Background: Staphylococcus aureus utilizes heme as an iron source during an infection.
Results: An oxidoreductase, IruO, can supply electrons to IsdI and IsdG for heme degradation and iron extraction.
Conclusion: IruO is likely the in vivo reductant for heme degradation to the staphylobilins.
Significance: Heme degradation is a potential target for anti-S. aureus therapeutics.
Keywords: FAD, Heme, Iron, Reductase, Staphylococcus aureus, Staphylobilins
Abstract
Staphylococcus aureus is a common hospital- and community-acquired bacterium that can cause devastating infections and is often multidrug-resistant. Iron acquisition is required by S. aureus during an infection, and iron acquisition pathways are potential targets for therapies. The gene NWMN2274 in S. aureus strain Newman is annotated as an oxidoreductase of the diverse pyridine nucleotide-disulfide oxidoreductase (PNDO) family. We show that NWMN2274 is an electron donor to IsdG and IsdI catalyzing the degradation of heme, and we have renamed this protein IruO. Recombinant IruO is a FAD-containing NADPH-dependent reductase. In the presence of NADPH and IruO, either IsdI or IsdG degraded bound heme 10-fold more rapidly than with the chemical reductant ascorbic acid. Varying IsdI-heme substrate and monitoring loss of the heme Soret band gave a Km of 15 ± 4 μm, a kcat of 5.2 ± 0.7 min−1, and a kcat/Km of 5.8 × 103 m−1 s−1. From HPLC and electronic spectra, the major heme degradation products are 5-oxo-δ-bilirubin and 15-oxo-β-bilirubin (staphylobilins), as observed with ascorbic acid. Although heme degradation by IsdI or IsdG can occur in the presence of H2O2, the addition of catalase and superoxide dismutase did not disrupt NADPH/IruO heme degradation reactions. The degree of electron coupling between IruO and IsdI or IsdG remains to be determined. Homologs of IruO were identified by sequence similarity in the genomes of Gram-positive bacteria that possess IsdG-family heme oxygenases. A phylogeny of these homologs identifies a distinct clade of pyridine nucleotide-disulfide oxidoreductases likely involved in iron uptake systems. IruO is the likely in vivo reductant required for heme degradation by S. aureus.
Introduction
Staphylococcus aureus is a Gram-positive pathogen that causes a diverse range of infections from skin and soft tissue infections to necrotizing pneumonia and fasciitis using many virulence factors (1, 2). S. aureus can be acquired either in the community or nosocomially, and many pathogenic strains are multidrug resistant, leaving a limited number of treatment options available (3). Furthermore, drug-resistant strains have spread throughout the world (4), leading to a need for the characterization of S. aureus pathways required for infectivity as a foundation to new human therapies.
Like almost all bacteria, S. aureus requires a source of iron for bacterial metabolism and growth. Within mammalian hosts, the concentration of iron freely available to S. aureus is negligible as iron is found either intracellularly as protein cofactors or complexed by host proteins such as transferrin and lactoferrin (5). This iron sequestration is a form of nutritional immunity that protects mammals from infection (6). Consequently, S. aureus has evolved multiple strategies for iron acquisition (7). S. aureus produces two siderophores, staphyloferrin A (8, 9) and staphyloferrin B (10), and has a transport system that can co-opt hydroxamate-type siderophores produced by other bacteria (11). S. aureus can also obtain heme from host heme-containing proteins hemoglobin and haptoglobin, transport it across the bacterial cell envelope, cleave the porphyrin ring, and release iron for use by the cell with the well characterized iron-regulated surface determinant (Isd)3 system (12). A series of cell wall-anchored proteins (IsdA, IsdB, IsdC, and IsdH) bind host heme-containing proteins, extract heme, and shuttle it to the bacterial membrane (13–19). There an ABC transporter consisting of IsdE, IsdF, and possibly IsdD moves heme across the membrane and into the cytoplasm (14, 20, 21). Once in the cytoplasm, two paralogous (64% amino acid sequence identity) but differentially regulated proteins (IsdG and IsdI) have the ability to cleave the porphyrin ring of heme and release iron (22–24). The Isd pathway is important for the pathogenesis of S. aureus as heme may be the preferred iron source (25), and IsdB and IsdE have both been implicated in systemic infections of mice (17, 26).
In vitro cleavage of the porphyrin ring by IsdG or IsdI requires molecular oxygen and a source of electrons, and ascorbic acid or non-S. aureus reductase proteins have typically been used as the electron donor (22). IsdG and IsdI cleave the porphyrin ring at either the δ-meso or β-meso carbons, resulting in two different products, 5-oxo-δ-bilirubin and 15-oxo-β-bilirubin, that are known as the staphylobilins. They are similar to but distinct from biliverdin, the product of heme degradation by conventional heme oxygenases such as human heme oxygenase (HO-1), suggesting that the reaction mechanism is different (27). Unlike HO-1, which generates CO during heme degradation, IsdG and IsdI generate formaldehyde (28). Heme bound to IsdG and IsdI is significantly distorted from planarity in a fashion described as ruffling (29, 30); IsdI amino acid variants with decreased heme ruffling capability have decreased heme degradation rates (31). Outstanding questions about heme degradation in S. aureus, include how the reaction differs from other heme degrading enzymes to generate these novel products, what is the intracellular fate of the staphylobilins, and what is the in vivo electron donor for the reaction?
Here, we show that a protein encoded by NWMN2274 in S. aureus strain Newman can act as a source of electrons for heme degradation by IsdG and IsdI in the presence of NADPH. In vitro heme degradation in the presence of this protein yields the same products as reactions with ascorbic acid as an electron donor. From the specificity of the reaction, NWMN2274 is proposed to be the biological reductase associated with IsdG or IsdI heme degradation within the cytoplasm of S. aureus, and we have named this protein the iron utilization oxidoreductase, or IruO.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals were obtained from Fisher unless noted below.
Cloning, Protein Expression, and Purification
Full-length NWMN2274 was PCR-amplified from S. aureus strain Newman chromosomal DNA with forward primer (5′-AGC GGC CTG GTG CCG CGC GGC AGC ATG AAA GAT GTT ACA ATC ATT GGT-3′) and reverse primer (5′-GCG GCC GCA AGC TTG TCG ACG GAG TTA CTA GTA TAA ATG TTT ATT TAC AAT-3′) to form a megaprimer PCR product. Underlined sequences represent homology to pET28a. The thermal cycling conditions were 98 °C for 1 min, 30 cycles of 98 °C (10 s), 58 °C (30 s), and 72 °C (30 s), and a final extension at 72 °C for 5 min. Megaprimer extension for increased homology to pET28a was performed with forward extension primer 5′-AGC AGC CAT CAT CAT CAT CAT CAC AGC AGC GGC CTG GTG CCG CGC GGC AGC-3′ and reverse extension primer 5′-TGG TGG TGG TGC TCG AGT GCG GCC GCA AGC TTG TCG ACG GAG TTA-3′. The thermal cycling conditions were 98 °C for 1 min, 25 cycles of 98 °C (10 s), 55 °C (20 s), and 72 °C (15 s), and a final extension at 72 °C for 5 min. Both reactions were performed with Phusion High-Fidelity DNA polymerase (New England Biolabs). Insertion of the NWMN2274 amplicon into pET28a was performed using a previously described whole plasmid PCR technique (32). The pET28a-NWMN2274 construct was introduced into Escherichia coli BL21(λDE3). Colonies containing pET28a-NWMN2274 were confirmed by DNA sequencing. A second PNDO-encoding gene, NWMN0732, was similarly cloned using the primers 5′-AGC GGC CTG GTG CCG CGC GGC AGC ATG ACT GAA ATA GAT TTT GA-3′ and 5′-GCG GCC GCA AGC TTG TCG ACG GAG TTA TTA AGC TTG ATC GTT TAA ATG TTC AAT-3′ to generate pET28a-NWMN0732.
For protein expression, E. coli BL21(λDE3) with pET28a-NWMN2274 was grown in 2× YT media supplemented with 25 mg/ml kanamycin at 30 °C to an optical density at 600 nm of ∼0.8. Cultures were then induced with 0.3 mm isopropyl β-d-thiogalactopyranoside and incubated for ∼16 h at 25 °C with shaking at 200 rpm. Cell pellets were collected by centrifugation at 4400 × g for 10 min, resuspended in 50 mm Tris (pH 8.0), 100 mm NaCl, 2 mm Tris(2-carboxyethyl)phosphine (TCEP) (Gold Biotechnology), and lysed at 10,000 p.s.i. with an EmulsiFlex-C5 homogenizer (Avestin). The supernatant was isolated after centrifugation at 39,000 × g for 45 min, and His6-NWMN2274 was purified using a 5-ml HisTrap HP column (GE Healthcare) with a linear imidazole gradient (0–500 mm). Protein fractions were dialyzed into 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, and 2 mm TCEP at 4 °C. The His6 tag was removed by thrombin (Hemotologic Technologies) digestion at a 1/500 (w/w) thrombin-to-protein ratio and incubated over 24 h at 4 °C followed by dialysis into 50 mm Tris-HCl (pH 8.0), 2 mm TCEP for 2 h at 4 °C. NWMN2274 was further purified by anion exchange chromatography using a Source 15Q column (GE Healthcare) equilibrated with 50 mm Tris-HCl (pH 8.0), 2 mm TCEP and eluted with a NaCl gradient (0–500 mm). NWMN2274 was dialyzed into 50 mm Tris (pH 8.0), 300 mm KCl, and 2 mm TCEP and concentrated to 20 mg/ml (Fig. 1D). Purified protein was protected from light and stored at 4 °C. NWMN0732 was purified by using the same protocol.
