SUMMARY
Merkel cell polyomavirus (MCV) causes an aggressive human skin cancer, Merkel cell carcinoma, through expression of small T (sT) and large T (LT) viral oncoproteins. MCV sT is also required for efficient MCV DNA replication by the multifunctional MCV LT helicase protein. We find that LT is targeted for proteasomal degradation by the cellular SCFFbw7 E3 ligase, which can be inhibited by sT through its LT stabilization domain (LSD). Consequently, sT also stabilizes cellular SCFFbw7 targets, including the cell cycle regulators c-Myc and cyclin E. Mutating the sT LSD decreases LT protein levels and eliminates synergism in MCV DNA replication as well as sT-induced cell transformation. SCFFbw7 knockdown mimics sT-mediated stabilization of LT, but this knockdown is insufficient to fully reconstitute the transforming activity of a mutant LSD sT protein. Thus, MCV has evolved a regulatory system involving SCFFbw7 that controls viral replication but also contributes to host cell transformation.
INTRODUCTION
Merkel cell carcinoma is one of the most aggressive human skin cancers, with a mortality rate exceeding melanoma (Lemos and Nghiem, 2007) and a tripled incidence between 1986 and 2001 (Hodgson, 2005). Approximately 80% of MCC are caused by the newly-discovered Merkel cell polyomavirus (MCV) (Feng et al., 2008), a small DNA virus that is an asymptomatic component of normal human skin flora (Tolstov et al., 2011). MCV-MCC tumors arise when the virus integrates into the host cell genome and mutations occur that inactivate the DNA-binding/helicase functions of MCV large T (LT) (Shuda et al., 2008). Xenomutation to commensal flora like MCV is an unexpected mechanism for human carcinogenesis (Arora et al., 2012).
Similar to other polyomaviruses, MCV LT and small T (sT) early antigens act in natural infections to promote virus replication. However, the expression of a replication-defective, C-terminally truncated MCV LT antigen promotes tumorigenesis in human Merkel cells. Both LT and sT antigens are required for MCV-positive MCC cell survival and proliferation (Houben et al., 2012; Houben et al., 2010; Shuda et al., 2011). In in vitro cell transformation assays, MCV sT is sufficient to transform rodent fibroblasts in culture whereas MCV LT cannot (Shuda et al., 2011). This is in marked contrast to the more well-known polyomavirus SV40, in which SV40 LT is the major transforming oncoprotein while sT promotes cell transformation (Hahn et al., 2002) but is not fully transforming on its own (Bikel et al., 1987).
A comparison of the cellular targets of SV40 and MCV viral sT proteins shows similarities as well as differences. Both MCV and SV40 sT target protein phosphatase 2A (PP2A) and heat-shock proteins through direct protein-protein interactions. SV40 sT prevents PP2A dephosphorylation of Akt whereas MCV sT has minimal effect at this node of the Akt-mTOR signaling pathway (Shuda et al., 2011). MCV sT, instead, acts downstream of mTOR to increase levels of hyper-phosphorylated 4E-BP1, a regulator of cap-dependent translation. MCV sT-induced cell transformation is independent of PP2A binding but can be inhibited by a constitutively-active mutant 4E-BP1 protein. In contrast, SV40 sT has been reported to diminish 4E-BP1 phosphorylation (Yu et al., 2005).
MCV sT also plays a key role in promoting MCV genome replication. MCV LT assembles on a 71 base pair stretch of the viral replication origin by recognizing specific pentanucleotide sequence repeats (Harrison et al., 2011; Kwun et al., 2009). For SV40, sT coexpression has minimal effect on SV40 LT-mediated replication. However, for MCV (Kwun et al., 2009) and JC polyomavirus (Prins and Frisque, 2001), sT coexpression markedly enhances LT-mediated viral DNA synthesis, although the mechanism for this effect is unknown. Like papillomaviruses, MCV is poorly transmissible in culture. Replication of genomic MCV clones (Feng et al., 2011; Neumann et al., 2011; Schowalter et al., 2011) can be markedly enhanced by coexpression of a MCV sT from a heterologous promoter (Feng et al., 2011).
While investigating the mechanism by which MCV sT synergistically enhances MCV LT-dependent replication, we found that MCV sT targets the cellular SCF (complex of Skp1, Cul1 and F-Box protein) ubiquitin ligase protein complex, SCFFbw7. Fbw7 (F-box and WD repeat domain-containing 7, also known as FBXW7, CDC4, AGO, and SEL10) serves as the substrate recognition component for this multiprotein cullin-RING ubiquitin ligase complex that includes Fbw7, Skp1, Cul1 and Rbx1 (Welcker and Clurman, 2008). The importance of Fbw7 in controlling cancer cell outgrowth has been highlighted by studies showing it to be dysregulated in breast cancer, colon cancer and T-cell acute lymphoblastic leukemia (Maser et al., 2007; Wood et al., 2007). Numerous cancer-associated mutations of Fbw7 also have been reported in various cancers (Akhoondi et al., 2007), and loss of Fbw7 function results in tumorigenesis and genetic instability (Mao et al., 2004; Rajagopalan et al., 2004; Rajagopalan and Lengauer, 2004). Fbw7 recognition promotes phosphorylation-dependent degradation in many proto-oncogenes, including cyclin E (Koepp et al., 2001; Strohmaier et al., 2001), c-Myc (Welcker et al., 2004; Yada et al., 2004), c-Jun (Nateri et al., 2004), Notch (Gupta-Rossi et al., 2001; Oberg et al., 2001; Wu et al., 2001), mTOR (Mao et al., 2008), MCL-1 (Inuzuka et al., 2011; Wertz et al., 2011), and NF-kB2 (p100/p52) (Arabi et al., 2012; Fukushima et al., 2012), indicating that Fbw7 is a critical nexus for diverse signaling pathways regulating cell proliferation and tumor suppression.
We show here that MCV LT oncoprotein level is regulated by Fbw7. MCV sT, through inhibition of Fbw7, enhances MCV genome replication by promoting accumulation of the MCV LT oncoprotein. This targeting of SCFFbw7 is mediated through an LT-stabilization domain (LSD) that maps to residues 91–95 and is predicted to be on the opposite molecular face of sT from its PP2A-targeting site. Mutation of the LSD not only abolishes MCV sT stabilization of MCV LT but eliminates sT-induced rodent cell transformation, inhibits viral replication and prevents MCV sT induction of several cellular oncoproteins, including c-Myc and cyclin E. These effects are independent of MCV sT binding to PP2A. In MCC tumor cells, having defective MCV genomes incapable of replication, MCV sT stabilizes and increases steady-state levels of viral and cellular oncoproteins. Fbw7 knockdown alone does not rescue the transforming phenotype for the MCV sT LSD mutant protein, however, suggesting that the LSD targeting domain also affects a broader range of cell signaling pathways.
