Background: GMF regulates Arp2/3 complex debranching.
Results: GMF binds preferentially to ADP-Arp2/3 complex. The phosphomimetic mutation S2E in GMF inhibits this interaction.
Conclusion: The preference of GMF for ADP-Arp2/3 complex might play a physiological role by promoting debranching of aged branch junctions without interfering with nucleation.
Significance: We show that GMF interaction with Arp2/3 complex obeys similar principles as ADF/cofilin interaction with actin.
Keywords: Actin, ADP, ATP, Isothermal Titration Calorimetry, Protein Phosphorylation, Arp2/3 Complex, GMF
Abstract
Glia maturation factor (GMF) is a member of the actin-depolymerizing factor (ADF)/cofilin family. ADF/cofilin promotes disassembly of aged actin filaments, whereas GMF interacts specifically with Arp2/3 complex at branch junctions and promotes debranching. A distinguishing feature of ADF/cofilin is that it binds tighter to ADP-bound than to ATP-bound monomeric or filamentous actin. The interaction is also regulated by phosphorylation at Ser-3 of mammalian cofilin, which inhibits binding to actin. However, it is unknown whether these two factors play a role in the interaction of GMF with Arp2/3 complex. Here we show using isothermal titration calorimetry that mammalian GMF has very low affinity for ATP-bound Arp2/3 complex but binds ADP-bound Arp2/3 complex with 0.7 μm affinity. The phosphomimetic mutation S2E in GMF inhibits this interaction. GMF does not bind monomeric ATP- or ADP-actin, confirming its specificity for Arp2/3 complex. We further show that mammalian Arp2/3 complex nucleation activated by the WCA region of the nucleation-promoting factor N-WASP is not affected by GMF, whereas nucleation activated by the WCA region of WAVE2 is slightly inhibited at high GMF concentrations. Together, the results suggest that GMF functions by a mechanism similar to that of other ADF/cofilin family members, displaying a preference for ADP-Arp2/3 complex and undergoing inhibition by phosphorylation of a serine residue near the N terminus. Arp2/3 complex nucleation occurs in the ATP state, and nucleotide hydrolysis promotes debranching, suggesting that the higher affinity of GMF for ADP-Arp2/3 complex plays a physiological role by promoting debranching of aged branch junctions without interfering with Arp2/3 complex nucleation.
Introduction
Glia maturation factor (GMF)2 is a 17-kDa protein conserved from yeast to human (1). Mammals express two GMF isoforms, GMFβ and GMFγ, which share 82% sequence identity but display different tissue distributions (2). GMFβ is expressed mainly in the brain and has been associated with nervous system development and degeneration (3). GMFγ is expressed in microvascular endothelial and inflammatory cells and has been implicated in promoting neutrophil and T cell migration (4, 5). GMF is a member of the actin-depolymerizing factor (ADF)/cofilin family (1). Thus, human GMFβ and GMFγ, which share 17.6 and 15.5% sequence identities with human cofilin-1, respectively, display a three-dimensional fold similar to that of other members of the ADF/cofilin family (6). Members of this family, including twinfilin, Abp1, drebrin, and coactosin, are generally implicated in regulation of actin cytoskeleton dynamics (1).
GMF is unique among ADF/cofilin family members in that it regulates the activity of Arp2/3 complex (7, 8). Arp2/3 complex mediates nucleation and branching of actin filaments at the leading edge of motile cells (9, 10). It consists of seven subunits, including the actin-related proteins Arp2 and Arp3 and subunits ARPC1–5. Multiple factors contribute to activating Arp2/3 complex, including ATP (11–13), pre-existing (mother) filaments (14), and nucleation-promoting factors (NPFs) (15, 16). Most NPFs contain a C-terminal WCA (WH2, central, and acidic domains) region featuring binding sites for actin (W) (17) and Arp2/3 complex (C and A) (14, 18). In this way, NPFs recruit actin and Arp2/3 complex and promote the formation of a branch filament that grows at a 70° angle relative to the mother filament (19, 20).
