Background: Heterotrimeric G proteins are essential for biological signaling; however, the mechanism of their biosynthesis remains poorly understood.
Results: Long and short splice isoforms of phosducin-like protein stimulate and inhibit production of G proteins in the cell.
Conclusion: Both G protein α and βγ functional units are subject to the regulation.
Significance: We describe a potential mechanism for regulating the cellular levels of G proteins.
Keywords: Chaperone, Chaperonin, Heterotrimeric G Proteins, Photoreceptors, Protein Assembly, Retina, RNA Splicing, Phosducin-like Protein
Abstract
Heterotrimeric G proteins play an essential role in cellular signaling; however, the mechanism regulating their synthesis and assembly remains poorly understood. A line of evidence indicates that the posttranslational processing of G protein β subunits begins inside the protein-folding chamber of the chaperonin containing t-complex protein 1. This process is facilitated by the ubiquitously expressed phosducin-like protein (PhLP), which is thought to act as a CCT co-factor. Here we demonstrate that alternative splicing of the PhLP gene gives rise to a transcript encoding a truncated, short protein (PhLPs) that is broadly expressed in human tissues but absent in mice. Seeking to elucidate the function of PhLPs, we expressed this protein in the rod photoreceptors of mice and found that this manipulation caused a dramatic translational and posttranslational suppression of rod heterotrimeric G proteins. The investigation of the underlying mechanism revealed that PhLPs disrupts the folding of Gβ and the assembly of Gβ and Gγ subunits, events normally assisted by PhLP, by forming a stable and apparently inactive tertiary complex with CCT preloaded with nascent Gβ. As a result, the cellular levels of Gβ and Gγ, which depends on Gβ for stability, decline. In addition, PhLPs evokes a profound and rather specific down-regulation of the Gα transcript, leading to a complete disappearance of the protein. This study provides the first evidence of a generic mechanism, whereby the splicing of the PhLP gene could potentially and efficiently regulate the cellular levels of heterotrimeric G proteins.
Introduction
Heterotrimeric G proteins are a conserved group of molecules involved in a great number of signaling processes in eukaryotes (1). Although much is known about G protein-mediated signaling, far less is known about G protein biogenesis. All heterotrimeric G proteins are composed of α, β, and γ subunits encoded by distinct genes. In humans, 16 genes code for Gα, 12 genes code for Gγ, and five genes code for Gβ. The mechanisms underlying the process of selection and assembly of these subunits into mature G proteins remain unknown.
Evidently, the folding of nascent G protein β polypeptides requires the direct assistance of chaperonin containing t-complex protein 1 (CCT2; also known as TRiC) (2, 3) and is finalized by a permanent association with Gγ (4); however, the order of these events has not been well defined. Chaperonin CCT is a large, cylindrically shaped ATPase complex composed of two stacked rings. Each ring is made of eight t-complex protein 1 (TCP-1) subunits, designated α–θ in mammals (5, 6). In its open conformation, the substrate proteins are allowed to enter the central cavity. Nucleotide binding occurs once the substrate is bound, and ATP hydrolysis triggers the conversion from the open to the closed conformation (5). The exact mechanism by which CCT folds the substrate proteins has not been clearly defined, but it is thought to involve a set of sequential, hierarchical steps. The CCT substrates identified thus far are all soluble proteins, diverse in functions and structures (7). Many of these substrates, exemplified by the G protein β subunits (8), possess WD-repeat domains that fold into a β-propeller structure (9–11). Nevertheless, due to the diversity of CCT substrates, it is impossible to predict which proteins are folded by CCT based on sequence alone (12). Plausibly, the CCT substrates are selected and directed to CCT by specific co-factors. The proteins from the phosducin family emerge as one such set of CCT co-factors that are involved in substrate guidance.
