scholar]
- 14.Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down Syndrome: An international clinical validation study. Genet Med. 2011;13:913–20. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 15.Chen EZ, Chiu RW, Sun H, et al. Non-invasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One. 2011;6:e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sehnert AJ, Rhees B, Comstock D, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem. 2011;57:1042–1049. doi: 10.1373/clinchem.2011.165910. [DOI] [PubMed] [Google Scholar]
- 17.Palomaki GE, Deciu C, Kloza EM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012 doi: 10.1038/gim.2011.73. doi:10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianchi DW, Platt LD, Goldberg JD. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 19.Sparks AB, Wang ET, Struble CA, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn. 2012;32:1–7. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sparks AB, Struble CA, Wang ET, et al. Non-invasive Prenatal Detection and Selective Analysis of Cell-free DNA Obtained from Maternal Blood: Evaluation for Trisomy 21 and Trisomy 18. Am J Ob Gyn. 2012;206:319e.1–9. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 21.Ashoor G, Syngelaki A, Wagner M, et al. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Ob Gyn. 2012;206:322.e1–5. doi: 10.1016/j.ajog.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Chiu RW, Sun H, Akolekar R, et al. Maternal plasma DNA analysis with massively parallel sequencing by ligations for non-invasive prenatal diagnosis of trisomy 21. Clin Chem. 2010;56:459–63. doi: 10.1373/clinchem.2009.136507. [DOI] [PubMed] [Google Scholar]
- 23.Jiang F, Ren J, Chen F, et al. Noninvasive Fetal Trisomy (NIFTY) test: an advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Medical Genomics. 2012;5:57. doi: 10.1186/1755-8794-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashoor G, Syngelaki A, Wang E, et al. Trisomy 13 detection in the first trimester of pregnancy using a chromosome-selective cell-free DNA analysis method. Ultrasound Obstet Gynecol. 2013;41:21–5. doi: 10.1002/uog.12299. [DOI] [PubMed] [Google Scholar]
- 25.Futch T, Spinosa J, Bhatt S, de Feo E, Rava RP, Sehnert AJ. Initial clinical laboratory experience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat Diagn. 2013 doi: 10.1002/pd.4123. doi:10.1002/pd.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazloom AR, Džakula Ž , Wang H, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat Diagn. 2013 doi: 10.1002/pd.4127. doi:10.1002/pd4127. [DOI] [PubMed] [Google Scholar]
- 27.Alkan C, Kidd JM, Marques-Bonet T, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–7. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dohm JC, Lottaz C, Borodina T, et al. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acid Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DS, Gemelos G, Baner J, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010;25:1066–75. doi: 10.1093/humrep/dep452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabinowitz M, Gemelos G, Banjevic M, et al. World Intellectual Property Organization; Patent application WO/2011/146632 Methods for non-invasive prenatal ploidy calling. 2011 Nov 24;
- 31.Rabinowitz M, Gemelos G, Banjevic M, et al. World Intellectual Property Organization; patent application WO/2012/108920 Methods for non-invasive prenatal ploidy calling. 2012 Aug 16;
- 32.Zimmermann B, Hill M, Gemelos G, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y using targeted sequencing of polymorphic loci. Prenat Diag. 2012;32:1–9. doi: 10.1002/pd.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolaides KH. Routine screening by maternal blood of DNA. Advances in Fetal Medicine Meeting; London, UK. December 2013. [Google Scholar]
- 34. http://www.ncbi.nlm.nih.gov/projects/genome/probe/doc/ProjHapmap.shtml.
- 35.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001 Jan 1;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson JL, Samango-Sprouse C. Prenatal diagnosis and 47,XXY. Am J Med Genet Part C Semin Med Genet. 2013;163C:64–70. doi: 10.1002/ajmg.c.31356. [DOI] [PubMed] [Google Scholar]
- 37.Linglart A, Cabrol S, Berlier P, et al. Growth hormone treatment before the age of 4 years prevents short stature in young girls with Turner syndrome. Eur J Endocrinol. 2011;164:891–897. doi: 10.1530/EJE-10-1048. [DOI] [PubMed] [Google Scholar]
- 38.Bondy CA. Care of girls and women with Turner syndrome: a guideline of the Turner Syndrome Study Group. J Clin Endocrinol Metab. 2007;92:10–25. doi: 10.1210/jc.2006-1374. [DOI] [PubMed] [Google Scholar]
- 39.Hanton L, Axelrod L, Bakalov V, Bondy CA. The importance of estrogen replacement in young women with Turner syndrome. J Womens Health. 2003;12:971–7. doi: 10.1089/154099903322643893. [DOI] [PubMed] [Google Scholar]
- 40.Ferlin A, Zuccarello D, Zuccarello B, Chirico MR, Zanon GF, Foresta C. Genetic alterations associated with cryptorchidism. JAMA. 2008;300:2271–2276. doi: 10.1001/jama.2008.668. [DOI] [PubMed] [Google Scholar]
- 41.Mehta A, Paduch DA. Klinefelter syndrome: an argument for early aggressive hormonal and fertility management. Fertil Steril. 2012;98:274–283. doi: 10.1016/j.fertnstert.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Liao GJ, Chan KC, Jiang P, et al. Non-invasive prenatal diagnosis of fetal trisomy 21 by allelic ratio analysis using targets massively parallel sequencing of maternal plasma DNA. PLoS ONE. 2012;7:e38154. doi: 10.1371/journal.pone.0038154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chadwick B, Willard HF. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Human Mol Genet. 2001;10:1101–1113. doi: 10.1093/hmg/10.10.1101. [DOI] [PubMed] [Google Scholar]
- 44.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 45.Chadwick BP, Willard HF. A novel chromatin proteins, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J Cell Biol. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bischoff FZ, Lewis DE, Simpson JL. Cell-free fetal DNA in maternal blood: kinetics, source and structure. Hum Reprod Update. 2005;11:59–67. doi: 10.1093/humupd/dmh053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: List of 189 samples with NATUS-generated copy number result when analyzed for autosomal aneuploidies and Monosomy X. The sample indicated in gray did not return a result when paternal samples were excluded from analysis; samples indicated in pink did not return a result for the sex chromosome trisomies.