RESULTS
MCV sT enhances LT-mediated MCV origin replication through a PP2A-independent mechanism
Using an MCV origin replication assay (Kwun et al., 2009), we tested various MCV sT constructs for their effect on MCV LT-dependent origin replication activity (Figure 1). Separate sT constructs and full length, wild type MCV LT (or empty vector) plasmids were cotransfected into 293 cells together with a plasmid containing the MCV replication origin (Ori339(97)) (Kwun et al., 2009). No replication of the origin plasmid occurs in the absence of MCV LT (Figure 1A, lane 1). Expression of MCV LT (Figure 1A, lane 2) activates baseline origin plasmid replication, which is amplified ~5-fold by cotransfection of wild-type MCV sT (Figure 1A, lane 3). This enhancement of MCV LT replication by sT is largely unaffected when wild-type MCV sT expression is substituted with expression of mutant MCV sT having alanine substitutions in amino acids R7 or L142 (Figure 1A, lanes 4 and 5) that inhibit PP2A interaction (Shuda et al., 2011), or an aspartate-asparagine substitution at D44 (Figure 1A, lane 6) in the MCV sT DnaJ domain eliminating heat-shock protein (Hsc70) interactions (Kwun et al., 2009).
Figure 1. MCV sT enhances LT-mediated MCV origin replication through an exon1A domain in a PP2A independent manor.
(A) An MCV sT exon1A-encoded domain enhances MCV LT-dependent origin replication. Diagram compares MCV and SV40 sT coding regions and sites of MCV point mutations used in replication assay. SV40 exon1A (83–174 a.a.) was replaced with MCV exon1A (80–186 a.a.) to make the chimeric construct, SV40/MCVsT. The obverse MCV exon1 and SV40 exon1A chimeric (MCV/SV40sT) could not be evaluated due to its protein instability. Dpn1-resistant MCV origin replication in 293 cells was assayed by Southern blotting (middle panel) in the absence of T antigens (lane 1), in the presence of LT alone (lane 2), or LT together with MCV or SV40 sT proteins (lanes 3–8). MCV sT point mutants that eliminate PP2A binding (lanes 4, 5) and Hsc70 binding (lane 6) have comparable activity to the wild-type sT protein (lane 3). SV40 sT did not induce enhanced LT-dependent MCV origin replication (lane 7) but the SV40/MCV sT chimeric protein possessing the C-terminal MCV peptide sequence (lane 8) restored origin replication activity. Corresponding immunoblots for LT (CM2B4) and sT proteins (mixture of anti-MCV sT (CM8E6) and anti-SV40 LT/sT (pAb419)) are shown (lower panels).
(B) Identification of an MCV sT LT-stabilization domain (LSD) responsible for enhanced LT-mediated origin replication. MCV origin replication was assayed in 293 cells by coexpression of LT together with sT mutants having sequential 5 amino acid deletions or 5 alanine substitutions in exon1A from 86 to 104 amino acids. Deletion or substitution at MCV sT amino acids 91 to 95 (LKDYM) ablated enhanced origin replication and increased steady-state MCV LT protein expression (lanes 5 and 6) compared to wild-type MCV sT (lane 2). Deletion (but not alanine substitutions) at residues 86–90 reduced sT protein stability as well as origin replication and LT expression. See also Figure S1.
MCV sT is generated by read-through of the splice-donor site in exon1 (a.a. 1–79) to produce a unique sT C-terminus peptide encoded by exon1A (a.a. 80–186) (Figure 1A) (Shuda et al., 2009; Shuda et al., 2008). SV40 sT is similarly generated from a 82 a.a. exon 1, which reads through splice site to generate a C-terminal region (a.a. 83–174). MCV sT and SV40 sT proteins are highly homologous to each other (33% a.a. identity by Basic Local Alignment Search Tool (BLAST) program). In contrast to MCV sT, SV40 sT does not cross-enhance MCV LT-mediated DNA replication despite conserved PP2A and Hsc domains (Figure 1A, lane 7). However, a chimeric sT comprised of SV40 sT exon 1 (a.a. 1–82) and MCV exon1A (a.a. 80–186) designated SV40/MCV sT (Figure 1A, lane 8) restored DNA replication activation, suggesting the responsible domain is encoded in the MCV sT C-terminus. The reverse chimeric construct (MCV/SV40 sT) comprised of N-terminus MCV exon1 (a.a. 1–79) and C terminus SV40 exon1A (a.a. 83–174) could not be evaluated due to protein instability. Notably, MCV sT constructs that activated MCV origin replication also correlatively enhanced levels of MCV LT protein expressed from a heterologous promoter (pcDNA6). SV40 sT, which increased MCV LT protein expression did not enhance MCV DNA replication (Figure 1A, lane 7). In line with our observation that MCV sT affects MCV LT protein levels, increased LT expression was consistently detected in populations of cells co-expressing sT but not in cells expressing only LT by immunofluorescence (Figure S1).
To map the MCV sT domain enhancing LT-dependent MCV DNA replication, we constructed sT mutants with sequential 5 amino acid deletions or 5 alanine substitutions in its C-terminal exon 1A. Deletion at residues 91–95 (sT.91-95Δ, Figure 1B, lane 5), as well as alanine substitution (sT.91-95A, Figure 1B, lane 6) ablated enhanced MCV DNA replication and reduced LT protein levels. Enhanced replication was also diminished compared to the wild-type sT protein (Figure 1B, lane 2) by deletion of residues 86–90 (sT.86–90Δ) (Figure 1B, lane 3). This, however, likely results from reduced stability of the mutant sT protein since that steady-state sT.86-90Δ protein level is reduced compared to wild-type sT. Alanine substitutions at a.a. 86–90 (Figure 1B, lane 4) fully restored this mutant’s protein stability and activation of MCV LT-mediated replication.
The high homology between MCV sT and SV40 sT allowed us to model MCV sT (I-TASSER) by threading its sequence onto the SV40 sT crystal structure (PDB ID:2PF4, 2PKG) (Chen et al., 2007; Cho et al., 2007). This predicts that the MCV sT amino acid 91–95 region, designated the LT-stabilization domain (LSD), forms a larger loop structure than the one present in the SV40 sT molecule (Figure 2 and Movie S1). The LSD is predicted to be on the opposing surface of the MCV sT from the PP2A-interaction region and distinct from the DnaJ domain (Movie S1), correlating with the mutation analysis in Figure 1A.
Figure 2. Predicted model of MCV sT structure.
MCV sT protein structure based on the SV40 sT crystal structure (I-TASSER server) shown in both space-filling (A) and ribbon (B) representations. The latter reveals the conserved structures with overlapping regions for SV40 (green) and MCV (turquoise). PP2A binding (gold) and DnaJ (magenta) domains conserved for both viruses are shown. The MCV LSD (red) is a large unstructured loop on the opposite molecular face from the PP2A-binding domain that is foreshortened in SV40 (black). Amino acid alignment between MCV sT and SV40 sT (32.52% sequence identity) indicates (|) identical, (:) similar and (.) dissimilar residues and shows loss of homology in the region corresponding to the LSD. See also Movie S1.