The phosducin gene family is divided into subgroups I–III (13). Subgroup I consists of phosducin and PhLP1, which both bind G protein β subunits with high affinity (2, 14). PhLP1 (often called PhLP) is ubiquitously expressed (15) and is shown to act as a CCT co-factor during the folding of G protein β (16–20). In contrast, phosducin is expressed predominantly in rod and cone photoreceptors, where it helps maintain high levels of and assists in the trafficking of the visual G protein transducin without any known connection to CCT (21–23). Subgroup II consists of PhLP2A and PhLP2B, which were both discovered in humans (13). The PhLP2 ortholog in Saccharomyces cerevisiae interacts with CCT and is essential for cell growth (24) due to its function in actin biogenesis (25, 26). In contrast to PhLP1, PhLP2 is not involved in Gβ folding. Subgroup III is composed of only PhLP3 (13), which is thought to participate in β-tubulin and possibly actin folding (27) but which binds Gβ poorly (24). Thus, one commonality among PhLP1 to -3 is that they all participate in protein folding by acting as co-chaperones of CCT (2).
Alternative splicing of the PhLP1 gene in rats gives rise to two distinct transcripts, 297 and 218 amino acids long. A product of a short transcript was originally designated as phosducin-like protein short isoform (PhLPs) (28). Essentially the same transcript was later detected in the human retina and named phosducin-like orphan protein 1 (PHLOP1) (29). The precise biological function of PhLPs has remained elusive; however, some evidence has suggested that it acts as a negative regulator of heterotrimeric G proteins (17, 30, 31). To gain a better understanding of this important function, we ectopically expressed PhLPs in mouse rods, photoreceptor neurons of the retina that maintain very high levels of the heterotrimeric G protein, transducin (Gαt1β1γ1). We found that PhLPs disrupted several steps in the biosynthesis of rod transducin, which could also be observed in cell culture models. PhLPs specifically targeted the chaperonin CCT preloaded with nascent Gβ1. In contrast to the full-length PhLP, which facilitated the assembly of the Gβ1γ1 heterodimer, PhLPs disrupted this process by trapping Gβ1 inside the chaperonin. In addition, PhLPs evoked a strong and specific transcriptional suppression of Gαt1. This study, thus, provides the first evidence that the short isoform of PhLP can act as a negative regulator of the synthesis and assembly of heterotrimeric G proteins in vivo.
EXPERIMENTAL PROCEDURES
Animal Models
Transgenic mice expressing Δ1–83PhLP-FLAG have been previously described (32). All analyses were performed using Δ1–83PhLP-FLAG+/− and Δ1–83PhLP-FLAG−/− littermates at postnatal day 10, according to the procedures approved by the animal care and use committees of West Virginia University.
Quantification of mRNA
The alternatively spliced PhLP transcripts were detected in the Human Total RNA Master Panel II (Clontech). One microgram from each RNA sample was reverse transcribed using random hexamers and RNase H(−) reverse transcriptase. The region of the phosducin-like protein gene (PDCL) containing exon 2 was amplified with primers located in exons 1 (PDCL_1-F, 6-FAM-TGACCCACTGCTCTTCCTCT) and 3 (PDCL_1-R, GATGGGACCTGCAAGTCATT). The forward primer carries a 6-carboxyfluorescein (6-FAM) fluorescent tag attached to its 5′-end. The resulting fluorescently labeled products were resolved using denaturing polyacrylamide electrophoresis and imaged on a Typhoon imager (GE Healthcare) in the fluorescence mode.