Both Arp2/3 complex and actin use nucleotide hydrolysis as a timer to regulate their transition in and out of filamentous networks. Thus, nucleotide hydrolysis on actin controls treadmilling, whereby polymerization of ATP-actin at the barbed end of the filament is followed by fast hydrolysis and slow phosphate release, resulting in the accumulation and subsequent dissociation of ADP-actin at the pointed end (21). Similarly, Arp2/3 complex nucleation occurs in the ATP state (11–13), and nucleotide hydrolysis promotes debranching (22–24). In actin, the nucleotide state also regulates its interactions with actin-binding proteins through subtle conformational changes (25). Specifically, most ADF/cofilin family members interact with both monomeric and filamentous actin with higher affinity in the ADP state than in the ATP state (26–30). In this way, the primary role of ADF/cofilin is to stimulate the depolymerization of aged ADP-containing actin filaments by promoting either filament severing (31–34) or monomer dissociation at the pointed end (35). This raises important questions. Is the interaction of GMF with Arp2/3 complex also stronger in the ADP state? If so, how does GMF inhibit the nucleation of Arp2/3 complex in the ATP state as suggested by some studies (7, 8, 36)? These questions are addressed here in an attempt to understand the role of GMF in Arp2/3 complex assembly dynamics.
Phosphorylation of a serine residue near the N terminus (Ser-3 in mammalian cofilin) inhibits the interactions of several ADF/cofilin family members with monomeric actin, as well as their filament disassembly activities (37–39). The structure of a complex of actin with the C-terminal ADF homology domain of twinfilin shows that the N-terminal region is directly involved in interactions with actin (40), explaining how phosphorylation at this site can play a regulatory role. GMF also contains conserved serine residues at positions 2 and 4. Phosphorylation of these two sites has been confirmed in cells, although only phosphorylation of Ser-2 appears to play a regulatory role (4). However, contrary to ADF/cofilin, it was initially reported that phosphorylation of Ser-2 increased the affinity of GMFγ for both Arp2/3 complex and F-actin (4). In contrast, another study found that GMFβ and GMFγ carrying the phosphomimetic mutation S2E inhibited yeast Arp2/3 complex nucleation to a lesser extent than the wild-type proteins, suggesting weaker affinity for the complex (8). A third study found that this mutation had no effect on yeast cell growth and did not affect the debranching activity of yeast GMF in vitro (36). Because of these conflicting results, we revisit here the role of N-terminal phosphorylation in the interaction of GMF with Arp2/3 complex.
EXPERIMENTAL PROCEDURES
Proteins
The cDNA encoding human GMFγ (UniProt O60234) was synthesized (GENEWIZ, Inc.) and cloned between the NdeI and SapI sites of the pTYB1 vector (New England Biolabs). This vector comprises a chitin-binding domain for affinity purification and an intein for precise self-cleavage of the purification tag such that no extra residues remain after purification that could interfere with GMF activity. Point mutants GMFγS2E and GMFγS2A were generated using the QuikChange mutagenesis kit (Qiagen). WCA fragments of mouse WAVE2 (residues 433–497; UniProt Q8BH43) and N-WASP (neural Wiskott-Aldrich syndrome protein; residues 426–501; UniProt Q91YD9) were cloned between the NdeI and EcoRI sites of the pTYB12 vector (New England Biolabs). Expression was carried out in BL21(DE3) cells (Invitrogen), grown in Terrific Broth medium at 37 °C for 6 h, and induced by the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside at 20 °C overnight. All proteins were first purified on a chitin affinity column (New England Biolabs). Affinity purification was followed either by HPLC purification on a reverse-phase C18 column using a CH3CN gradient of 0–90 and 0.1% TFA (WCA constructs) or by gel filtration on a HiLoad 26/600 Superdex 200 column (GMFγ). Arp2/3 complex was purified from bovine brain as described (41), but in the absence of nucleotide. Actin was purified from rabbit skeletal muscle (42).