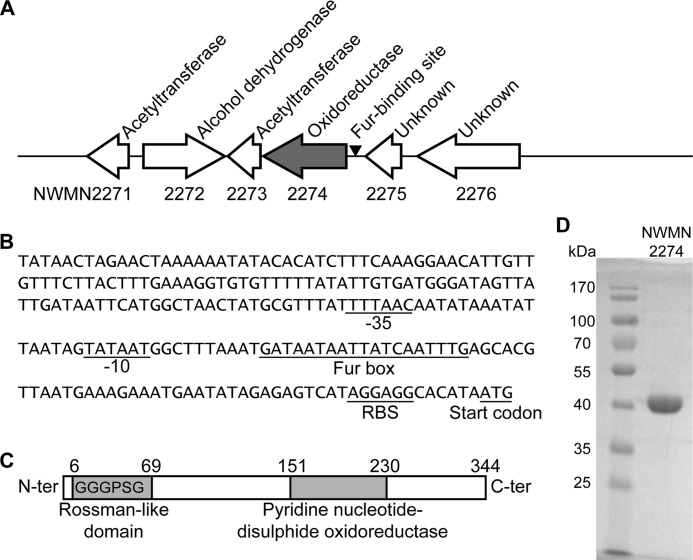
FIGURE 1.
Genomic context of NWMN2274 and predicted protein domains. A, NWMN2274 is the first gene of a predicted two gene operon with a putative Fur box (arrowhead) immediately upstream of it. Above the diagram are the predicted functions of the proteins encoded by these genes as determined by BLASTP analysis (35), and below the diagram are the open reading frame IDs from the genome sequence of S. aureus strain NEWMAN (49). B, the intergenic region between NWMN2274 and NWMN2275 contains predicted −10 and −35 sites for RNA polymerase binding, a Fur box, and a ribosome binding site (RBS) that were manually identified. C, NWMN2274 encodes a 344-amino acid protein with a Rossman fold (including the consensus GXGXXG motif beginning at the eighth amino acid) and a PNDO domain predicted by Pfam (57). D, NWMN2274 was purified to >95% homogeneity and is close to its predicted size (38 kDa) when separated by SDS-PAGE and stained with Coomassie.
IsdI and IsdG were expressed in E. coli BL21 (λDE3) cells from the plasmid pET15b, purified by His-tag affinity chromatography, and digested with the tobacco etch virus protease to remove the His tag as previously described (22). Tobacco etch virus protease was purified as previously described (33).
FAD Identification
Electronic spectra from 250 to 800 nm were measured with a Cary 50 Bio UV-visible spectrophotometer. Samples were NWMN2274 (as purified), NWMN2274 after heat denaturation and protein removal, and FAD, FMN, and riboflavin standards (Sigma). All samples were at 30 μm in a buffer of 50 mm Tris-HCl (pH 8.0), 100 mm NaCl. To heat-denature and remove NWMN2274 from the flavin molecule, the protein solution was boiled for 10 min and centrifuged at 21,000 × g for 3 min, and the supernatant was centrifuged through a Nanosep centrifugal device (PALL Life Sciences) with a molecular mass cut-off of 3 kDa as described previously (34).
Flavin removed from NWMN2274 as well as FAD, FMN, and riboflavin standards (all at 20 μm) were separated using an Infinity 1260 Quaternary high performance liquid chromatography (HPLC) system (Agilent) equipped with an Aqua 5-mm C18 column (Phenomenex). A previously established procedure was utilized (34) with modifications. A flow rate of 1 ml/min and a column temperature of 20 °C were maintained during the entire analysis. The column was equilibrated with 85% solvent A (10 mm ammonium acetate (pH 6.5)) and 15% solvent B (methanol) at sample injection. After a 5-min post-injection period, a linear gradient was developed over 20 min to 75% solvent B followed by a second linear gradient over 5 min to 100% solvent B. Flavins were detected by absorption at 264 nm using an Infinity 1260 multiple wavelength detector (Agilent).
Heme Degradation Assays
In vitro heme degradation assays were conducted as previously described (29). 10 mm stock solutions of porcine hemin (Sigma) in 0.1 m NaOH were prepared and kept at −20 °C. 10 μm IsdI and 10 μm porcine hemin (Sigma) were mixed in buffer containing 50 mm Tris-HCl (pH 7.5) and 150 mm NaCl and incubated at 4 °C for 1 h. Spectra from 300 to 800 nm were measured with a Cary 50 Bio UV-visible spectrophotometer. As noted below, some reactions were also supplemented with 200 μm NADPH (EMD Biosciences), 200 μm NADH (Roche), 5 μm bovine liver catalase (Sigma), or 4 units/ml bovine erythrocyte superoxide dismutase (Sigma). To initiate degradation reactions, 1 mm ascorbic acid, 1 μm NWMN2274, or NWMN0732, 20 μm to 2 mm H2O2, or 5 units/ml Aspergillus niger glucose oxidase (Sigma), and 1 mm glucose (Sigma) were added to cuvettes. Spectra were recorded either every minute or every 10 min for up to 90 min depending on the rate at which the reaction progressed.
For kinetic analysis, 600 μm NADPH and 0.1 μm NWMN2274 were added to reactions containing 1–25 μm IsdG-heme or IsdI-heme, and the decrease in the Soret peak at 412 nm was monitored every 0.1 s for 180 s. The concentration of IsdI-heme was determined from Soret absorbance measurements for 30 s starting at 10 s after the addition of the reductase using the reported extinction coefficients for IsdG-heme (131 mm−1 cm−1) and IsdI-heme (126 mm−1 cm−1) (22). Slopes determined by linear regressions of these data were taken as initial reaction rates. Initial steady-state rates were plotted against the concentration of IsdG-heme or IsdI-heme, and a non-linear regression of the data were calculated to fit Michaelis-Menten kinetics. Linear and non-linear regressions were calculated using GraphPad Prism 6.
Heme Degradation Product Analyses
For these experiments, 1-ml reactions containing 100 μm IsdI-heme were prepared, and 2 mm ascorbic acid, 2 mm H2O2, 2 units/ml glucose oxidase, and 5 mm glucose or 1 mm NADPH and 5 μm NWMN2274 or NWMN0732 were added to initiate reactions. Reactions were monitored, and once complete, heme degradation products were purified as previously described (27) with one important modification. Purification includes sample filtration through a Nanosep centrifugal device. The presence of the larger NWMN2274 protein blocked the pores of spin columns with a molecular mass cutoff of 3 kDa and prevented samples from passing easily through the column. Pore size was increased to a cutoff of 10 kDa, which improved product purification significantly, but lower yields were typically obtained from the NWMN2274/NADPH reactions than from the ascorbic acid reactions.
HPLC separation of the degradation products was completed as previously described (27) using either a Waters 2695 separation module with a Waters 2996 photodiode array detector or an Agilent Infinity 1260 multi-wavelength detector. With either setup the flow rate was 0.5 ml/min, and a Waters XTerra C18 column was used.
Bioinformatics Analyses
Homologs of NWMN2274 were identified with BLASTP (35) searches of the annotated genomes of S. aureus, Listeria monocytogenes, Bacillus subtilis, Bacillus anthracis, and Mycobacterium tuberculosis. Only hits with E-values equal to or less than 0.001 and covering 40% or more of the query sequence were considered significant. Multiple sequence alignments were generated with T-Coffee (36, 37). Maximum likelihood phylogenetic trees were built with the PhyML method in SeaView Version 4 (38) using the LG model and bootstrapping with 100 replicates.
The following S. aureus proteins (strain names in parentheses) all share >97% amino acid sequence identity to NWMN2274: SACOL2369 (SHY97–3906), SA2162 (N315), SAUSA300_2319 (USA300), SAOUHSC_02654 (NCTC 832), and MW2294 (MW2). Genes corresponding to these proteins were assumed to be orthologs of NWMN2274 in the analysis of microarray papers from various S. aureus strains.