To measure the levels of mRNA in the retina, RNA was isolated from single retinas using the Absolutely RNA Miniprep kit (Agilent Technologies), according to the manufacturer's protocol for small samples and including two DNase treatments. The final RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Using a two-step quantitative RT-PCR process, cDNA from each retina was generated from 5 ng of total RNA using the AffinityScript qPCR cDNA synthesis kit (Agilent Technologies) and oligo(dT) primers. Negative controls included no reverse transcriptase and no RNA template. Quantitative real-time PCR experiments were performed in triplicate with 0.5 ng of cDNA using the Brilliant II SYBR® Green qPCR master mix (Agilent Technologies), reference dye, and a 200 nm concentration of each primer. Reactions were incubated at 95 °C for 10 min and then cycled 27 times at 95 °C for 30 s, 55 °C for 60 s, and 72 °C for 30 s using a Stratagene Mx3000PTM real-time PCR system. A melting curve analysis was added at the end to verify a single product from each reaction, and the fluorescence was recorded during every qPCR cycle at both the annealing step (55 °C) and the extension step (72 °C). Fluorescence values were analyzed using MxPro qPCR software version 4.10, and amplification thresholds were normalized to those of GAPDH.
Primers for mouse Gnat1, Gnb1, Gnb5, and Gngt1 were designed using GenBankTM mouse mRNA sequences and PrimerQuest software and selected to amplify products crossing exon-exon boundaries. The primers were synthesized and HPLC-purified by Integrated DNA Technologies (Coralville, IA). Primer concentrations were optimized for each reaction prior to running the quantitative RT-PCR experiments. The primer sequences used were as follows: Gnat1, 5′-TGC CAT CAT CTA CGG CAA CAC TCT-3′ (forward) and 5′-CTT GGG CAT TGT GCC TTC CTC AAT-3′ (reverse); Gnb1, 5′-AGA ATC CAA ATG CGG ACC AGG AGA-3′ (forward) and 5′-ACC ACA GGC CAC ATA ATT CCC AGA-3′ (reverse); Gnb5, 5′- ACC AGA AGG ACC CTC AAA GG-3′ (forward) and 5′-GCA TGT CAG AGT TGG TGA AGC-3′ (reverse); Gngt1, 5′-TGC CAG TGA TCA ACA TCG AAG ACC-3′ (forward) and 5′-TCA CAC AGC CTC CTT TGA GTT CCT-3′ (reverse); GAPDH, 5′-GAC TTC AAC AGC AAC TCC CAC-3′ (forward) and 5′-TCC ACC ACC CTG TTG CTG TA-3′ (reverse).
Western Blotting
Retinas were collected at postnatal day 10 and frozen on dry ice. Each retina was homogenized in 0.2 ml of buffer containing 125 mm Tris/HCl, pH 6.8, 4% SDS, and 6 m urea by short ultrasonic pulses, and the resulting extract was cleared by centrifugation. The total protein concentration was determined on a Nanodrop ND-1000 spectrophotometer. To prepare SDS-PAGE samples, the protein concentration in each of the compared samples was adjusted to the same value, and bromphenol blue tracking dye and 5% β-mercaptoethanol were added. Typically, 10-μl samples from Tg(+) and Tg(−) littermates were separated side by side on a 10–20% Tris/HCl acrylamide gel (Bio-Rad). Quantification of the specific bands was performed using an Odyssey infrared imaging system (LI-COR Biosciences) according to the manufacturer's protocols. Data from three independent measurements utilizing at least three mice of each genotype each time were averaged.
Microarray Analysis
Total RNA were isolated from the retina using the Absolutely RNA Miniprep kit, and its quality was assessed with Bioanalyzer (both from Agilent Technologies). Samples were processed using an Affymetrix GeneChip whole transcript sense target labeling assay and hybridized to Affymetrix Mouse Exon 1.0 ST microarrays according to the manufacturer's instructions. Microarrays were analyzed using GeneBase software as described previously (33, 34). GeneBase estimates the gene expression levels based on probes with highly correlated signals across multiple samples, thus reducing the bias from cross-hybridizing probes or probes located in alternative exons. To take advantage of the probe selection algorithm of GeneBase, we combined our mouse retina arrays with the Affymetrix mouse exon 1.0 ST mouse tissue panel, which contains 33 arrays hybridized to samples from 11 tissues. p values were adjusted using Benjamini-Hochberg's procedure.