Preparation of Different Nucleotide States of Arp2/3 Complex and Actin
Tissue-purified Arp2/3 complex is in a nucleotide-free state (43). After purification, the complex was dialyzed for 72 h against Arp buffer (20 mm HEPES (pH 7.5), 100 mm KCl, 1 mm MgCl2, 1 mm EGTA, and 1 mm DTT) supplemented with 0.2 mm ADP (or ATP). In one of the experiments, ATP-bound Arp2/3 complex was converted to the ADP state in two dialysis steps: first, against Arp buffer supplemented with a high amount of ADP (2 mm) to ensure complete exchange, followed by dialysis against 0.2 mm ADP (as in other experiments). Actin is a very slow ATPase and is purified as ATP-bound actin. To obtain the ADP-bound state, actin was dialyzed against 5 mm HEPES (pH 7.5), 0.2 mm CaCl2, and 1 mm DTT supplemented with 0.2 mm ADP, with the addition of 20 units ml−1 hexokinase and 1 mm glucose, as described previously (44).
Isothermal Titration Calorimetry (ITC)
ITC measurements were performed on a MicroCal VP-ITC calorimeter at 20 °C. The duration of each injection was 7 s, with an interval of 200 s between injections. Arp2/3 complex (ADP- or ATP-bound) in the cell (1.44 ml at 8–10 μm concentration) was titrated in 7-μl injections with a 14-fold molar excess of GMFγ. The same conditions were used in titrations of GMFγ into ADP- or ATP-actin (1.44 ml at 13–15 μm concentration), which was kept monomeric with the addition of latrunculin B. Each experiment was corrected for the small exothermic heat of injection resulting from the titration of GMFγ into buffer. Data were analyzed using the MicroCal Origin program.
Actin Polymerization Assay
Actin polymerization was measured as the fluorescence increase resulting from the incorporation of pyrene-labeled actin into filaments using a Cary Eclipse fluorescence spectrophotometer (Varian). Prior to data acquisition, 2 μm MgATP-actin (6% pyrene-labeled) was mixed with different components, including Arp2/3 complex, WCA, and GMF (as indicated in Fig. 2) in 5 mm Tris (pH 8.0), 1 mm MgCl2, 50 mm KCl, 1 mm EGTA, 0.1 mm NaN3, 0.02 mg ml−1 BSA, and 0.2 mm ATP. Data acquisition started 10 s after mixing. All measurements were done at 25 °C. Control experiments were carried out with the addition of buffer alone. Polymerization rates were calculated as the slope at 50% polymerization and converted to nm s−1 (nm monomers adding to filaments s−1), assuming a total concentration of polymerizable actin of 1.9 μm (45).
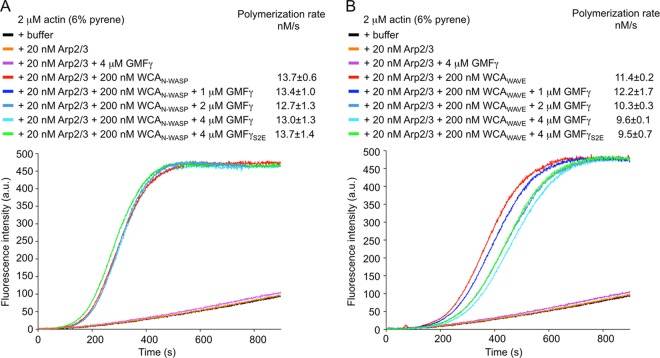
FIGURE 2.