RESULTS
Identification of NWMN2274 as a Putative Electron Donor to IsdG and IsdI
The heme-degrading proteins IsdG and IsdI require a source of electrons for porphyrin cleavage and iron release (22). We sought to identify a protein that could act as the in vivo source of electrons when IsdI and IsdG degrade heme within the cytoplasm of S. aureus. No candidate reductases are located within the two operons that encode the Isd system of S. aureus or in the regions immediately upstream or downstream. We hypothesized that the candidate reductase would be annotated in the genome as a reductase and that mRNA expression would be co-regulated with other Isd genes.
Data from several published microarray studies demonstrating expression changes in the Isd genes (39–46) were analyzed for changes in expression of uncharacterized genes encoding predicted reductases. Using a combination of cell culture and infection models with a bovine mastitis S. aureus isolate (strain SHY97–3906), Allard et al. (39) found that SACOL2369 was up-regulated under both iron-restricted growth conditions in cell culture and in tissue cages embedded in mice abdomens. SACOL2369 is annotated as a pyridine nucleotide-disulfide oxidoreductase (PNDO), and the authors of this study confirmed the microarray results by real time PCR. Identified upstream of the homologous gene (SA2162) in S. aureus strain N315 is a putative Fur box, the operator sequence to which the ferric-uptake regulator (Fur) binds in the promoter of iron-regulated genes (47, 48). Most other Isd genes were also up-regulated under the conditions of this study. Furthermore, Malachowa et al. (42) showed that isdA-I were all highly up-regulated (in some cases up to 200-fold) in S. aureus strain USA300 when grown in human blood or serum compared with standard media and that SAUSA300_2319 (the USA300 gene orthologous to SACOL2369) was also up-regulated in serum and blood, although not nearly to the same extent.
The Isd system has been extensively characterized in S. aureus strain Newman, and the orthologous gene of SACOL2369 is NWMN2274 (Fig. 1A). The genomic context of NWMN2274 (49) is similar to that of SACOL2369 (50) and SAUSA300_2319 (51), and all three genes encode nearly identical proteins (>97% amino acid identity). NWMN2274 is in a predicted two-gene operon with NWMN2273 that encodes a putative GCN5-like N-acetyltransferase. The 231-base pair intergenic region between NWMN2274 and NWMN2275 includes probable −10 and −35 sites for RNA polymerase binding and a predicted Fur box identical to that identified by Allard et al. (39) for SA2162 (Fig. 1B). The gene encodes a 344-amino acid protein (Fig. 1C) with a predicted Rossman fold domain for NAD(P) binding and a predicted PNDO domain (49).
A BLASTP search of NWMN2274 against the sequences from the Protein Data Bank found distantly related homologs (<35% amino acid sequence identity). These include a ferredoxin-NADP+ oxidoreductase (YumC) of B. subtilis (52), thioredoxin reductases from bacterial (53), yeast (54), and plant species (55), and Escherichia coli alkylhydroperoxide reductase (56). Pfam (57) predicts that these proteins are all PNDOs.
NWMN2274 Binds FAD
Purified NWMN2274 in solution is a dark yellow color consistent with binding of a flavin group. To determine the identity of the unknown flavin, we used HPLC to compare it to FAD, FMN, and riboflavin standards. The unknown flavin isolated from NWMN2274 had the same retention time by HPLC analysis as FAD and the retention time differed significantly from FMN and riboflavin (Fig. 2). In addition, the unknown flavin had absorption maxima at 376 and 450 nm and a spectrum nearly identical to that of a FAD standard and differed from that of FMN or riboflavin standards that both have absorption maxima at 375 and 447 nm (data not shown). The absorption maxima of FAD at 450 nm and FMN at 447 nm are consistent with previous studies and can be used to distinguish between the two molecules (34). Together these data indicate that NWMN2274 is an FAD-binding protein.
FIGURE 2.
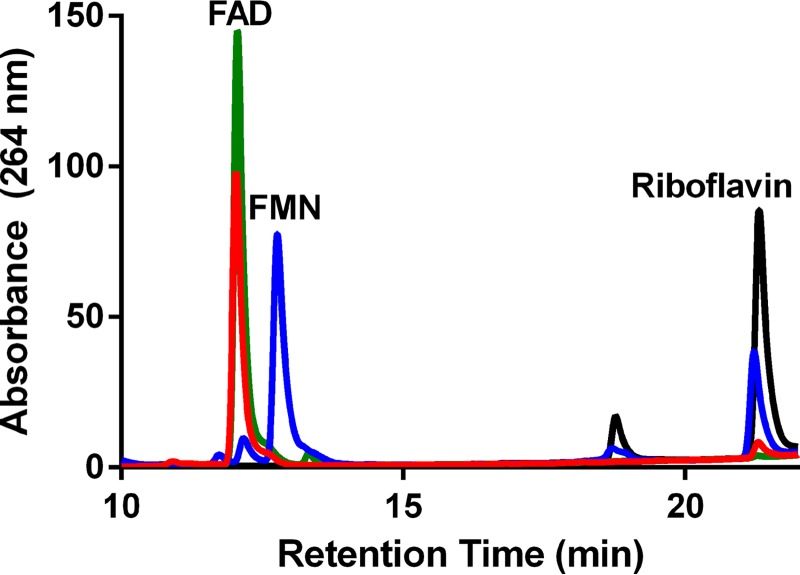
NWMN2274 binds FAD. HPLC separation of unknown flavin removed from NWMN2274 (red), FAD (green), FMN (blue), and riboflavin (black) is shown.
NWMN2274 and NADPH Are an Electron Source for Heme Degradation by IsdI or IsdG
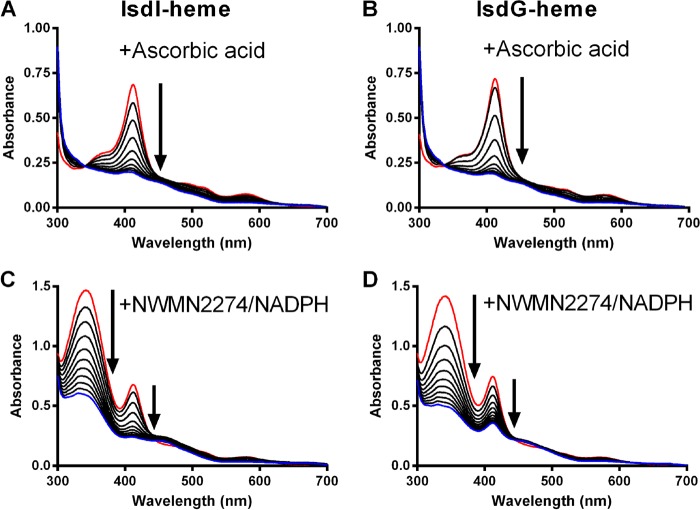
As previously demonstrated (29), upon the addition of ascorbic acid to IsdI-heme or IsdG-heme, the Soret peak at 412 nm decreases over the course of 90 min, indicative of heme degradation (Fig. 3, A and B). When a 20-fold excess of NADPH was added to IsdI-heme or IsdG-heme and purified NWMN2274 (Fig. 1D) was added to initiate the reaction, there were rapid decreases in both absorption at 340 nm, indicating that NADPH was being oxidized to NADP+, and at 412 nm, indicating that heme was being degraded (Fig. 3, C and D). Heme degradation occurred more rapidly under these conditions with the reactions nearly complete by 10 min. The data in Fig. 3, C and D, suggest that, at least qualitatively, the reaction with IsdG-heme is slower than with IsdI-heme. NWMN2274 or NADPH added alone to either IsdI-heme or IsdG-heme did not initiate heme degradation (data not shown). Similar experiments were also conducted with NADH substituted for NADPH, and in the presence of NWMN2274 these reactions progressed but at rates slower than the control reactions in the presence of ascorbic acid (data not shown). Taken together, these experiments indicate that NWMN2274 can act as a reducing agent in the presence of NADPH and provides electrons for heme degradation by IsdI and IsdG.
FIGURE 3.
NWMN2274 and NADPH can substitute for ascorbic acid as an electron source for heme degradation by IsdI or IsdG. UV-visible spectra of 10 μm IsdI-heme (left panels) or IsdG-heme (right panels) were obtained in the presence of 1 mm ascorbic acid (A and B) or 1 μm IruO and 200 μm NADPH (C and D). Spectra were recorded every 10 min for 90 min (A and B) or every minute for 10 min (C and D). Arrows indicate spectral changes over time, and red and blue lines indicate the first and last time points, respectively.
The stoichiometry of heme degradation by IsdG and IsdI is unknown, including the number of electrons required. Some uncoupled oxidation of NADPH to yield H2O2 and superoxide is likely to occur in these in vitro reactions, and thus the equivalents of NADPH consumed does not necessarily equate to reduction equivalents for heme degradation.