Mass Spectrometry
Δ1–83PhLP-FLAG was captured from the retinas with anti-FLAG-agarose and vacuum-dried, as described previously (32). A pull-down from 70 retinas was sent to Applied Biomics, Inc. (Hayward, CA), which conducted two-dimensional PAGE separation, protein identification in the spots by LC/MS/MS, and data analysis. In another experiment, a pull-down from 150 retinas was dissolved in buffer containing 125 mm Tris/HCl, pH 6.8, 4% SDS, 6 m urea, 5% β-mercaptoethanol, and bromphenol blue tracking dye and subjected to a standard SDS-PAGE separation on a 12.5% Tris/HCl gel (Bio-Rad). Following a short run, the gel was stained with Colloidal Coomassie (LC 6025, Invitrogen), and the entire lane was excised from the gel and cut into identical fragments. The proteins in each gel fragment were digested with trypsin and analyzed by microcapillary reverse-phase HPLC nanoelectrospray tandem mass spectrometry (μLC/MS/MS) on the Thermo LTQ-Orbitrap mass spectrometer at the Harvard Mass Spectrometry and Proteomic Resource Laboratory (Cambridge, MA). The acquired MS/MS spectra were correlated with known sequences using programs developed in this laboratory (35) and the Sequest algorithm (40). The data were analyzed to reach an estimated false discovery rate of ∼1% using a reverse database strategy. When a peptide sequence could be assigned to more than one protein, the assignment was made to the protein that had accumulated the most peptide spectra; thus, no peptide spectrum was ever assigned to more than one protein.
Preparation of cDNA Constructs
Mouse phosducin-like protein with a C-terminal FLAG tag (PhLP-FLAG) was prepared by reverse transcription of total retinal RNA from a 129/SV mouse using the gene-specific primer 5′-ACT AAA TGA GAC TAC AA, followed by PCR with the following primers: forward primer, 5′-GAG ACC ATG GCA ATG ACA ACC CTG GAT GAT AAG TTA CTG; reverse primers, 5′-CTT GTC ATC GTC GTC CTT GTA ATC ATC TAT TTC CAG ATC GCT GTC TTC and 5′-GCC TGG ATC CCT ACT ACT TGT CAT CGT CGT CCT TGT AAT C. The product was cut with NcoI and BamHI restriction endonucleases and inserted into the pTriEx4 expression vector (Novagen) at these sites. To utilize the potential for a Kozak sequence at the NcoI site and 5′-end of PhLP-FLAG, a few additional bases needed to be added, thereby expressing PhLP-FLAG with an additional methionine and alanine at the N terminus. PhLPs (Δ1–83PhLP-FLAG) was subcloned from pRhop4.4k-Δ1–83PhLP (32) using PCR and the following primers to add a C-terminal FLAG tag: forward primer, 5′-GAG ACC ATG GAG CGG CTG ATC AAA AAG CTG TC T ATG AG; reverse primer, 5′-GCC TGG ATC CCT ACT ACT TGT CAT CGT CGT CCT TGT AAT C. The product was cut with NcoI and BamHI restriction endonucleases and inserted into the pTriEx4 vector at these sites. The coding sequence of mouse Pdc was subcloned from another vector in our laboratory, pSC-A Kozak/Pdc/FLAG, using PCR and the following primers: forward primer, 5′-GAG ACC ATG GAA GAA GCC GCC AGC CAA AGC; reverse primer, 5′-GGG CCT CTC GAG TTC AAT GTC CTC GTC TTC CAT GTT G. The product and pTriEx4 vector were digested with NcoI and XhoI restriction endonucleases and ligated at these sites to generate a Pdc construct that would be expressed with a C-terminal His6 epitope tag. Human rod transducin β (Gβ1) cDNA was amplified from a human retinal cDNA library (a kind gift from Dr. Jeremy Nathans, Johns Hopkins University) and subcloned into the EcoRI and NotI restriction sites in the pCMV-myc vector to create Gβ1 with an N-terminal myc epitope tag (Clontech). Similar to the transducin β subunit, human rod transducin Gγ1 with an N-terminal HA epitope tag was generated using the NcoI (partial digestion) and XhoI sites in the pCMV-HA vector (Clontech). The integrity of all constructs was confirmed by sequence analysis.