GMFγ does not inhibit actin polymerization by Arp2/3 complex. Shown are the time courses of the fluorescence increase upon polymerization of 2 μm actin (6% pyrene-labeled) alone (black line) and with addition of the indicated proteins (colored lines). A, effect of different concentrations of GMFγ on actin polymerization induced by 20 nm Arp2/3 complex activated by 200 nm WCAN-WASP. Polymerization rates at 50% polymerization are shown. The lag time (measured as the time to 10% polymerization) was 190 s. B, effect of different concentrations of GMFγ on actin polymerization induced by 20 nm Arp2/3 complex activated by 200 nm WCAWAVE. Lag times were 229, 245, 277, and 292 s for 0, 1, 2, and 4 μm GMF, respectively. Each measurement was performed three times; one representative curve is shown. Errors are reported as S.E. a.u., arbitrary units.
RESULTS
The Interaction of GMF with Arp2/3 Complex Depends on the State of the Nucleotide
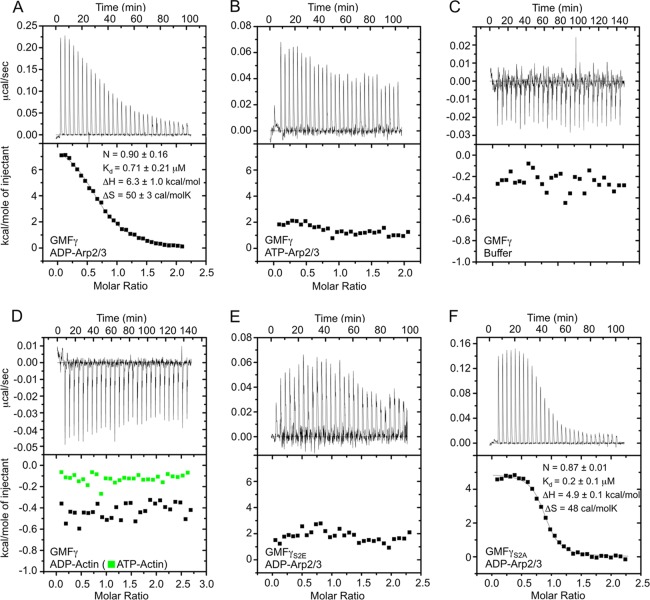
Previous studies have reported binding of yeast GMF to yeast Arp2/3 complex with widely diverging affinities: 1.0 μm (8) or 13 nm (36). One of these studies additionally reported that there were two binding sites for GMF on Arp2/3 complex (36). Moreover, these studies did not consider the nucleotide state of Arp2/3 complex, which, by analogy with other ADF/cofilin family members, should play a critical role in the interaction. Here, we used ITC to analyze the interaction of mammalian GMFγ with mammalian Arp2/3 complex in the ADP- and ATP-bound states. We found that ADP-Arp2/3 complex bound GMFγ with 0.7 μm affinity (Fig. 1A). In contrast, GMFγ interacted very weakly with ATP-Arp2/3 complex, and the data could not be fit to a binding isotherm (Fig. 1B). Note, however, that the titration of GMFγ into ATP-Arp2/3 complex had an endothermic profile, whereas that of GMFγ into buffer was exothermic (Fig. 1, compare B and C), suggesting some binding, albeit very weak. Of note, the heats of titration for ATP-Arp2/3 complex were similar to those observed at saturation for ADP-Arp2/3 complex. The parameters shown in Fig. 1A are the average of four different experiments, including one experiment in which ATP-Arp2/3 complex was recovered after the titration, converted to ADP-Arp2/3 complex (see “Experimental Procedures”), and re-titrated with GMFγ, which resulted in very similar binding. In contrast to a previous study (36), we observed only a single binding site for GMFγ on Arp2/3 complex, which is consistent with a recent crystal structure of this complex (46). The interaction was also specific for Arp2/3 complex because GMFγ did not bind to either ADP- or ATP-bound actin (Fig. 1D).
FIGURE 1.