NWMN2274 Kinetic Parameters
To determine steady-state kinetic parameters for NWMN2274 with IsdI-heme or IsdG-heme as the substrate, initial velocities were plotted against the concentration of IsdG-heme or IsdI-heme, and data were fit by nonlinear regression to the Michaelis-Menten equation (Fig. 4). From these data the kcat of the overall reaction in the presence of IsdI-heme was determined to be 0.09 ± 0.01 s−1. The Km of NWMN2274 for IsdI-heme was calculated to be 15 ± 4 μm. The specificity constant (kcat/Km) for NWMN2274 catalyzed heme degradation was 5.8 × 103 m−1 s−1. Our data from reactions at concentrations below the Km fit the theoretical curve well, whereas above the Km there is greater error. This may be due to heme degradation by IsdI being a multistep process. At high concentrations of IsdI-heme some heme may only be partially degraded and not contribute to decreased Soret peak height. As mentioned above, some uncoupling of NWMN2274 from IsdI may be present and could alter the observed kinetics. The potential effect of uncoupling is mitigated by the presence of excess of IsdI-heme to NWMN2274 (10–250-fold) and the monitoring of the heme Soret peak to define the rate. Additionally, as shown below (see Fig. 7), the addition of catalase and superoxide dismutase did not significantly alter heme degradation reactions.
FIGURE 4.
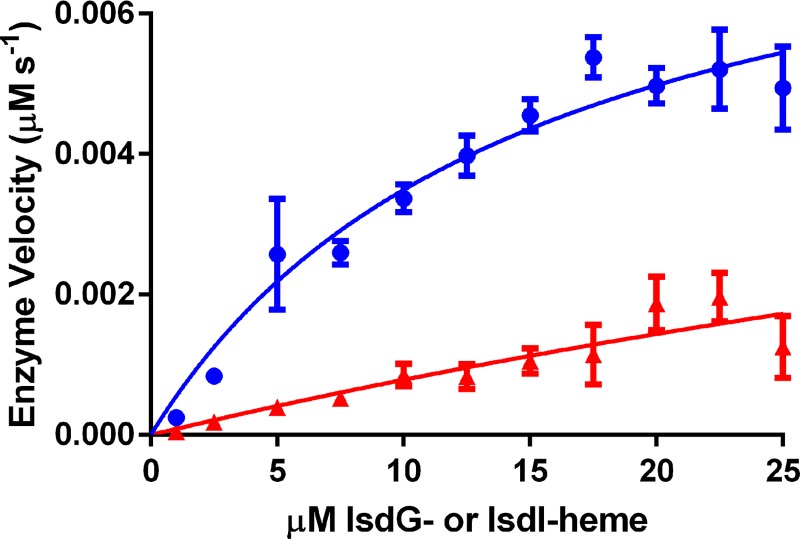
Michaelis-Menten reaction kinetics of NWMN2274 with IsdI-heme or IsdG-heme as the substrate. Reactions were conducted with 0.1 μm NWMN2274, 600 μm NADPH, and 1–25 μm IsdG-heme or IsdI-heme. Initial enzyme velocities were calculated for 30 s of the reaction and plotted against the concentration of IsdG-heme (red) or IsdI-heme (blue). Non-linear regression was used to fit data to Michaelis-Menten kinetics. Data points and error bars represent the mean S.E. for reactions from four (IsdG-heme) or five (IsdI-heme) independent experiments.
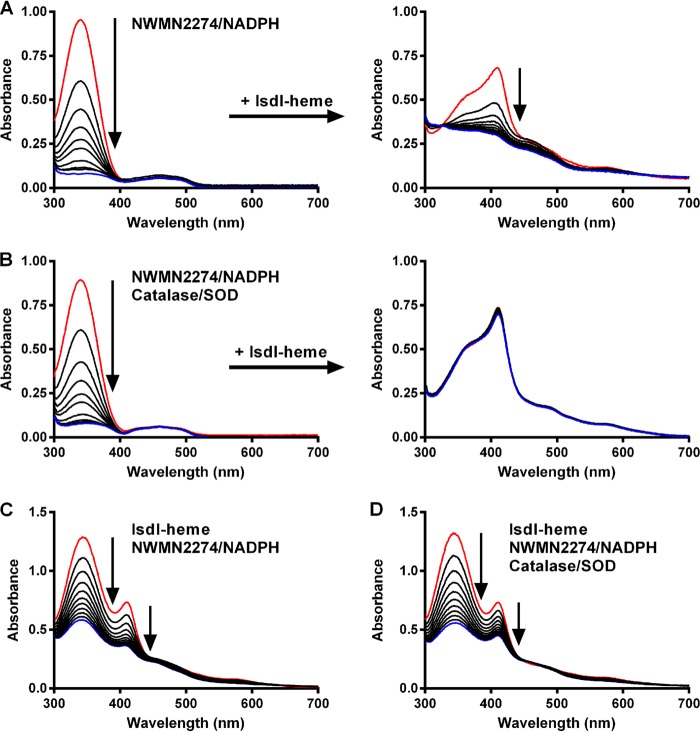
FIGURE 7.
NWMN2274 can act as an electron donor to IsdI for in vitro heme degradation either with or without the generation of reactive oxygen species. UV-visible spectra of 200 μm NADPH in the absence (A, left panel) or presence (B, left panel) of catalase and superoxide dismutase were measured for 30 min after the addition of 10 μm NWMN2274 to initiate NADPH oxidation. Lines represent spectra at the time points of 0, 2, 4, 6, 8, 10, 15, 20, 25, and 30 min. For A and B, after 30 min 10 μm IsdI-heme was added to the cuvettes (right panels), and spectra were taken every 2 min for 20 min. Bottom panels represent coupled reactions containing all components from the outset either in the absence (C) or presence (D) of catalase and superoxide dismutase. For C and D, spectra were taken every minute for 10 min. Arrows indicate spectral changes over time, and red and blue lines indicate the first and last time points, respectively.
Reactions with IsdG-heme were slower and, at the concentrations of IsdG-heme tested, increases in enzyme velocity were still in the linear range (Fig. 4). We were, therefore, unable to calculate reliable kinetic parameters for this reaction. This observation leaves open the possibility that IsdG may preferentially use an alternative reductase or interact with some other component in vivo.
IsdI-Heme Reactions with NWMN2274 and NADPH Generate the Staphylobilins
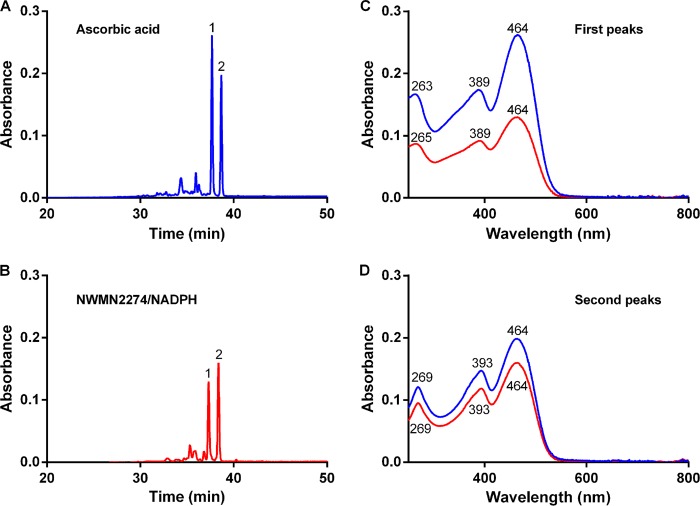
Heme degradation products derived from IsdI with either ascorbic acid or NWMN2274/NADPH were analyzed by HPLC. As previously reported (27), reactions with ascorbic acid resulted in two major products that absorb at 465 nm (Fig. 5A). Two products with similar HPLC retention times (Fig. 5B) and electronic spectra (Fig. 5, C and D) were also obtained from reactions with NWMN2274 and NADPH. Spectra for these peaks are highly similar to those previously published for the staphylobilins (27). These spectra differ from the IsdI/heme degradation spectra (Fig. 3) due to alterations in the molecular environment. Heme and products are removed from the protein to a solution containing 50% acetonitrile and 0.1% trifluoroacetic acid. Note that the products are also at least 10 times more concentrated than in the degradation reaction spectra (Fig. 3). These data confirm that the same reaction products are derived when IsdI cleaves heme with either ascorbic acid or NWMN2274/NADPH as a reductant. Thus, the staphylobilins identified in in vitro degradation assays are likely produced by IsdI cleavage of heme within an S. aureus cell as well.
FIGURE 5.
IsdI-heme reaction products with characteristics of the staphylobilins are produced from either the ascorbic acid or NWMN2274/NADPH reactions. Reaction products were separated from proteins and analyzed by HPLC, and the elution profiles at 465 nm are shown for the ascorbic acid (A) and NWMN2274/NADPH (B) reactions. In both cases there are two major peaks that absorb at 465 nm and are labeled 1 and 2. Optical spectra for peaks 1 (C) and peaks 2 (D) from ascorbic acid (blue lines) and NWMN2274/NADPH (red lines) reactions are shown with wavelengths of maximum absorbance shown for each spectra either above or below the lines.