Cell Culture
HEK 293 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM)/Ham's F-12 (50:50 mixture) medium (Mediatech, Inc.), supplemented with 10% defined fetal bovine serum (HyClone) and 1× penicillin-streptomycin-l-glutamine (Invitrogen) or 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.292 mg/ml l-glutamine. For the transfection, cells were plated in 10-cm plates, so they would be at least 50% confluent the following day. 9.6 μg of total plasmid DNA was transfected per plate using FuGENE® 6 transfection reagent (Roche Applied Science and Promega) at a 6:1 FuGENE 6 reagent/plasmid DNA ratio. The total amount of cDNA was kept constant by adding empty pTriEx-4 vector to make up any differences in the co-transfections, and the transfections were performed in triplicate. Cells transfected with empty pTriEx 4 vector only were used as a negative control. All cells were collected in Hanks' balanced salt solution (HBSS), supplemented with 1 mm EDTA, 48 h after transfection, and pull-down assays were performed in either radioimmune precipitation assay buffer (R0278, Sigma) or buffer containing 20 mm MES, pH 6.5, 5 mm EDTA, and 1.5% Nonidet P-40 using anti-FLAG M2 and anti-c-Myc-agarose affinity gels (A2220 and A7470, Sigma) and anti-HA-agarose (26181, Pierce). Captured proteins were eluted with FLAG, HA, or Myc peptides in 1 mm Tris/HCl (pH 7.4), vacuum-dried, and reconstituted in USB buffer for Western blot analyses.
Antibodies
Proteins were detected using antibodies against FLAG (600-401-383, Rockland), Myc (M4439, Sigma), Gβ (sc-378, Santa Cruz Biotechnology, Inc.), Gγ1 (sc-373, Santa Cruz Biotechnology), TCP-1α (ab109126, Abcam), TCP-1β (sc-47717, Santa Cruz Biotechnology), TCP-1γ (sc-33145, Santa Cruz Biotechnology), TCP-1ϵ (MCA2178, Serotec), visual arrestin (PA1–731, Thermo Scientific), or phosducin (21). Antibody against Gβ5 was a generous gift from Ching-Kang Jason Chen (Virginia Commonwealth University).
RESULTS
Identification of the PhLPs Transcript in Human Tissues
The skipping of exon 2 in the phosducin-like protein gene gives rise to two alternative transcripts encoding full-length PhLP and N-terminally truncated PhLPs (Fig. 1A). To determine the expression profile of these transcripts, we quantified their relative abundance in 20 human tissues by RT-PCR (Fig. 1B). Both transcripts could be detected in all tested tissues, with PhLP being the predominant splice isoform. The highest levels of PhLPs were found in neural tissue, where it comprised ∼10% of the total PhLP mRNA. Interestingly, expressed sequence tag evidence suggests that this PhLP splicing does not take place in mice. As such, there are 82 spliced expressed sequence tags and six full-length mRNAs that align to the genomic region containing exon 2 of the mouse PhLP (Pdcl) gene. All of these sequences include exon 2. The alignment can be viewed at the University of California Santa Cruz genome browser. Subsequently, we were unable to detect the PhLPs transcript by RT-PCR in mouse retina and brain (not shown). Thus, the alternative splicing of the PhLP gene, which produces the truncated PhLPs protein, appeared to be widespread in human tissues although not conserved evolutionarily in mice. Due to the low expression levels of PhLP and lack of adequate antibodies, our attempts to detect endogenous PhLP isoforms by Western blotting were unsuccessful.
FIGURE 1.