Analysis by ITC of the binding of GMFγ to Arp2/3 complex. Experiments were conducted at 20 °C. Arp2/3 complex (or actin-latrunculin B) in the cell at 8–10 μm (or 13–15 μm) was titrated with a 14-fold molar excess of GMFγ in 7-μl injections (7-s injections, with an interval of 200 s between injections). A, titration of GMFγ into ADP-Arp2/3 complex. The data were fit to a binding isotherm derived from the integrated heats of binding plotted against the molar ratio of ligand (GMFγ) added to ADP-Arp2/3 complex in the cell after subtracting the heat of dilution. The best fit parameters (solid black line) correspond to a one-site binding model with a dissociation constant of 0.7 μm. B, titration of GMFγ into ATP-Arp2/3 complex. The data could not be fit to a binding isotherm. C, titration of GMFγ into buffer (control experiment). D, titration of GMFγ into ADP-actin (black squares) and ATP-actin (green squares). Note that these two titrations look similar to that of GMFγ into buffer, indicating a complete lack of interaction. E, titration of GMFγS2E into ADP-Arp2/3 complex. The data could not be fit to a binding isotherm. F, titration of GMFγS2A into ADP-Arp2/3 complex. Each titration was repeated at least two times, and four times for that shown in A. In A, errors are reported as S.E., whereas for the other titrations, errors were derived from curve fitting.
The Phosphomimetic Mutation S2E Inhibits Binding of GMFγ to ADP-Arp2/3 Complex
Previous studies have produced conflicting results regarding the role of N-terminal phosphorylation in the interaction of GMF with Arp2/3 complex (4, 8, 36). These studies used the phosphomimetic mutation S2E. By ITC, we found that this mutation strongly inhibited binding to ADP-Arp2/3 complex (Fig. 1E), resulting in a titration profile similar to that observed with ATP-Arp2/3 complex (Fig. 1, compare B and E). In contrast, the GMFγS2A mutant bound ADP-Arp2/3 complex with similar affinity to wild-type GMFγ (Fig. 1F), suggesting that the inhibitory effect of the S2E mutation was due to the additional charge and not to the removal of the serine side chain.
GMFγ Does Not Interfere with the Nucleation Activity of Mammalian Arp2/3 Complex
Previous studies have suggested that both yeast and mammalian GMFs inhibit yeast and mammalian Arp2/3 complex nucleation (7, 8, 36). Arp2/3 complex requires ATP for nucleation (11–13), and polymerization assays are always conducted in the presence of ATP. In light of our finding that GMFγ has very low affinity for ATP-Arp2/3 complex, we questioned whether it could interfere with nucleation. We conducted pyrene-actin (6% pyrene-labeled) polymerization assays using mammalian Arp2/3 complex and the WCA regions of two NPFs, N-WASP and WAVE2, in the absence or presence of increasing concentrations of GMFγ (Fig. 2). Compared with control experiments with actin alone, polymerization was strongly stimulated in the presence of 20 nm Arp2/3 complex and 200 nm WCAN-WASP. The addition of increasing concentrations of GMFγ or GMFγS2E (up to 4 μm) had no effect on this activity (Fig. 2A). In the case of WCAWAVE, we observed a small inhibitory effect at high concentrations of GMFγ, as reflected by somewhat lower polymerization rates and increased lag times of polymerization (Fig. 2B).
DISCUSSION
Together, the results presented here show a parallel between the ways in which GMF interacts with Arp2/3 complex and ADF/cofilin interacts with actin. We have demonstrated for the first time that GMFγ has a clear preference for ADP-bound versus ATP-bound Arp2/3 complex, which could have important physiological implications. Indeed, in vitro branches formed by Arp2/3 complex persist for hundreds of seconds (47), whereas branch turnover in cells occurs within a few seconds (48), implying the existence of cellular factors that accelerate debranching. One mechanism for debranching has been proposed to involve cofilin-mediated disassembly of the mother filament (49). However, by interacting directly with Arp2/3 complex, GMF acts in a more specific manner, inducing debranching at low concentrations (7, 36). While Arp2/3 complex nucleation requires ATP (11–13), nucleotide hydrolysis occurs almost immediately after nucleation and promotes debranching (23, 24). It thus appears that the higher affinity of GMF for ADP-Arp2/3 complex is specifically tailored for disassembly of older ADP-containing branches. The weak affinity for ATP-Arp2/3 complex is equally important, as it reduces the likelihood of GMF interfering with the nucleation step, i.e. the formation of new branches. ATP binding to Arp2/3 complex may also provide a mechanism for GMF dissociation, freeing both Arp2/3 complex and GMF for new rounds of nucleation and debranching.