MCV sT synergistically enhances MCV LT-dependent virus replication
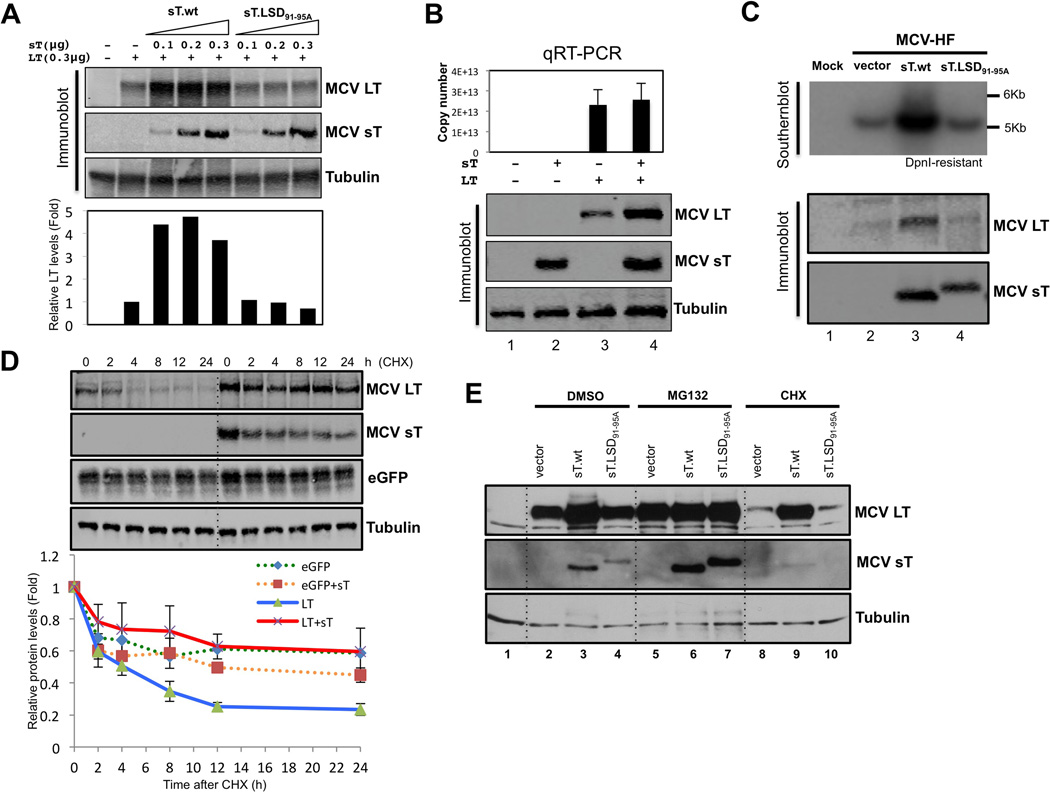
We next sought to determine if increased MCV LT protein accumulation by MCV sT contributes to MCV replication. Cotransfection of small amounts of wild type sT expression plasmid (0.1 µg) maximally activated LT expression (Figure 3A) and LT-dependent origin replication (Figure S2A). This is not due to enhanced transcription of the LT gene since sT does not increase LT mRNA levels (Figure 3B). These effects can be demonstrated with whole MCV genome replication as well. MCV sT expressed in trans markedly activates viral replication from the MCV-HF plasmid clone (Figure 3C) (Feng et al., 2011). The effect is lost using MCV sT having alanine substitutions in the LSD (MCV sT.LSD91-95A). In the MCV-positive MCC cell line MKL-2, knockdown of MCV sT was correlated with reduction in LT protein expression (Figure S2B).
Figure 3. MCV sT inhibits proteasomal degradation of MCV LT.
(A) MCV sT expression increases LT expression through a domain encompassed by amino acids 91–95. Extremly low levels of wild type small T (sT.wt) expression (0.1 µg transfected plasmid) results in fully saturated (5–7 folds) LT stability. In contrast, the titration of mutant (sT.LSD91-95A) expression, even over a broad range of levels, has no effect on LT stability. Quantitative LICOR immunoblotting for LT expression was determined using CM2B4 antibody in triplicate, with a representative shown. (B) MCV sT expression does not transcriptionally regulate MCV LT expression. 293 cells were transfected with empty vector (lane 1, negative control), sT alone (lane 2), LT alone (lane 3), LT and sT together (lane 4), and quantitative RT-PCR was performed to detect LT mRNA transcription. Corresponding levels of sT and LT protein expression by immunoblotting are shown. Error bars represent SEM; n = 3. (C) sT expression in trans increases MCV replication. MCV-HF (0.3 µg) was cotransfected with empty vector (lane 2), sT wild type (lane 3), sT.LSD91-95A mutant (lane 4) (0.3 µg each) into 293 cells and replication was assayed by Southern blotting. Wild type MCV sT cotransfection markedly activates viral replication (upper panel) and LT expression (lower panel) from the MCV-HF clone, whereas the MCV sT LSD mutant does not. (D) MCV sT specifically inhibits LT protein turnover. LT protein turnover was measured by a cycloheximide (CHX) chase assay using quantitative immunoblot analysis. LT (0.3 µg) and eGFP (0.3 µg) constructs were cotransfected together with either empty vector or sT plasmid (0.3 µg). Cells were treated with CHX (0.1 mg/ml) 24 hours after transfection and harvested at each time point indicated. Protein expression was quantified in triplicate using an LI-COR IR imaging system (bottom). Coexpression of sT extended the half-life of LT from ~3–4 h up to >24 h, but did not significantly affect eGFP protein turnover. Error bars represent SEM; n = 3. .(E) MCV sT inhibits proteasomal degradation of LT protein. 293 cells were transfected with LT with either wild-type sT or sT.LSD91-95A. Cells were treated with MG132 (10 µM) or CHX (0.1 mg/ml) 24 h after transfection. Comparable levels of LT accumulation are seen with empty vector or sT.LSD91-95A expression to those seen with wild-type sT expression during MG132 treatment. See also Figure S2
MCV sT inhibits proteasomal degradation of MCV LT
To measure sT effects on LT stability, quantitative immunoblotting (Figures 3D and S2C) was used to measure LT abundance in the presence of the translation inhibitor cycloheximide (CHX). In the absence of sT, LT had a rapid turnover with a half-life (t1/2) of ~3–4 hrs and diminished to low levels within 24 hours after CHX addition. Coexpression of sT together with LT increased the t1/2 for LT to >24 hours but did not significantly alter stability of a control protein (eGFP). Additional evidence that sT inhibits LT turnover was seen after treatment with the proteasome inhibitor MG132 (Figure 3E). MG132 treatment did not increase LT levels above those found with sT coexpression alone (Figure 3E, lanes 5 and 6), consistent with both sT and MG132 acting to prevent LT proteolysis. In this same experiment, wild-type sT expression led to accumulation of LT protein after 24 hours of 0.1 mg/ml CHX treatment (Figure 3E, lanes 8 and 9) which was not seen during expression of sT.LSD91-95A (Figure 3E, lane 10). MG132 markedly increases sT and sT.LSD91-95A levels (Figure 3E, lanes 6 and 7) indicating that sT itself is subject to rapid proteasomal turnover.