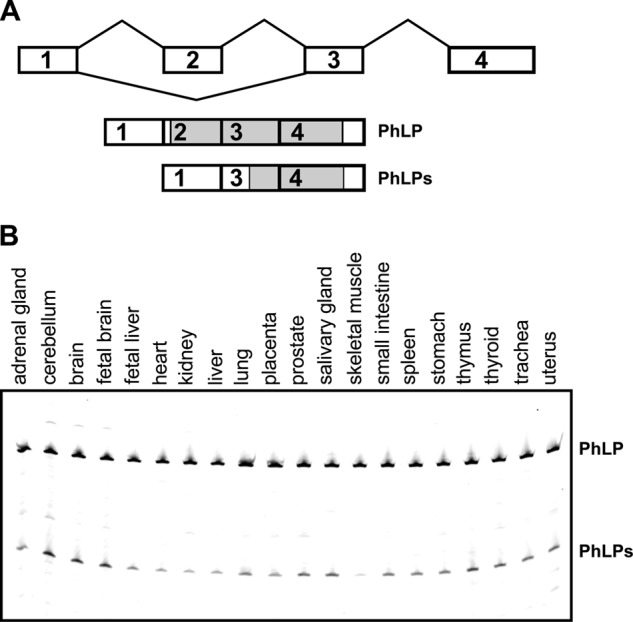
PhLP exon 2 splicing and isoform expression. A, diagram of the PhLP gene structure, showing the splicing pattern of exon 2 and the mRNA isoforms generated as a result of exon 2 alternative splicing. The open reading frame of each mRNA is indicated in gray. B, the truncated PhLPs transcript is expressed in multiple tissues. Shown is RT-PCR analysis of PhLP exon 2 splicing in 20 human tissues. Transcripts skipping exon 2 (PhLPs) were present in all examined tissues. Brain samples express the highest levels of the PhLPs variant.
PhLPs Disrupts the Expression of Heterotrimeric G Proteins in Mouse Photoreceptors
Having found that mice lack endogenous PhLPs, we used this model to explore the function of this protein. Toward this goal, a transgene encoding epitope-tagged PhLPs was expressed in mice. This transgene, designated here as Δ1–83PhLP-FLAG, encodes truncated PhLP that starts from Met-84, and is identical to PhLPs. Driven by a 4.4-kb rhodopsin promoter, the transgene was turned on at around postnatal day 5, and its expression was primarily restricted to the rod photoreceptors of the retina. We found that although the majority of rods survived until postnatal day 10 (Fig. 2A), these cells did not elaborate their light-sensing compartments, the outer segments, and failed to express heterotrimeric G proteins (Fig. 2, B and C). As such, the Gαt1 subunit of the abundant rod G protein, transducin, was undetectable on the protein level, and two other transducin subunits, Gβ1 and Gγ1, were reduced by 63 ± 4 and 95 ± 1%, respectively. The photoreceptor-specific long isoform, Gβ5L, was also significantly reduced by 84 ± 3%, whereas the short isoform, Gβ5S, expressed in the other retinal cell types, remained unaffected. The protein levels of photoreceptor-specific visual arrestin and ubiquitously expressed β-tubulin were also affected insignificantly. With the exception of Gαt1, whose transcript, Gnat1, was down-regulated 10-fold, the mRNA levels of Gβ1, Gγ1, and Gβ5L were reduced by less than 50%, which is insufficient to account for the observed reduction in these proteins. Hence, it appeared that all Gβ and Gγ affected by Δ1–83PhLP-FLAG were destabilized primarily on a posttranslational level.
FIGURE 2.