Another way in which GMF might be prevented from interfering with the nucleation step is by competition with NPFs. Indeed, a recent crystal structure shows that GMF binds at the barbed end of Arp2 (46), whereas various studies have suggested that NPFs deliver an actin monomer at the barbed end of Arp2 during the first steps of nucleation (41, 50, 51). Thus, although our results generally contrast with previous reports of strong inhibition of nucleation by GMF (8, 36), we observed some inhibition of nucleation induced by WCAWAVE but not WCAN-WASP, which can be explained by competition. Indeed, actin-WCAWAVE has 50-fold lower affinity for the site on Arp2 than actin-WCAN-WASP.3 Similarly, differences in affinity between Arp2/3 complex and NPFs and/or GMF may also explain why the yeast system analyzed previously (8, 36) is more susceptible to inhibition by GMF. It is important to point out, however, that the slight inhibition observed with mammalian WCAWAVE occurred at high GMF concentrations, which are probably irrelevant in the cellular context, particularly considering the marked preference of GMF for ADP-Arp2/3 complex.
We also found that GMFγ binds to ADP-Arp2/3 complex with 1:1 stoichiometry, not 2:1 as suggested by a recent study (36). The crystal structure of GMFγ bound to Arp2/3 complex shows a single binding site at the barbed end of Arp2, consistent with our results. The protein-protein contacts appear to be highly specific for GMF-Arp2 and cannot be reproduced on Arp3, where the corresponding binding interface is very different. We also note that this structure was determined in the ATP state, which we found binds very weakly to GMFγ. However, the interaction is made possible by the high protein concentration used in crystallization. Curiously, the only existing structure of a complex of actin with a member of the ADF/cofilin family was also determined in the ATP-bound state (40). Therefore, there is a clear need for a structure of a complex showing the higher affinity ADP-bound state.
Finally, through the study of the S2E phosphomimetic mutant (and a control S2A mutant), we found that phosphorylation at the N terminus of GMF inhibits its interaction with ADP-Arp2/3 complex. We thus conclude that striking parallels exist between GMF and ADF/cofilin, both in the way they select for their ADP-bound partners and in the way they are regulated by phosphorylation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM073791.