SCFFbw7 binds to and promotes turnover of MCV LT
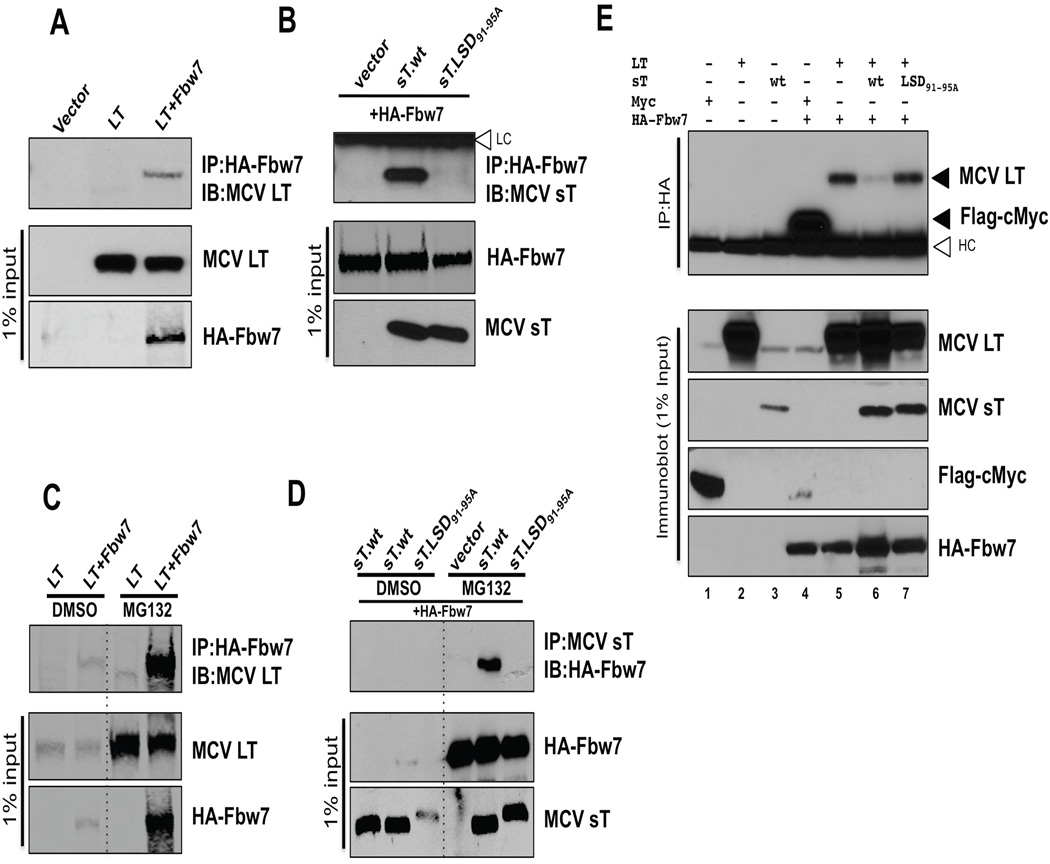
SV40 LT is a pseudo-substrate for Fbw7 (Welcker and Clurman, 2005) and so we examined potential interaction between MCV LT and Fbw7 by immunoprecipitation (Figure 4A and 4C). When MCV LT (Figure 4A) or sT (Figure 4B) are expressed in U2OS cells, specific coimmunoprecipitation with HA-Fbw7 was readily detectable with each viral protein. For sT, mutation within the LSD eliminated the sT-Fbw7 interaction. MCV LT, however, is a poly-phosphoprotein with multiple potential consensus F-box binding motifs (S/TPXX) that could serve as recognition sequences for SCFFbw7. Mutation analyses to define the Fbw7 recognition site(s) on LT are ongoing, and are complicated by multiple phosphorylation-dependent turnover pathways for the LT protein (data not shown), but preliminary analyses suggest that this domain is likely to be conserved in most tumor-derived MCV strains. In 293 cells, Fbw7 was rapidly turned over and required MG132 treatment for robust detection. Consistent with this, LT and sT interaction with Fbw7 was only revealed in the presence of MG132 (Figures 4C and 4D).
Figure 4. sT targets SCFFbw7 complex to stabilize LT.
(A) LT interacts with the Fbw7 E3 ligase complex. HA-tagged Fbw7 (6 µg) was cotransfected with MCV LT (6 µg) into U2OS cells and immunoprecipitated with anti-HA antibody. LT interaction was detected with CM2B4 antibody. Negative controls for immunoprecipitation include transfection with LT alone or an empty vector. (B) sT interacts with the Fbw7 E3 ligase complex. HA-Fbw7 (6 µg) was cotransfected with wild type (sT.wt) or mutant (sT.LSD91-95A) or empty vector (6 µg), followed HA immunoprecipitation and immunoblotting for sT using CM8E6 antibody. MCV sT, but not mutant sT (sT.LSD91-95A) interacted with overexpressed HA-Fbw7. White arrowhead indicates immunoglobulin light chains (LC) at 25 kDa. (C) LT interaction with the Fbw7 E3 ligase complex is accentuated with MG132 treatment. Binding of MCV LT to Fbw7 was examined with lysates treated with MG132 (10 µM, 24 h). Both MCV LT and Fbw7 proteins are stabilized in 293 cells treated with MG132, and protein interactions are more readily detected as compared to vehicle treatment control. (D) sT interaction with the Fbw7 E3 ligase complex is also revealed with MG132 treatment. Fbw7 was cotransfected into 293 cells with wild type (sT.wt) or mutant sT (sT.LSD91-95A) or empty vector. Cells were treated with MG132 (10 µM, 24 h) 24 hours after transfection, then harvested. CM8E6 was used for sT immunoprecipitation followed by Fbw7 detection using HA antibody. Fbw7 in cell lysates as well as following immunoprecipitation with sT is more readily detectable by MG132 treatment. (E) MCV sT inhibits Fbw7 interaction with MCV LT. LT, sT, Flag-c-Myc and HA-Fbw7 proteins, were expressed in 293 cells treated with MG132 (10 µM) for 24 hours. Cell lysates were then immunoprecipitated with anti-HA antibody and immunoblotted for LT or Flag-c-Myc. No immunoprecipitation is seen in negative control lanes transfected with HA expressing vector without Fbw7 (lane 1–3) but coimmunoprecipitation of Flag-tagged c-Myc (lane 4) and LT (lane 5) by HA-Fbw7 is evident. LT-Fbw7 interaction is inhibited when both LT and sT are coexpressed with HA-Fbw7 (lane 6), but not when LT is coexpressed with the LSD mutant (lane 7). White arrowhead indicates the immunoglobulin heavy chains (HC) at 50 kDa. See also Figure S3.
Fbw7 targeting of MCV LT is inhibited by coincident expression of MCV sT protein as shown in Figure 4E. 293 cells expressing HA-tagged Fbw7 were treated with MG132 and then immunoprecipitated using an HA-tagged antibody. LT immunoprecipitated with HA-Fbw7 (Figure 4E, lane 5) but this interaction is markedly diminished by expression of sT together with LT (Figure 4E, lane 6), which can be reversed by mutation to the sT LSD (Figure 4E, lane 7). Fbw7 immunoprecipitation of c-Myc protein (Yada et al., 2004), was performed as an immunoprecipitation control (Figure 4E, lane 4). In addition, an arginine mutant of human Fbw7 R465C, which is defective in binding to its substrate (Akhoondi et al., 2007) was examined for LT binding. MCV LT interacts with wild type (wt) Fbw7 but it loses its binding with R465C mutant (Figure S3).