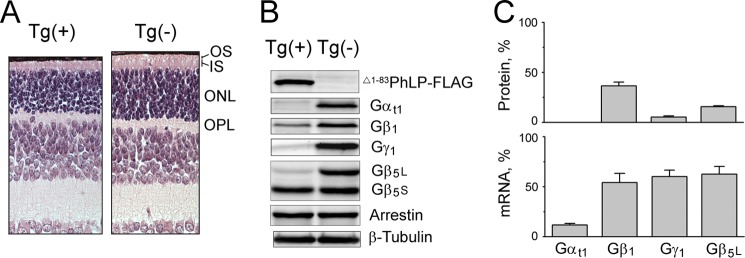
The effect of Δ1–83PhLP-FLAG on heterotrimeric G proteins of photoreceptors. A, paraffin-embedded retina cross-sections were stained with hematoxylin and eosin to demonstrate the retinal morphology of 10-day-old transgene-positive Tg(+) and transgene-negative Tg(−) littermates. OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer. B, retinas as in A were homogenized in SDS sample buffer, and equal aliquots were analyzed by Western blotting using antibodies against the indicated proteins. C, top graph, Gαt1, Gβ1, Gγ1, Gβ5L, and Gβ5S bands from B were quantified and expressed as a percentage fraction in Tg(+) versus Tg(−) retinas. The values are given under “Results” (means ± S.E. (error bars), n = 6, p < 0.05; as determined by paired t test). Bottom graph, the same operation was performed to compare the levels of corresponding mRNA determined by quantitative RT-PCR. The levels of mRNA are 12 ± 2% for Gαt1, 60 ± 7% for Gγ1, 54 ± 9% for Gβ1, and 63 ± 8% for Gβ5 (mean ± S.E., n = 3).
The observed significant down-regulation of the Gnat1 gene prompted us to examine the global impact of Δ1–83PhLP-FLAG on gene expression. We used an Affymetrix GeneChip whole transcript exon array to identify the changes in mRNA expression in the retinas of Δ1–83PhLP-FLAG-positive mice, as compared with their littermate controls at postnatal day 10. The change in gene expression was considered significant if the -fold change was greater than 2.00 at a false discovery rate below 0.05. We identified nine significantly down-regulated genes, including Gnat1, which was down-regulated the most (Table 1). Besides phosducin-like protein, which was overexpressed from the transgene, the only significantly up-regulated gene was activating transcription factor 3 (Atf3). Forty-three additional transcripts showed a statistically significant (false discovery rate <0.05), albeit small in magnitude, change in expression levels (supplemental Table 1). Of these, the majority (38 transcripts) were down-regulated, which is consistent with the general repression of transcription by Atf3. Thus, the early transcriptional responses of rods evoked by Δ1–83PhLP-FLAG appeared to be specifically focused on the suppression of the Gnat1 gene, encoding the Gαt1 subunit of transducin, and on the activation of the stress response via Atf3.
TABLE 1.
Genes significantly affected by Δ1–83PhLP-FLAG
| Description | Gene name | Tg(+)/Tg(−) ratio | p value |
|---|---|---|---|
| Guanine nucleotide-binding protein, subunit α-1 | Gnat1 | 0.234 | 0.039 |
| Sarcoglycan, γ (dystrophin-associated glycoprotein) | Sgcg | 0.369 | 0.046 |
| Low density lipoprotein receptor-related protein 4 | Lrp4 | 0.380 | 0.026 |
| RAB guanine nucleotide exchange factor (GEF) 1 | Rabgef1 | 0.421 | 0.022 |
| Predicted gene 9909 | Gm9909 | 0.427 | 0.039 |
| Protein phosphatase, Mg2+/Mn2+-dependent, 1N (putative) | Ppm1n | 0.456 | 0.035 |
| Solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | Slc24a1 | 0.460 | 0.039 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | Pfkfb2 | 0.466 | 0.039 |
| Guanylate cyclase 2f | Gucy2f | 0.490 | 0.035 |
| Activating transcription factor 3 | Atf3 | 3.747 | 0.043 |
| Phosducin-like protein | Pdcl | 5.954 | 0.022 |