M. Boczkowska, G. Rebowski, and R. Dominguez, unpublished data.
- GMF
- glia maturation factor
- ADF
- actin-depolymerizing factor
- NPF
- nucleation-promoting factor
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Poukkula M., Kremneva E., Serlachius M., Lappalainen P. (2011) Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton 68, 471–490 [DOI] [PubMed] [Google Scholar]
- 2. Inagaki M., Aoyama M., Sobue K., Yamamoto N., Morishima T., Moriyama A., Katsuya H., Asai K. (2004) Sensitive immunoassays for human and rat GMFB and GMFG, tissue distribution and age-related changes. Biochim. Biophys. Acta 1670, 208–216 [DOI] [PubMed] [Google Scholar]
- 3. Stolmeier D., Thangavel R., Anantharam P., Khan M. M., Kempuraj D., Zaheer A. (2013) Glia maturation factor expression in hippocampus of human alzheimer's disease. Neurochem. Res. 38, 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ikeda K., Kundu R. K., Ikeda S., Kobara M., Matsubara H., Quertermous T. (2006) Glia maturation factor-γ is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ. Res. 99, 424–433 [DOI] [PubMed] [Google Scholar]
- 5. Aerbajinai W., Lee K., Chin K., Rodgers G. P. (2013) Glia maturation factor-γ negatively modulates TLR4 signaling by facilitating TLR4 endocytic trafficking in macrophages. J. Immunol. 190, 6093–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goroncy A. K., Koshiba S., Tochio N., Tomizawa T., Sato M., Inoue M., Watanabe S., Hayashizaki Y., Tanaka A., Kigawa T., Yokoyama S. (2009) NMR solution structures of actin depolymerizing factor homology domains. Protein Sci. 18, 2384–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gandhi M., Smith B. A., Bovellan M., Paavilainen V., Daugherty-Clarke K., Gelles J., Lappalainen P., Goode B. L. (2010) GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr. Biol. 20, 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakano K., Kuwayama H., Kawasaki M., Numata O., Takaine M. (2010) GMF is an evolutionarily developed Adf/cofilin-super family protein involved in the Arp2/3 complex-mediated organization of the actin cytoskeleton. Cytoskeleton 67, 373–382 [DOI] [PubMed] [Google Scholar]
- 9. Pollard T. D., Beltzner C. C. (2002) Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 12, 768–774 [DOI] [PubMed] [Google Scholar]
- 10. Mullins R. D., Heuser J. A., Pollard T. D. (1998) The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 95, 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dayel M. J., Holleran E. A., Mullins R. D. (2001) Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad. Sci. U.S.A. 98, 14871–14876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goley E. D., Rodenbusch S. E., Martin A. C., Welch M. D. (2004) Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol. Cell 16, 269–279 [DOI] [PubMed] [Google Scholar]
- 13. Le Clainche C., Didry D., Carlier M. F., Pantaloni D. (2001) Activation of Arp2/3 complex by Wiskott-Aldrich syndrome protein is linked to enhanced binding of ATP to Arp2. J. Biol. Chem. 276, 46689–46692 [DOI] [PubMed] [Google Scholar]
- 14. Higgs H. N., Blanchoin L., Pollard T. D. (1999) Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38, 15212–15222 [DOI] [PubMed] [Google Scholar]
- 15. Machesky L. M., Mullins R. D., Higgs H. N., Kaiser D. A., Blanchoin L., May R. C., Hall M. E., Pollard T. D. (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 96, 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Welch M. D., Rosenblatt J., Skoble J., Portnoy D. A., Mitchison T. J. (1998) Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science 281, 105–108 [DOI] [PubMed] [Google Scholar]
- 17. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Panchal S. C., Kaiser D. A., Torres E., Pollard T. D., Rosen M. K. (2003) A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat. Struct. Biol. 10, 591–598 [DOI] [PubMed] [Google Scholar]
- 19. Blanchoin L., Amann K. J., Higgs H. N., Marchand J. B., Kaiser D. A., Pollard T. D. (2000) Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007–1011 [DOI] [PubMed] [Google Scholar]
- 20. Rouiller I., Xu X. P., Amann K. J., Egile C., Nickell S., Nicastro D., Li R., Pollard T. D., Volkmann N., Hanein D. (2008) The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dominguez R. (2010) Structural insights into de novo actin polymerization. Curr. Opin. Struct. Biol. 20, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le Clainche C., Pantaloni D., Carlier M. F. (2003) ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc. Natl. Acad. Sci. U.S.A. 100, 6337–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin A. C., Welch M. D., Drubin D. G. (2006) Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol. 8, 826–833 [DOI] [PubMed] [Google Scholar]