To examine whether the ubiquitylation status of LT is Fbw7-dependent, LT and a vector containing CMV promoter-driven HA-ubiquitin were cotransfected in HCT116 wt and Fbw7 (−/−) cells. Immunoprecipitation of LT followed by immunoblot analysis of the HA-ubiquitin showed efficient LT ubiquitylation found in cells preserving a functional Fbw7 gene and reduced ubiquitylation in Fbw7 (−/−) cells (Figure S4A). Ubiquitylation of LT is also reduced by sT overexpression. Proteasome inhibition using MG132 increased the amount of ubiquitin chains bound to LT and is reduced by sT suggesting that MCV sT inhibits LT ubiquitylation by prohibition of Fbw7 E3 ligase activity.
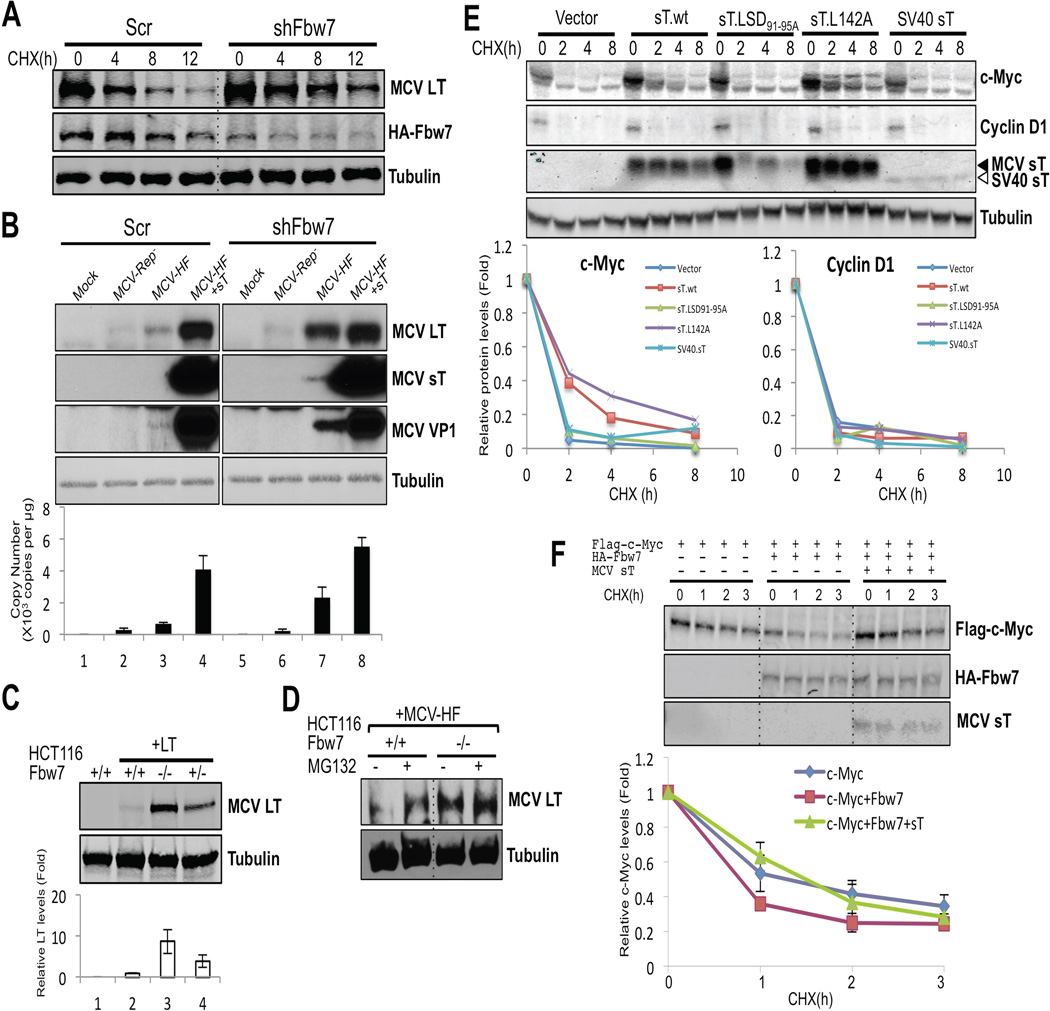
Consistent with sT targeting Fbw7 to enhance steady-state LT expression, knockdown of cellular Fbw7 by lentiviral transduction also decreased turnover rates of LT protein (Figure 5A). In these experiments, we were unable to measure endogenous Fbw7 levels using commercial antibodies and so knockdown efficacy was monitored by ectopic HA-Fbw7 expression. Whole virus replication was also enhanced by Fbw7 knockdown (Figure 5B). Cells transduced by scrambled (Scr) or shFbw7 lentiviruses were transfected with MCV-HF genome and MCV replication was assessed. Fbw7 knockdown alone enhanced virally-expressed LT protein and VP1 protein expression and virus genome replication. Coexpression of sT protein (Figure 5B, lanes 4) markedly increased viral replication that was further increased by simultaneous knockdown of Fbw7 (Figure 5B, lanes 8). In comparison, MCV-Rep− (Figure 5B, lanes 2 and 6) is a replication defective control virus having a point mutation in its replication origin (Feng et al., 2011). As expected, MCV-Rep− showed no late protein or genome replication after Fbw7 knockdown but knockdown did increase MCV-Rep− LT protein levels indicating that this effect was not dependent on virus replication.
Figure 5. SCFFbw7 inhibits MCV replication and increases turnover of viral and cellular oncoproteins.
(A) Knockdown of Fbw7 reduces LT protein turnover. HA-Fbw7 and LT were cotransfected together with either scrambled shRNA (Scr) or Fbw7 shRNA (shFbw7.2) constructs into 293 cells. 3 days after transfection cells were treated with CHX, and LT and Fbw7 protein expressions were measured by immunoblotting. (B) Knockdown of Fbw7 promotes MCV replication and protein expression. MCV genomic clone early and late protein expression (top panels) and genome copy number (bottom panel) are increased in 293 cells transduced with Fbw7 shRNA compared to scrambled shRNA. MCV-Rep− is a replication defective clone of MCV-HF having a point mutation in its replication origin. Each cell line was mock transfected or transfected with a genomic clone for 5 days prior to harvesting. In lanes 4 and 8, an sT expression vector was cotransfected in trans with MCV-HF. Replication efficiency was measured in triplicate by qPCR. Error bars represent SEM; n = 3. (C) Genetic deletion of Fbw7 increases ectopic LT expression in HCT116 cells. LT expression was examined by immunoblotting with CM2B4 antibody in HCT116 cells null (−/−), heterozygous (+/−) or wild-type (+/+) for the Fbw7 gene. Quantification was performed with LI-COR. Error bars represent SEM; n = 3. (D) Knockout of Fbw7 decreases proteasomal degradation of MCV LT from MCV-HF virus. MCV-HF was transfected into HCT116 wild-type and Fbw7-null cells. 5 days after transfection, cells were treated with MG132 or DMSO for 12 h and LT protein levels were measured by immunoblotting. In wild-type cells, virus-generated LT protein was increased by proteasome inhibition. In Fbw7 null cells, LT protein expression was elevated and unchanged by MG132 treatment. (E) sT stabilizes a proto-oncogene c-Myc but not cyclin D1 in Rat-1 cells. Rat-1 cells were stably transduced with vector, wild type sT, sT.LSD91-95A, L142A and SV40 sT and turnover of c-Myc and cyclin D1 were measured by CHX treatment. c-Myc protein was stabilized by either MCV sT.wt or sT.L142A but not by sT.LSD91-95A or SV40 sT. MCV sT did not affect turnover of cyclin D1. Mixture of CM8E6 and pAb419 antibodies was used to detect viral sTs. (F) sT decreases the degradation rate of c-Myc. Flag-c-Myc expression construct was cotransfected with either empty vector or HA-Fbw7, with or without sT, and turnover was measured in a CHX chase assay. Fbw7 overexpression destabilized exogenous c-Myc protein and this was reversed by coexpression of MCV sT. Error bars represent SEM; n = 3. See also Figure S4.