The Presence of H2O2 Also Leads to Heme Degradation but with an Altered Product
Based on previous attempts to crystallize IsdI or IsdG with heme for structural determination, it was observed that red crystals containing protein bound to heme were only obtained with either inactive variants of IsdG (29) or for wild-type IsdI crystallized at 4 °C (27). We hypothesized that at room temperature and with active protein, a component in the crystallization solution leads to heme degradation and a resulting failure to crystallize protein bound to intact heme. In fact when enzyme assays similar to those above are conducted in conditions that mimic the crystallization conditions (0.1 m Bis-tris (pH 5.5), 0.2 m MgCl2, 25% polyethylene glycol-3350) there is slow heme degradation at room temperature but not at 4 °C (data not shown), and we believe that peroxides generated by the polyethylene glycol-3350 may be the cause. Under our standard reaction conditions, but with H2O2 added to initiate the reaction in place of ascorbic acid or NWMN2274/NADPH, heme was degraded by IsdI (Fig. 6A).
FIGURE 6.
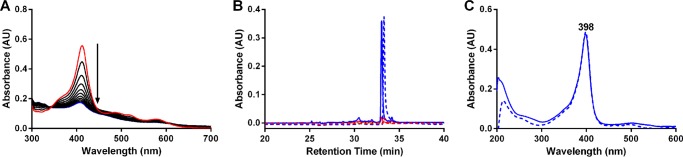
Addition of H2O2 to IsdI-heme leads to heme degradation with an altered byproduct. A, UV-visible spectra of 10 μm IsdI-heme with 20 μm H2O2 were measured every 2 min for 20 min. Arrows indicate spectral changes over time. Red and blue lines are the first and last time points, respectively. B, shown is HPLC analysis of products extracted from either untreated IsdI-heme (dashed lines) or IsdI-heme treated with H2O2 (solid lines) monitored at 405 nm (blue lines) and 465 nm (red lines). C, shown are spectra of the major peaks from B with dashed and solid lines for untreated and H2O2-treated IsdI-heme, respectively.
The product of the IsdI-heme reaction with H2O2 was analyzed by HPLC and consisted of one major peak that absorbs at 405 nm but not 465 nm (Fig. 6B). The retention time of the peak is very similar to that of heme extracted from untreated IsdI-heme and analyzed by HPLC (Fig. 6B). Spectra of both are similar with an absorbance maximum at 398 nm (Fig. 6C). Similar results were obtained for heme degradation reactions by IsdG in the presence of H2O2 (data not shown). These data indicate that reaction of IsdI-heme or IsdG-heme with H2O2 leads to loss of the Soret peak, a heme degradation product that is similar to heme and, importantly, no production of the staphylobilins.
Catalase and Superoxide Dismutase Do Not Inhibit Heme Degradation by NWMN2274/NADPH
Based on these observations, we thought that degradation of heme by IsdI or IsdG in the presence of NWMN2274/NADPH could occur via two pathways. One, NWMN2274 could oxidize NADPH to NADP+ and provide electrons directly to IsdI or IsdG for heme degradation. Or two, NWMN2274 could oxidize NADPH to NADP+ and transfer electrons to mediators in the reaction mixture, such as dissolved dioxygen, forming superoxide or hydrogen peroxide that may cause heme degradation by IsdI or IsdG.
To address these issues we conducted a series of experiments in which we either uncoupled the reaction, premixing NWMN2274 and NADPH for a period of time before the addition of IsdI-heme and/or added catalase and superoxide dismutase to reactions to see if they had a significant impact on heme degradation. First, we mixed NWMN2274 and NADPH in our standard reaction buffer but without IsdI-heme and followed the spectrum of the reaction for 30 min (Fig. 7A). In the absence of IsdI-heme, the NADPH absorption peak decreased rapidly, indicating that NWMN2274 still oxidized NADPH to NADP+ in the absence of IsdI-heme. Then IsdI-heme was added, and the reaction was monitored for an additional 20 min. In this uncoupled reaction, heme degradation still occurs despite the absence of NADPH. When this uncoupled reaction was conducted in the presence of catalase and superoxide dismutase, NADPH was oxidized to NADP+ over the first 30 min, but upon the addition of IsdI-heme to the reaction, very little heme degradation was observed (Fig. 7B). Thus, free superoxide or hydrogen peroxide, generated when NWMN2274 oxidizes NADPH in the absence of IsdI-heme, can serve as a mediator to transfer electrons from NWMN2274 to IsdI-heme.
Next, we conducted coupled experiments where IsdI-heme and NADPH were mixed in the standard reaction buffer and NWMN2274 was added to initiate the reaction. In the absence of catalase and superoxide dismutase (Fig. 7C), the reaction progressed as previously demonstrated (compare with Fig. 3G). The addition of catalase and superoxide dismutase (Fig. 7D) did not significantly alter the rate of heme degradation. The same result was obtained with IsdG-heme (data not shown). These latter data support a model of direct electron transfer from NWMN2274 to IsdG- or IsdI-heme. Because NWMN2274, IsdG, and IsdI are available to interact in the cell, this model is a much more likely scenario. These results agree with those of Skaar et al. (22) who found that catalase did not alter heme degradation by IsdG and IsdI in reactions initiated with ascorbic acid.
As negative controls, we also analyzed the results of IsdG- or IsdI-heme reactions with either an alternative PNDO (NWMN0732) and NADPH or glucose oxidase and glucose (which generates H2O2 upon oxidation of glucose) in the presence or absence of catalase and superoxide oxidase. In the absence of catalase and superoxide dismutase, the reactions progressed, and heme was degraded; however, unlike for NWMN2274, the addition of catalase and superoxide dismutase prevented heme degradation (data not shown). Furthermore, HPLC analysis of heme degradation products from reactions initiated with either NWMN0732/NADPH or glucose oxidase/glucose gave profiles similar to H2O2-initiated reactions, with no indication of staphylobilin formation (data not shown). These data further support the hypothesis that NWMN2274 specifically interacts with IsdG and IsdI for heme degradation.
Based on the whole of our experimental evidence, we believe that NWMN2274 is an in vivo electron donor for IsdG and IsdI-mediated heme degradation to the staphylobilins, and we propose naming NWMN2274 IruO, for iron utilization oxidoreductase.
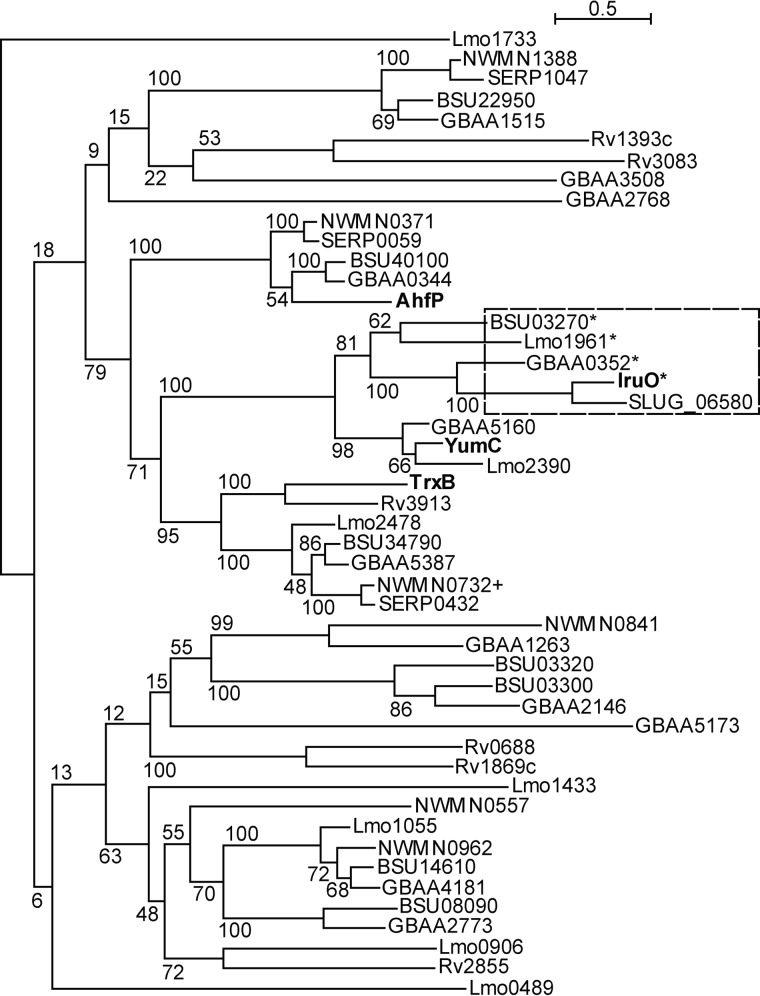
IruO Is an Archetype for a Family of PNDOs
Finally, we searched for other potential IruO proteins in Gram-positive bacteria with IsdG-family proteins. The sequences of IruO and three related PNDOs that have been structurally characterized, E. coli thioredoxin reductase (TrxB) (53, 58), B. subtilis YumC, a ferredoxin reductase (52), and E. coli alkylhydroperoxide reductase (AhpF) (56), were used to search the genomes of S. aureus strain Newman (49), B. subtilis strain 168 (59), B. anthracis strain Ames “Ancestor” (60), L. monocytogenes strain EGD-e (61), and M. tuberculosis strain H37Rv (62). Between 6 and 12 homologs were retrieved per organism, subjected to multiple sequence alignment, and organized into a phylogenetic tree (Fig. 8). IruO groups with a number of PNDOs have also been implicated by microarray and proteomic studies to be iron- and/or Fur-regulated. B. subtilis BSU03270 and Lmo1961 of L. monocytogenes are both Fur- and iron-regulated (63, 64), whereas GBAA0352 of B. anthracis is up-regulated during iron starvation (65) and during culture in macrophages (66). Of the genomes searched, only that of M. tuberculosis has no immediately obvious IruO ortholog. Among the other PNDOs of S. aureus identified through this analysis, NWMN0732 is most closely related to IruO; however, in vitro the staphylobilins are not generated with NWMN0732 as an electron donor.