- 24. Ingerman E., Hsiao J. Y., Mullins R. D. (2013) Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J. Cell Biol. 200, 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dominguez R., Holmes K. C. (2011) Actin structure and function. Annu. Rev. Biophys. 40, 169–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maciver S. K., Weeds A. G. (1994) Actophorin preferentially binds monomeric ADP-actin over ATP-bound actin: consequences for cell locomotion. FEBS Lett. 347, 251–256 [DOI] [PubMed] [Google Scholar]
- 27. Blanchoin L., Pollard T. D. (1998) Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J. Biol. Chem. 273, 25106–25111 [DOI] [PubMed] [Google Scholar]
- 28. Carlier M. F., Laurent V., Santolini J., Melki R., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D. (1997) Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vartiainen M. K., Mustonen T., Mattila P. K., Ojala P. J., Thesleff I., Partanen J., Lappalainen P. (2002) The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell 13, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yeoh S., Pope B., Mannherz H. G., Weeds A. (2002) Determining the differences in actin binding by human ADF and cofilin. J. Mol. Biol. 315, 911–925 [DOI] [PubMed] [Google Scholar]
- 31. Okreglak V., Drubin D. G. (2007) Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178, 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andrianantoandro E., Pollard T. D. (2006) Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 [DOI] [PubMed] [Google Scholar]
- 33. Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C. J., Martiel J. L., Blanchoin L. (2007) Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17, 825–833 [DOI] [PubMed] [Google Scholar]
- 34. McCullough B. R., Grintsevich E. E., Chen C. K., Kang H., Hutchison A. L., Henn A., Cao W., Suarez C., Martiel J. L., Blanchoin L., Reisler E., De La Cruz E. M. (2011) Cofilin-linked changes in actin filament flexibility promote severing. Biophys. J. 101, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carlier M. F., Ressad F., Pantaloni D. (1999) Control of actin dynamics in cell motility. Role of ADF/cofilin. J. Biol. Chem. 274, 33827–33830 [DOI] [PubMed] [Google Scholar]
- 36. Ydenberg C. A., Padrick S. B., Sweeney M. O., Gandhi M., Sokolova O., Goode B. L. (2013) GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr. Biol. 23, 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morgan T. E., Lockerbie R. O., Minamide L. S., Browning M. D., Bamburg J. R. (1993) Isolation and characterization of a regulated form of actin depolymerizing factor. J. Cell Biol. 122, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Agnew B. J., Minamide L. S., Bamburg J. R. (1995) Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 270, 17582–17587 [DOI] [PubMed] [Google Scholar]
- 39. Ressad F., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D., Carlier M. F. (1998) Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J. Biol. Chem. 273, 20894–20902 [DOI] [PubMed] [Google Scholar]
- 40. Paavilainen V. O., Oksanen E., Goldman A., Lappalainen P. (2008) Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 182, 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boczkowska M., Rebowski G., Petoukhov M. V., Hayes D. B., Svergun D. I., Dominguez R. (2008) X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure 16, 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pardee J. D., Spudich J. A. (1982) Purification of muscle actin. Methods Enzymol. 85, 164–181 [DOI] [PubMed] [Google Scholar]
- 43. Nolen B. J., Littlefield R. S., Pollard T. D. (2004) Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc. Natl. Acad. Sci. U.S.A. 101, 15627–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pollard T. D. (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harris E. S., Higgs H. N. (2006) Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 406, 190–214 [DOI] [PubMed] [Google Scholar]
- 46. Luan Q., Nolen B. J. (2013) Structural basis for regulation of Arp2/3 complex by GMF. Nat. Struct. Mol. Biol. 10.1038/nsmb.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blanchoin L., Pollard T. D., Mullins R. D. (2000) Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 48. Lai F. P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H. G., Stradal T. E., Dunn G. A., Small J. V., Rottner K. (2008) Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chan C., Beltzner C. C., Pollard T. D. (2009) Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Padrick S. B., Doolittle L. K., Brautigam C. A., King D. S., Rosen M. K. (2011) Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. U.S.A. 108, E472–E479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ti S. C., Jurgenson C. T., Nolen B. J., Pollard T. D. (2011) Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 108, E463–E471 [DOI] [PMC free article] [PubMed] [Google Scholar]