These Fbw7 knockdown results were confirmed using HCT116 cells engineered for deletion of the Fbw7 gene (Rajagopalan et al., 2004). Transient LT protein was barely detected in homozygous Fbw7 wild-type (+/+) cells but increased 10-fold when the same amount of LT DNA was transfected into homozygous (−/−) Fbw7 null cells (Figure 5C). Intermediate LT protein levels were detected in cells with hemizygous (+/−) Fbw7 status. Similarly, LT expression from the MCV-HF virus was minimal in wild-type HCT116 cells but markedly increased in Fbw7-null HCT116 cells (Figure 5D). MG132 proteasome inhibition increased LT expression from virus in wild-type but not in Fbw7-null HCT116 cells.
The effect of sT expression on the cellular Fbw7 oncoprotein target c-Myc was examined in Rat-1 fibroblasts transduced with empty vector, sT or sT.LSD91-95A, sT.L142A, SV40 sT-expressing lentiviruses, and treated with CHX (Figure 5E). Both sT and sT.L142A increased c-Myc expression and prolonged its turnover, an effect which is lost for sT.LSD91-95A. Cyclin D1, a target of SCFSkp2, SCFFbx4 and SCFFbw8 (Lin et al., 2006; Okabe et al., 2006; Yu et al., 1998), was unaffected by sT expression. Overexpressed HA-Fbw7 increased turnover of Flag-labeled c-Myc in 293 cells and this could be fully reversed by simultaneous expression of MCV sT (Figure 5F). Cyclin E turnover is also reduced by sT but not sT.LSD91-95A expression (Figure S4B).
MCV sT activates 4E-BP1 hyperphosphorylation required for MCV sT-induced transformation (Shuda et al., 2011), but unlike c-Myc, we did not find evidence that 4E-BP1 is regulated by Fbw7 (Figure S4C) suggesting that sT effects on 4E-BP1 hyperphosphorylation are independent of Fbw7 targeting.
The sT LSD domain is required for sT-induced, PP2A-independent Rat-1 cell transformation
Focus formation and soft-agar colony growth assays were used to determine cell transformation of Rat-1 (Figure 6A) and NIH3T3 cells (Figure S5) after lentiviral transduction with various sT genes (Shuda et al., 2011). Only wild-type MCV sT and sT.L142A reproducibly formed colonies after 3 weeks of growth in soft agar. In contrast, the MCV sT.LSD91-95A mutation ablated transforming activity. SV40 sT had no transforming activity, as previously described (Chang et al., 1984). These differences occur in the setting of comparable levels of sT expression for the different constructs (Figure 6B). As seen in Figure 6C, the sT.LSD91-95A mutation did not affect sT binding to endogenous PP2Ac in Rat-1 cells whereas the sT.L142A mutation eliminated PP2Ac interaction reaffirming that MCV sT transforms cells through a PP2A-independent mechanism (Shuda et al., 2011). We did not, however, find that SCFFbw7 targeting by MCV sT is sufficient to transform cells, and LSD-mutant sT protein was not complemented by lentiviral knockdown of Fbw7 protein (Figure S6) in rodent cell transformation (Figure 7).
Figure 6. MCV small T antigen exon1A LSD domain is required for sT-induced transformation in a PP2A-independent manor.
(A) LSD domain is critical for sT-induced transformation. Rat-1 cells were stably transduced with vector, wild type sT, mutants (sT.LSD91-95A, L142A), and SV40 sT. Both wild-type MCV sT and sT.L142A reproducibly formed colonies after 3 weeks of growth in soft agar, whereas the MCV sT.LSD91-95A mutation ablated transforming activity. Cells were stained with crystal violet (upper). Transformation-associated foci were photographed (×40) (middle) and colony number per high-power field was counted (bottom). All assays were performed in triplicate. Error bars represent SEM; n = 3. (B) All sT constructs show similar levels of expression in Rat-1 cell lines from transformation assays. Stable sT expressing cell lines used for soft agar assay were tested by immunoblotting with mixed antibodies, CM8E6 and pAb419 for MCV and SV40 sT detection, respectively. (C) LSD mutation does not affect sT binding to PP2A. Immunoprecipitation analysis was performed to measure PP2A binding capacity of sT mutants stably expressed in Rat-1. Either MCV sT antibody (CM8E6) or SV40 sT antibody (pAb419) was used for immunoprecipitation of sT and endogenous PP2A was detected. The non-transforming LSD mutant sT interacts with PP2A, similar to wild type sT. A PP2A binding mutant L142A ablates its binding to PP2A but retains its transforming activity (Figure 6A). These data indicate that the sT transforming activity is PP2A-independent. SV40 sT was used as a positive control of PP2A binding and a negative control for the transformation. See also Figure S5.
Figure 7. Fbw7 knockdown is insufficient for MCV sT-induced transformation.
Rat-1 cells were stably transduced with two lentiviral shRNAs (sh Fbw7.1 and shFbw7.2) targeting the rat specific Fbw7. Empty vector lentivirus, sT.LSD91-95A or wild type MCV sT (sT.wt) were individually transduced into Fbw7 knockdown cells and subjected to transformation assays. Transformation-induced foci were stained with crystal violet (upper). Colonies formed in soft-agar (middle) were counted to quantitate transformation activity (bottom). Error bars represent SEM; n = 3. The SCFFbw7 targeting by Fbw7 knockdown is not sufficient for rodent cell transformation but decreases soft agar growth induced by MCV sT. Fbw7 knockdown level was documented in Figure S6.
DISCUSSION
MCV sT is a viral oncoprotein that causes cell transformation as measured by focus formation assays and anchorage-independent growth of NIH3T3 and Rat-1 cells. This effect is independent of MCV LT (Shuda et al., 2011). MCV sT also enhances MCV LT replication functions (Feng et al., 2011; Kwun et al., 2009; Schowalter et al., 2011). We show here that MCV achieves this by targeting the SCFFbw7 holoenzyme through the LSD encoded by sT exon1A.