FIGURE 8.
IruO is related to iron- and Fur-regulated reductases from other Gram-positive bacteria with IsdG-family proteins. Proteins used as query sequences to identify other putative PNDOs are shown in bold. Genes that are transcriptionally regulated by either iron and/or Fur are marked with an asterisk. A dashed box outlines a clade of PNDOs that are proposed to be involved in iron reduction. Numbers represent the bootstrap values for each branch. + denotes NWMN0732, which was also purified and used in some experiments.
We also searched for IruO proteins in two other staphylococcal species. Staphylococcus lugdunensis contains an Isd heme import system similar to that of S. aureus (67) and has an experimentally characterized IsdG protein (68) that shares >60% amino acid sequence identity with S. aureus IsdG and IsdI. The genome of S. lugdunensis (69) contains a PNDO (SLUG_06580) that shares 68% amino acid sequence identity with S. aureus IruO, is similarly organized in a two-gene operon with an acetyltransferase, and is the protein most similar to S. aureus IruO in our phylogenetic analysis (Fig. 8). In contrast, in the genome of Staphylococcus epidermidis strain (50), no Isd heme import system is detected, and the one IsdG-family protein present is distantly related, sharing only ∼35% identity with S. aureus IsdG and IsdI. BLASTP analysis of the S. epidermidis proteome with S. aureus IruO returns three hits that matched our search criteria SERP0059, SERP0432, and SERP1047. However, these proteins share >80% amino acid sequence identity with NWMN0371, NWMN0732, and NWMN1388, respectively, and group with them in our phylogenetic tree (Fig. 8).
Thus, the boxed clade in Fig. 8 represents a family of PNDOs used by these organisms to provide electrons to heme degrading enzymes, allowing iron to be released for bacterial use. These proteins could also potentially be used to reduce iron for its release from bacterial siderophores, although that remains to be experimentally demonstrated. Interestingly, this clade clusters with a second clade that contains YumC, a ferredoxin reductase (70). The IsdG and IsdI homodimers possess ferredoxin-like folds (23), and the associated reductase might be expected to resemble a ferredoxin reductase.
DISCUSSION
We show here that the protein encoded by NWMN2274 of S. aureus strain Newman catalyzes the transfer of electrons from NADPH to IsdI and IsdG for heme degradation. Likely some reactive oxygen species are generated through uncoupled oxidation of NADPH by NWMN2274; however, heme degradation to the staphylobilins is a consequence of a coupled enzymatic reaction between NWMN2274 and IsdI or IsdG bound to heme. From the determined kinetic parameters, the specificity constant of NWMN2274 for IsdI-heme is 5800 m−1 s−1. Finally, degradation products from reactions with NWMN2274 and NADPH are the same as those from reactions with ascorbic acid. Based on these data, we have named the protein IruO for iron utilization oxidoreductase. We acknowledge the possibility that other reductases could act as sources of electrons for cytoplasmic heme degradation by S. aureus and that it may be possible to resolve this issue through characterization of an S. aureus iruO mutant.
A second PNDO from S. aureus (NWMN0732) does not induce heme degradation leading to the staphylobilins, suggesting that there is specificity between IruO and IsdG and IsdI. Human heme oxygenase utilizes cytochrome P450 reductase as an electron donor for during heme degradation, and Skaar et al. (22) demonstrated that human cytochrome P450 reductase could serve as an electron donor for IsdG- and IsdI-mediated heme degradation in vitro. The possibility exists that the putative cytochrome P450 reductase of S. aureus (NWMN2518) might also be an in vivo electron donor for heme degradation, particularly by IsdG. IruO may act as a reductase for other S. aureus iron acquisition pathways, a possibility that we are currently exploring. Our data indicate that there may be redundancies in these systems as B. subtilis, B. anthracis, and L. monocytogenes all have additional PNDOs (YumC, GBAA5160, and Lmo2390, respectively) that group in a clade near that of IruO (Fig. 8).
We identified iruO through examination of a number of microarray papers to identify a potential reductase protein whose gene expression was similar to other isd genes under iron limitation conditions. Other studies also show that a correlation exists between expression changes in previously identified isd genes and iruO, including treatment with peracetic acid (43), nitric oxide (45), and ortho-phenylphenol (40). However, correlation between gene expression changes in iruO and other isd genes is not absolute. IsdA-G but not iruO were up-regulated in the presence of hydrogen peroxide (41). A ΔmurF strain with defective cell wall biosynthesis decreased transcription of isdC-G but not of iruO (46). Finally, a Δclp mutant with impaired degradation of misfolded proteins increased expression of iruO and some other Fur-regulated genes but not the rest of the isd genes (44). These studies show that other regulatory elements may also tune the response of these genes to conditions the bacteria encounter. Our strategy of mining microarray papers for evidence of coordinated gene transcription changes could help to identify the pathways to which other IruO paralogs in S. aureus belong.
IruO is homologous to E. coli TrxB, an FAD binding enzyme that reduces thioredoxin (TrxA), which in turn reduces disulfide bonds in numerous cellular targets (71–74). TrxB is thought to transfer electrons in multiple steps, first from NADPH to FAD, then from FAD to the protein disulfide active site, and finally to TrxA (75). IruO also binds FAD and favors NADPH over NADH as a source of electrons that are eventually funneled to IsdI-heme. The Km of IruO for IsdI-heme that we determined (14.9 μm) is similar to the Km (3.0 μm) of E. coli TrxB for TrxA; however, our measured kcat (0.09 s−1) is orders of magnitude lower than that of TrxB (23 s−1) (74). Kinetic parameters for TrxB were measured in an assay that monitored the reduction of DTNB by TrxA. Heme degradation by IsdI in the presence of IruO is a much slower, but also more complex reaction by comparison. Although low, the kcat value for heme degradation reported here is comparable to other reported values, as bovine and chicken heme oxygenases have turnover numbers of 0.058 and 0.32 s−1, respectively (76, 77).
Orthologs of IruO (Fig. 8) appear to also be present in other Gram-positive bacteria with IsdG-family heme oxygenases. B. subtilis, B. anthracis, and L. monocytogenes all possess IsdG-family proteins and have homologs of IruO that are regulated in either iron- and/or Fur-dependent fashions (63–66). These observations and our phylogenetic analysis indicate that the function we have attributed here to IruO may be of significance to the lifestyle of other Gram-positive bacteria that obtain iron from heme.
Predicted IsdG orthologs are found across diverse classes of bacteria (68), but evidence suggests that other types of reductases may be used under some circumstances. For example, IsdG and IsdI share structural similarity to Streptomyces monooxygenases used in antibiotic synthesis, such as ActVA of Streptomyces coelicolor (23). However, the oxidoreductase ActVB oxidizes NADH to NAD+, whereas it reduces FMN and then transfers reduced FMN to ActVA, which in turn re-oxidizes FMN during catalysis (78). No homolog of ActVB could be identified by a BLASTP analysis of the genome of S. aureus strain Newman. The highest ranked hit has an E-value of 2.0 with 30% amino acid sequence identity over only 24% percent of the query sequence. The heme degradation protein of M. tuberculosis, MhuD, shares sequence and structural similarity to IsdG and IsdI but has both distinct heme binding properties (79) and degradation products (80), and a candidate IruO homolog is not as evident. Finally, S. epidermidis, which has no Isd heme uptake system and a more distantly related IsdG homolog, has no IruO ortholog (Fig. 8).