We have previously shown that sT induces hyper-phosphorylated 4E-BP1, a regulator of cap-dependent translation, during transformation (Shuda et al., 2011). This is not dependent on Fbw7 targeting by sT (Figure S4C) and so the relationship between these two sT-regulated transformation pathways remains unclear. Fbw7 knockdown alone fails to transform Rat-1 cells expressing sT.LSD91-95A and so it is likely that LSD targeting has broader effects than inhibition of SCFFbw7 alone. Further, a survey of other known cellular Fbw7 targets such as Mcl-1 (data not shown) did not reveal consistent turnover inhibition, making it likely that the role of sT in transformation are more complex than Fbw7 targeting alone.
The LT phosphorylation sites that serve as recognition motifs for SCFFbw7 have not yet been mapped but seem to be retained in many of the tumor-derived LT proteins (data not shown). Physiological substrates of Fbw7 hold the phosphodegron with a wide range of binding affinities and retain multiple low-affinity degrons in some case, for example, Sic1 (Nash et al., 2001), which may allow fine-tuning of protein proteolysis. Comparing c-Myc, MCV LT binding to Fbw7 is predicted to be weak and low affinitive, presumably with multiple binding sites (Figure 4). Dimerization of Fbw7 is known to be required for the turnover of low-affinity substrates, possibly facilitating the degradation by multi-recruitment of E2 ubiquitin conjugase (Welcker and Clurman, 2007), which might be the case in LT degradation. Given the large number of potential phosphorylation sites present in LT, degradation might be triggered by multisite sequential phosphorylation events to induce binding to the SCF ubiquitin ligase complex, SCFFbw7. Both MCV LT and sT interact with Fbw7 but we cannot exclude the possibility that this might be a bridging effect mediated by direct sT targeting of other SCF complex proteins (e.g., Skp1, Cul-1 or Rbx-1) or an indirect inhibitory effect by sT on the recruitment of Fbw7 dimers to LT. Further, it is possible that LT not only has complex interaction with Fbw7 but is regulated by other E3 ligases affected by sT: LT is ubiquitylated in Fbw7(−/−) cells and sT inhibits the ubiquitylation of LT in Fbw7(−/−) cells (Figure S4A). Although LT is degraded as a target of SCFFbw7, sT readily interacts with Fbw7 through its LSD domain without itself being targeted by Fbw7 at this site.
MCV hijacking of the SCFFbw7 circuitry nonetheless has important consequences since this may increase expression of viral LT as well as other cellular oncoproteins such as c-Myc and cyclin E. The reasons why MCV has evolved such a complex mechanism to autoregulate LT levels through SCFFbw7 targeting remain enigmatic. sT is an alternatively spliced isoform from LT and is expected to be expressed together with LT under most cellular conditions. sT targeting of SCFFbw7 can be expected to perpetuate phosphorylated forms of LT that would otherwise be marked for rapid turnover. This is functionally reminiscent of SV40 LT, which interacts with Fbw7 to competitively interfere with substrate turnover (Welcker and Clurman, 2005) to achieve similar molecular end results. Other viral proteins, including HPV E7 (Oh et al., 2004), KSHV vIRF3 (LANA2) (Baresova et al., 2012), EBV EBNA3C (Knight et al., 2005), HPV E2 (Bellanger et al., 2010) and adenovirus E1A (Isobe et al., 2009) inhibit SCFFbw7 or related target F-box protein complexes, suggesting that this is a common theme among persistent DNA viral infections.
PP2A targeting is another common feature of polyomavirus early proteins (Campbell et al., 1995; Pallas et al., 1990; Sontag et al., 1993) and is critical in defined gene transformation of human cells using SV40 early proteins (Hahn et al., 1999; Hahn et al., 2002). PP2A actually represents a large class of protein phosphatases, comprising hundreds of different phosphatase holoenzymes that use different combinations of PP2A A, B and C subunits to target substrate for dephosphorylation. MCV retains PP2A interacting domains but these domains are predicted to be distinct from the region targeting SCFFbw7. The LSD, required for MCV sT to transform cells, is on the opposite molecular surface to the PP2A docking domain in MCV sT (Figure 2 and Movie S1). This loop is absent from SV40 sT, potentially explaining the inability of SV40 sT to synergistically promote MCV LT-mediated replication (Figure 2 and Movie S1).
Merkel cell carcinoma is a dead-end for the MCV lifecycle since the clonally-integrated virus is no longer capable of producing infectious virions from tumor cells. MCV targeting of SCFFbw7 through the MCV sT LSD appears to be a key feature in the normal viral lifecycle that allows the virus to persist and replicate as a harmless viral skin infection (Tolstov et al., 2011). Under conditions in which MCV initiates human tumor cell transformation, however, sT LSD-mediated targeting of viral and cellular oncoproteins may contribute to promotion of an aggressive human malignancy.
EXPERIMENTAL PROCEDURES
Plasmids
Codon-optimized, commercially synthesized MCV LT and sT antigen sequences were cloned into pcDNA6/V5/His vector (Invitrogen) with a modified multiple cloning site (MCS). All LT and sT constructs used are untagged. HA-Fbw7 and Flag-cMyc (Yada et al., 2004) plasmids were kindly provided by Dr. Nakayama (Kyushu University, Japan). See also supplemental experimental procedures.
Cell lines
293 or U2OS cells were cultured in DMEM with 10% fetal bovine serum (FBS) from Sigma. MCV positive Merkel cell carcinoma (MCC) cell line MKL-2 was cultured in RPMI with 10% FBS. HCT116 cells (hCDC4 +/+, +/−, −/−), kindly provided by Dr. Bert Vogelstein and obtained from Johns Hopkins University Genetic Resources Core Facility, were grown in McCoy’s 5A medium with 10% FBS. Rat-1 cells and NIH3T3 cells were maintained in DMEM with 5% FBS or 10% calf serum, respectively.
MCV Origin Replication Assay
The MCV replication origin assay was described previously (Feng et al., 2011; Kwun et al., 2009). Briefly, 293 cells were transfected with T antigen expression vector (LT/sT, 0.3 µg), pMCV-Ori339(97) (0.3 µg) by Lipofectamine 2000 (Invitrogen) in 12-well plates. Forty-eight hours after transfection, episomal DNA was collected by salt-precipitation. DNA was double-digested with BamHI and DpnI, then subjected to Southern hybridization or qRT-PCR.
Structural modeling
The model of MCV sT structure was generated using the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) based on SV 40 sT homolog structures (PDB ID: 2PF4, 2PKG). The figures were generated using PyMOL (http://www.pymol.org/).
Quantitiative Real-time RT-PCR Analysis
293 cells were transfected with LT or sT expression (0.3 µg) constructs and total RNA was isolated 2 days after transfection using TRIzol LS reagent (Invitrogen). qRT-PCR was carried out with total RNA (0.1 µg) and iScript one-step RT-PCR kit (Bio-Rad) using a SmartCycler (Cepheid) according to the manufacturer’s protocol. Primer sequences used for LT and GAPDH cDNA detection are described in Table S1.
Immunoprecipitation (IP) and immunoblotting
Cells were lysed in IP buffer (Tris-HCl (pH 7.4), 150 mM NaCl, 2% Triton X-100) freshly supplemented with protease inhibitor cocktail (Roche), 1 mM PMSF, 5 mM NaF, and 2.5 mM NaVO3. Lysates were incubated with specific antibody and followed by immunoblotting to detect interacting proteins. See also supplemental experimental procedures.