Heme acquisition and utilization likely play an important role in S. aureus pathogenesis as heme iron has been proposed to be preferred by S. aureus to transferrin iron during an infection (25). A S. aureus mutant lacking both IsdG and IsdI grows poorly when heme is the only iron source (24, 68) Furthermore, S. aureus mutants lacking components of the Isd system have been shown to be defective in lung, heart, and kidney colonization during systemic murine infections (17, 26). These data make the Isd system a target for the development of new anti-staphylococcal drugs, and the identification of new members of this system provides additional targets for therapeutic development. Future directions include determining how the loss of IruO impacts S. aureus biology, particularly its ability to grow on heme as a sole source of iron and cause disease in experimental animal models. Also unknown is the nature of the physical interaction that occurs between IruO and IsdI or IsdG and how this interaction impacts the multistep reaction required for S. aureus heme degradation to the staphylobilins.
Acknowledgments
We thank members of the Murphy Laboratory for helpful advice and constructive criticism.
This work was supported by a Canadian Institutes of Health Research Grant MOP-49597 (to M. E. P. M.).
- Isd
- iron-regulated surface determinant
- IruO
- iron utilization oxidoreductase
- PNDO
- pyridine nucleotide-disulfide oxidoreductase
- TCEP
- Tris(2-carboxyethyl)phosphine
- Fur
- ferric-uptake regulator
- Bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Trx
- thioredoxin reductase.
REFERENCES
- 1. Gordon R. J., Lowy F. D. (2008) Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46, S350–S359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 3. Thurlow L. R., Joshi G. S., Richardson A. R. (2012) Virulence strategies of the dominant USA300 lineage of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). FEMS Immunol. Med. Microbiol. 65, 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nimmo G. R. (2012) USA300 abroad. Global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 18, 725–734 [DOI] [PubMed] [Google Scholar]
- 5. Bullen J. J., Rogers H. J., Spalding P. B., Ward C. G. (2005) Iron and infection. The heart of the matter. FEMS Immunol. Med. Microbiol. 43, 325–330 [DOI] [PubMed] [Google Scholar]
- 6. Weinberg E. D. (1977) Infection and iron metabolism. Am. J. Clin. Nutr. 30, 1485–1490 [DOI] [PubMed] [Google Scholar]
- 7. Hammer N. D., Skaar E. P. (2011) Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 65, 129–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beasley F. C., Vinés E. D., Grigg J. C., Zheng Q., Liu S., Lajoie G. A., Murphy M. E., Heinrichs D. E. (2009) Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol. Microbiol. 72, 947–963 [DOI] [PubMed] [Google Scholar]
- 9. Cotton J. L., Tao J., Balibar C. J. (2009) Identification and characterization of the Staphylococcus aureus gene cluster coding for Staphyloferrin A. Biochemistry 48, 1025–1035 [DOI] [PubMed] [Google Scholar]
- 10. Cheung J., Beasley F. C., Liu S., Lajoie G. A., Heinrichs D. E. (2009) Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 74, 594–608 [DOI] [PubMed] [Google Scholar]
- 11. Sebulsky M. T., Hohnstein D., Hunter M. D., Heinrichs D. E. (2000) Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182, 4394–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grigg J. C., Ukpabi G., Gaudin C. F., Murphy M. E. (2010) Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 104, 341–348 [DOI] [PubMed] [Google Scholar]
- 13. Muryoi N., Tiedemann M. T., Pluym M., Cheung J., Heinrichs D. E., Stillman M. J. (2008) Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 283, 28125–28136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. M., Schneewind O. (2003) Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909 [DOI] [PubMed] [Google Scholar]
- 15. Torres V. J., Pishchany G., Humayun M., Schneewind O., Skaar E. P. (2006) Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188, 8421–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vermeiren C. L., Pluym M., Mack J., Heinrichs D. E., Stillman M. J. (2006) Characterization of the heme binding properties of Staphylococcus aureus IsdA. Biochemistry 45, 12867–12875 [DOI] [PubMed] [Google Scholar]
- 17. Pishchany G., Dickey S. E., Skaar E. P. (2009) Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect. Immun. 77, 2624–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tiedemann M. T., Heinrichs D. E., Stillman M. J. (2012) The multi-protein heme shuttle pathway in Staphylococcus aureus. Isd cog-wheel kinetics. J. Am. Chem. Soc. 134, 16578–16585 [DOI] [PubMed] [Google Scholar]
- 19. Dryla A., Gelbmann D., von Gabain A., Nagy E. (2003) Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49, 37–53 [DOI] [PubMed] [Google Scholar]
- 20. Grigg J. C., Vermeiren C. L., Heinrichs D. E., Murphy M. E. (2007) Heme coordination by Staphylococcus aureus IsdE. J. Biol. Chem. 282, 28815–28822 [DOI] [PubMed] [Google Scholar]
- 21. Pluym M., Vermeiren C. L., Mack J., Heinrichs D. E., Stillman M. J. (2007) Heme binding properties of Staphylococcus aureus IsdE. Biochemistry 46, 12777–12787 [DOI] [PubMed] [Google Scholar]
- 22. Skaar E. P., Gaspar A. H., Schneewind O. (2004) IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279, 436–443 [DOI] [PubMed] [Google Scholar]
- 23. Wu R., Skaar E. P., Zhang R., Joachimiak G., Gornicki P., Schneewind O., Joachimiak A. (2005) Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 280, 2840–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reniere M. L., Skaar E. P. (2008) Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 69, 1304–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Skaar E. P., Humayun M., Bae T., DeBord K. L., Schneewind O. (2004) Iron-source preference of Staphylococcus aureus infections. Science 305, 1626–1628 [DOI] [PubMed] [Google Scholar]
- 26. Mason W. J., Skaar E. P. (2009) Assessing the contribution of heme-iron acquisition to Staphylococcus aureus pneumonia using computed tomography. PLoS ONE 4, e6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reniere M. L., Ukpabi G. N., Harry S. R., Stec D. F., Krull R., Wright D. W., Bachmann B. O., Murphy M. E., Skaar E. P. (2010) The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 75, 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsui T., Nambu S., Ono Y., Goulding C. W., Tsumoto K., Ikeda-Saito M. (2013) Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry 52, 3025–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee W. C., Reniere M. L., Skaar E. P., Murphy M. E. (2008) Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 283, 30957–30963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takayama S.-i. J., Ukpabi G., Murphy M. E., Mauk A. G. (2011) Electronic properties of the highly ruffled heme bound to the heme degrading enzyme IsdI. Proc. Natl. Acad. Sci. 108, 13071–13076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ukpabi G., Takayama S.-i. J., Mauk A. G., Murphy M. E. (2012) Inactivation of IsdI heme oxidation by an active site substitution that diminishes heme ruffling. J. Biol. Chem. 287, 34179–88179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacPherson I. S., Rosell F. I., Scofield M., Mauk A. G., Murphy M. E. (2010) Directed evolution of copper nitrite reductase to a chromogenic reductant. Protein Eng. Des. Sel. 23, 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tropea J. E., Cherry S., Waugh D. S. (2009) Expression and purification of soluble His6-tagged TEV protease. Methods Mol. Biol. 498, 297–307 [DOI] [PubMed] [Google Scholar]
- 34. Aliverti A., Curti B., Vanoni M. A. (1999) Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol. Biol. 131, 9–23 [DOI] [PubMed] [Google Scholar]
- 35. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 36. Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.-M., Taly J.-F., Notredame C. (2011) T-Coffee. A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Notredame C., Higgins D. G., Heringa J. (2000) T-coffee. A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 38. Gouy M., Guindon S., Gascuel O. (2010) SeaView version 4. A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 [DOI] [PubMed] [Google Scholar]
- 39. Allard M., Moisan H., Brouillette E., Gervais A. L., Jacques M., Lacasse P., Diarra M. S., Malouin F. (2006) Transcriptional modulation of some Staphylococcus aureus iron-regulated genes during growth in vitro and in a tissue cage model in vivo. Microbes Infect. 8, 1679–1690 [DOI] [PubMed] [Google Scholar]
- 40. Jang H. J., Nde C., Toghrol F., Bentley W. E. (2008) Microarray analysis of toxicogenomic effects of ortho-phenylphenol in Staphylococcus aureus. BMC genomics 9, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palazzolo-Ballance A. M., Reniere M. L., Braughton K. R., Sturdevant D. E., Otto M., Kreiswirth B. N., Skaar E. P., DeLeo F. R. (2008) Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180, 500–509 [DOI] [PubMed] [Google Scholar]