Lentiviral/retroviral infection for short hairpin RNA knockdown
Short hairpin RNAs (shRNA) targeting Fbw7 were designed (Table S1) and cloned into lentiviral vector pLKO.1 (Addgene) using AgeI and EcoRI. A control shRNA construct was obtained from Addgene (1864). shRNA transfected cells were selected with puromycin (2 µg/ml) for 4 days after infection. See also supplemental experimental procedures.
Soft agar colony formation assay
Rat-1 stably expressing small T antigens were seeded over a 0.6% agar layer in 6-well plate (2.5×104 cells/well) and grown for 3 weeks. Colonies were photographed and counted after 0.05% crystal violet staining as described previously (Shuda et al., 2011). All experiments were performed in triplicate.
Supplementary Material
HIGHLIGHTS.
-
-
MCV sT prevents turnover of the MCV helicase LT to enhance viral DNA replication.
-
-
The MCV sT domain, LSD, inhibits MCV LT turnover by targeting the cellular ligase SCFFbw7.
-
-
MCV sT inhibition of SCFFbw7 also attenuates the degradation of cellular oncoproteins.
-
-
MCV sT LSD mediates rodent cell transformation.
ACKNOWLEDGMENTS
We thank Bert Vogelstein for providing the HCT116 cells, and Keiichi I Nakayama and Robert Weinberg for providing HA-Fbw7, Flag-c-Myc and SV40 plasmids. We thank Gregg Allman and Erik Satie for help with this manuscript. This study was supported by National Institutes of Health grants CA136363 and CA120726 to P. S. Moore and Y. Chang, who are also supported as American Cancer Society Research Professors. UPCI shared resources (LI-COR IR Imaging Facility) used in this study is supported in part by award P30CA047904.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- Arabi A, Ullah K, Branca RM, Johansson J, Bandarra D, Haneklaus M, Fu J, Aries I, Nilsson P, Den Boer ML, et al. Proteomic screen reveals Fbw7 as a modulator of the NF-kappaB pathway. Nat. Commun. 2012;3:976. doi: 10.1038/ncomms1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Chang Y, Moore PS. MCV and Merkel cell carcinoma: a molecular success story. Curr. Opin. Virol. 2012;2:489–498. doi: 10.1016/j.coviro.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baresova P, Pitha PM, Lubyova B. Kaposi sarcoma-associated herpesvirus vIRF-3 protein binds to F-box of Skp2 protein and acts as a regulator of c-Myc protein function and stability. J. Biol. Chem. 2012;287:16199–16208. doi: 10.1074/jbc.M111.335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger S, Tan CL, Nei W, He PP, Thierry F. The human papillomavirus type 18 E2 protein is a cell cycle-dependent target of the SCFSkp2 ubiquitin ligase. J. Virol. 2010;84:437–444. doi: 10.1128/JVI.01162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikel I, Montano X, Agha ME, Brown M, McCormack M, Boltax J, Livingston DM. SV40 small t antigen enhances the transformation activity of limiting concentrations of SV40 large T antigen. Cell. 1987;48:321–330. doi: 10.1016/0092-8674(87)90435-1. [DOI] [PubMed] [Google Scholar]
- Campbell KS, Auger KR, Hemmings BA, Roberts TM, Pallas DC. Identification of regions in polyomavirus middle T and small t antigens important for association with protein phosphatase 2A. J. Virol. 1995;69:3721–3728. doi: 10.1128/jvi.69.6.3721-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LS, Pater MM, Hutchinson NI, di Mayorca G. Transformation by purified early genes of simian virus 40. Virology. 1984;133:341–353. doi: 10.1016/0042-6822(84)90400-8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Xu Y, Bao Q, Xing Y, Li Z, Lin Z, Stock JB, Jeffrey PD, Shi Y. Structural and biochemical insights into the regulation of protein phosphatase 2A by small t antigen of SV40. Nat. Struct. Mol. Biol. 2007;14:527–534. doi: 10.1038/nsmb1254. [DOI] [PubMed] [Google Scholar]
- Cho US, Morrone S, Sablina AA, Arroyo JD, Hahn WC, Xu W. Structural basis of PP2A inhibition by small t antigen. PLoS Biol. 2007;5:e202. doi: 10.1371/journal.pbio.0050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS ONE. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Matsumoto A, Inuzuka H, Zhai B, Lau AW, Wan L, Gao D, Shaik S, Yuan M, Gygi SP, et al. SCF(Fbw7) Modulates the NFkappaB Signaling Pathway by Targeting NFkappaB2 for Ubiquitination and Destruction. Cell Rep. 2012;1:434–443. doi: 10.1016/j.celrep.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J. Mol. Biol. 2011;409:529–542. doi: 10.1016/j.jmb.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson NC. Merkel cell carcinoma: changing incidence trends. J. Surg. Oncol. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int. J. Cancer. 2012;130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe T, Hattori T, Kitagawa K, Uchida C, Kotake Y, Kosugi I, Oda T, Kitagawa M. Adenovirus E1A inhibits SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 2009;284:27766–27779. doi: 10.1074/jbc.M109.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J. Virol. 2009;83:12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Nghiem P. Merkel cell carcinoma: more deaths but still no pathway to blame. J. Invest. Dermatol. 2007;127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol. Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- Neumann F, Borchert S, Schmidt C, Reimer R, Hohenberg H, Fischer N, Grundhoff A. Replication, gene expression and particle production by a consensus Merkel Cell Polyomavirus (MCPyV) genome. PLoS ONE. 2011;6:e29112. doi: 10.1371/journal.pone.0029112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Oh KJ, Kalinina A, Wang J, Nakayama K, Nakayama KI, Bagchi S. The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin 1- and Skp2-containing E3 ligase. J. Virol. 2004;78:5338–5346. doi: 10.1128/JVI.78.10.5338-5346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe H, Lee SH, Phuchareon J, Albertson DG, McCormick F, Tetsu O. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS ONE. 2006;1:e128. doi: 10.1371/journal.pone.0000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- Prins C, Frisque RJ. JC virus T' proteins encoded by alternatively spliced early mRNAs enhance T antigen-mediated viral DNA replication in human cells. J. Neurovirol. 2001;7:250–264. doi: 10.1080/13550280152403290. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. hCDC4 and genetic instability in cancer. Cell Cycle. 2004;3:693–694. doi: 10.4161/cc.3.6.940. [DOI] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Buck CB. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011;7:e1002161. doi: 10.1371/journal.ppat.1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int. J. Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Tolstov YL, Knauer A, Chen JG, Kensler TW, Kingsley LA, Moore PS, Chang Y. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg. Infect. Dis. 2011;17:1371–1380. doi: 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J. Biol. Chem. 2005;280:7654–7658. doi: 10.1074/jbc.M413377200. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Kudchodkar SB, Alwine JC. Effects of simian virus 40 large and small tumor antigens on mammalian target of rapamycin signaling: small tumor antigen mediates hypophosphorylation of eIF4E-binding protein 1 late in infection. J. Virol. 2005;79:6882–6889. doi: 10.1128/JVI.79.11.6882-6889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex regulates p21(CIP1/WAF1) and cyclin D proteins. Proc. Natl. Acad. Sci. USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.