- 42. Malachowa N., Whitney A. R., Kobayashi S. D., Sturdevant D. E., Kennedy A. D., Braughton K. R., Shabb D. W., Diep B. A., Chambers H. F., Otto M., DeLeo F. R. (2011) Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6, e18617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chang W., Toghrol F., Bentley W. E. (2006) Toxicogenomic response of Staphylococcus aureus to peracetic acid. Environ. Sci. Technol. 40, 5124–5131 [DOI] [PubMed] [Google Scholar]
- 44. Michel A., Agerer F., Hauck C. R., Herrmann M., Ullrich J., Hacker J., Ohlsen K. (2006) Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Richardson A. R., Dunman P. M., Fang F. C. (2006) The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61, 927–939 [DOI] [PubMed] [Google Scholar]
- 46. Sobral R. G., Jones A. E., Des Etages S. G., Dougherty T. J., Peitzsch R. M., Gaasterland T., Ludovice A. M., de Lencastre H., Tomasz A. (2007) Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189, 2376–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heinrichs J. H., Gatlin L. E., Kunsch C., Choi G. H., Hanson M. S. (1999) Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J. Bacteriol. 181, 1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiong A., Singh V. K., Cabrera G., Jayaswal R. K. (2000) Molecular characterization of the ferric-uptake regulator, Fur, from Staphylococcus aureus. Microbiology 146, 659–668 [DOI] [PubMed] [Google Scholar]
- 49. Baba T., Bae T., Schneewind O., Takeuchi F., Hiramatsu K. (2008) Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes. Polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190, 300–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gill S. R., Fouts D. E., Archer G. L., Mongodin E. F., Deboy R. T., Ravel J., Paulsen I. T., Kolonay J. F., Brinkac L., Beanan M., Dodson R. J., Daugherty S. C., Madupu R., Angiuoli S. V., Durkin A. S., Haft D. H., Vamathevan J., Khouri H., Utterback T., Lee C., Dimitrov G., Jiang L., Qin H., Weidman J., Tran K., Kang K., Hance I. R., Nelson K. E., Fraser C. M. (2005) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., Lin F., Lin J., Carleton H. A., Mongodin E. F., Sensabaugh G. F., Perdreau-Remington F. (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 [DOI] [PubMed] [Google Scholar]
- 52. Komori H., Seo D., Sakurai T., Higuchi Y. (2010) Crystal structure analysis of Bacillus subtilis ferredoxin-NADP+ oxidoreductase and the structural basis for its substrate selectivity. Protein Sci. 19, 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kuriyan J., Krishna T. S., Wong L., Guenther B., Pahler A., Williams C. H., Jr., Model P. (1991) Convergent evolution of similar function in two structurally divergent enzymes. Nature 352, 172–174 [DOI] [PubMed] [Google Scholar]
- 54. Oliveira M. A., Discola K. F., Alves S. V., Medrano F. J., Guimarães B. G., Netto L. E. (2010) Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeastthioredoxin system. Biochemistry 49, 3317–3326 [DOI] [PubMed] [Google Scholar]
- 55. Kirkensgaard K. G., Hägglund P., Finnie C., Svensson B., Henriksen A. (2009) Structure of Hordeum vulgare NADPH-dependent thioredoxin reductase 2. Unwinding the reaction mechanism. Acta Crystallogr. D. Biol. Crystallogr. 65, 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bieger B., Essen L.-O. (2001) Crystal structure of the catalytic core component of the alkylhydroperoxide reductase AhpF from Escherichia coli. J. Mol. Biol. 307, 1–8 [DOI] [PubMed] [Google Scholar]
- 57. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lennon B. W., Williams C. H., Jr., Ludwig M. L. (2000) Twists in catalysis. Alternating conformations of Escherichia coli thioredoxin reductase. Science 289, 1190–1194 [DOI] [PubMed] [Google Scholar]
- 59. Kunst F., Ogasawara N., Moszer I., Albertini A. M., Alloni G., Azevedo V., Bertero M. G., Bessières P., Bolotin A., Borchert S., Borriss R., Boursier L., Brans A., Braun M., Brignell S. C., Bron S., Brouillet S., Bruschi C.V., Caldwell B., Capuano V., Carter N. M., Choi S. K., Codani J. J., Connerton I. F., Danchin A. (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390, 249–256 [DOI] [PubMed] [Google Scholar]
- 60. Ravel J., Jiang L., Stanley S. T., Wilson M. R., Decker R. S., Read T. D., Worsham P., Keim P. S., Salzberg S. L., Fraser-Liggett C. M., Rasko D. A. (2009) The complete genome sequence of Bacillus anthracis Ames “Ancestor.” J. Bacteriol. 191, 445–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Charbit A., Chetouani F., Couvé E., de Daruvar A., Dehoux P., Domann E., Domínguez-Bernal G., Duchaud E., Durant L., Dussurget O., Entian K. D., Fsihi H., García-del Portillo F., Garrido P., Gautier L., Goebel W., Gómez-López N., Hain T., Hauf J., Jackson D., Jones L. M., Kaerst U., Kreft J., Kuhn M., Kunst F., Kurapkat G., Madueno E., Maitournam A., Vicente J. M., Ng E., Nedjari H., Nordsiek G., Novella S., de Pablos B., Pérez-Diaz J. C., Purcell R., Remmel B., Rose M., Schlueter T., Simoes N., Tierrez A. (2001) Comparative genomics of Listeria species. Science 294, 849–852 [DOI] [PubMed] [Google Scholar]
- 62. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 63. Ollinger J., Song K. B., Antelmann H., Hecker M., Helmann J. D. (2006) Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 188, 3664–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ledala N., Sengupta M., Muthaiyan A., Wilkinson B. J., Jayaswal R. K. (2010) Transcriptomic response of Listeria monocytogenes to iron limitation and Fur mutation. Appl. Environ. Microbiol. 76, 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Carlson P. E., Jr., Carr K. A., Janes B. K., Anderson E. C., Hanna P. C. (2009) Transcriptional profiling of Bacillus anthracis Sterne (34F2) during iron starvation. PLoS ONE 4, e6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bergman N. H., Anderson E. C., Swenson E. E., Janes B. K., Fisher N., Niemeyer M. M., Miyoshi A. D., Hanna P. C. (2007) Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect. Immun. 75, 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zapotoczna M., Heilbronner S., Speziale P., Foster T. J. (2012) Iron-regulated surface determinant (Isd) proteins of Staphylococcus lugdunensis. J. Bacteriol. 194, 6453–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Haley K. P., Janson E. M., Heilbronner S., Foster T. J., Skaar E. P. (2011) Staphylococcus lugdunensis IsdG liberates iron from host heme. J. Bacteriol. 193, 4749–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Heilbronner S., Holden M. T., van Tonder A., Geoghegan J. A., Foster T. J., Parkhill J., Bentley S. D. (2011) Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 322, 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Seo D., Kamino K., Inoue K., Sakurai H. (2004) Purification and characterization of ferredoxin-NADP+ reductase encoded by Bacillus subtilis yumC. Arch. Microbiol. 182, 80–89 [DOI] [PubMed] [Google Scholar]
- 71. Laurent T. C., Moore E. C., Reichard P. (1964) Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 239, 3436–3444 [PubMed] [Google Scholar]
- 72. Moore E. C., Reichard P., Thelander L. (1964) Enzymatic synthesis of deoxyribonucleotides. V. Purification and properties of thioredoxin reductase from Escherichia coli B. J. Biol. Chem. 239, 3445–3452 [PubMed] [Google Scholar]
- 73. Russel M., Model P. (1985) Direct cloning of the trxB gene that encodes thioredoxin reductase. J. Bacteriol. 163, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Navarro J. A., Gleason F. K., Cusanovich M. A., Fuchs J. A., Meyer T. E., Tollin G. (1991) Kinetics of electron transfer from thioredoxin reductase to thioredoxin. Biochemistry 30, 2192–2195 [DOI] [PubMed] [Google Scholar]
- 75. Williams C. H., Arscott L. D., Müller S., Lennon B. W., Ludwig M. L., Wang P.-F., Veine D. M., Becker K., Schirmer R. H. (2000) Thioredoxin reductase. Eur. J. Biochem. 267, 6110–6117 [DOI] [PubMed] [Google Scholar]
- 76. Yoshinaga T., Sassa S., Kappas A. (1982) Purification and properties of bovine spleen heme oxygenase. Amino acid composition and sites of action of inhibitors of heme oxidation. J. Biol. Chem. 257, 7778–7785 [PubMed] [Google Scholar]
- 77. Bonkovsky H. L., Healey J. F., Pohl J. (1990) Purification and characterization of heme oxygenase from chick liver. Eur. J. Biochem. 189, 155–166 [DOI] [PubMed] [Google Scholar]
- 78. Valton J., Mathevon C., Fontecave M., Nivière V., Ballou D. P. (2008) Mechanism and regulation of the two-component FMN-dependent monooxygenase ActVA-ActVB from Streptomyces coelicolor. J. Biol. Chem. 283, 10287–10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chim N., Iniguez A., Nguyen T. Q., Goulding C. W. (2010) Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J. Mol. Biol. 395, 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nambu S., Matsui T., Goulding C. W., Takahashi S., Ikeda-Saito M. (2013) A new way to degrade heme. The Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. J. Biol. Chem. 288, 10101–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]