Identification of the Protein Interactions of PhLPs by Mass Spectrometry
To understand the mechanism whereby Δ1–83PhLP-FLAG is suppressing G proteins, we identified its protein interactions in rods using a pull-down assay with anti-FLAG-agarose. As a first approach, the proteins in pull-downs were separated by two-dimensional gel electrophoresis, and the resulting protein spots were excised from the gel and identified by MALDI/MS/MS (Fig. 3). We identified seven major spots, present only in the Tg(+) preparation, as the TCP-1α, TCP-1β, TCP-1γ, TCP-1ϵ, TCP-1ζ, TCP-1η, and TCP-1θ subunits of the cytosolic chaperonin CCT. As a second approach, the pull-downs from Tg(+) and Tg(−) preparations were separated side by side by a short run on a conventional one-dimensional SDS gel, and then the entire lane was excised from the gel and cut into progressive fragments. The protein content of each gel fragment was analyzed by LC/MS/MS, using spectral counts to quantify the relative abundance of each target. In order to reduce error, of 4048 proteins identified by this approach, we selected those with minimal spectral counts of 20 and a Tg(+)/Tg(−) ratio greater than 2 (Table 2). Again, we found that eight core subunits of CCT were the most abundant proteins in the pull-downs. Combined, these data demonstrate that the chaperonin CCT, implicated in the posttranslational processing of G proteins (2), is a primary binding target of Δ1–83PhLP-FLAG in rods.
FIGURE 3.

Δ1–83PhLP-FLAG was captured with anti-FLAG agarose from Tg(+) retinas, and the resulting pull-down was separated by two-dimensional gel electrophoresis. The indicated proteins were later excised from the gel and identified by MALDI/MS/MS as TCP-1α (1), TCP-1β (2), TCP-1γ (3), TCP-1ϵ (5), TCP-1ζ (6), TCP-1η (7), and TCP-1θ (8). Nonspecific spots also present in Tg(−) preparations were γ-actin (a) and antibody chain (b).
TABLE 2.
Proteins identified in the pull-downs by LC/MS/MS
| Protein | Symbol | Spectral counts |
|
|---|---|---|---|
| Tg(+) | Tg(−) | ||
| Phosducin-like protein | Pdcl | 167 | 11 |
| T-complex protein 1 subunit α | TCP-1α | 459 | 121 |
| T-complex protein 1 subunit β | TCP-1β | 594 | 191 |
| T-complex protein 1 subunit γ | TCP-1γ | 393 | 106 |
| T-complex protein 1 subunit δ | TCP-1δ | 430 | 94 |
| T-complex protein 1 subunit ϵ | TCP-1ϵ | 361 | 134 |
| T-complex protein 1 subunit ζ | TCP-1ζ | 451 | 133 |
| T-complex protein 1 subunit η | TCP-1η | 554 | 85 |
| T-complex protein 1 subunit θ | TCP-1θ | 530 | 148 |
| Integrin β-1 | Itgb1 | 87 | 29 |
| Sorting nexin 12 | Snx12 | 61 | 24 |
| Paralemmin-1 | Palm | 54 | 25 |
| Oxysterol-binding protein 2 | Osbp2 | 46 | 7 |
| DNA-directed RNA polymerase I subunit RPA1 | Polr1a | 43 | 0 |
| KLRAQ motif-containing protein 1 | Klraq1 | 38 | 12 |
| Destrin | Dstn | 37 | 8 |
| Peripheral plasma membrane protein CASK | Cask | 37 | 18 |
| WD repeat-containing protein 82 | Wdr82 | 33 | 15 |
| Phosphomannomutase 1 | Pmm1 | 32 | 7 |
| Exocyst complex component 1 (SEC3) | Exoc1 | 29 | 4 |
| Fructose-bisphosphate aldolase C | Aldoc | 29 | 10 |
| Mage-d2 | Maged2 | 29 | 8 |
| 26 S proteasome non-ATPase regulatory subunit 6 | Psmd6 | 27 | 8 |
| Sorting nexin-1 | Snx1 | 27 | 13 |
| 26 S proteasome non-ATPase regulatory subunit 11 | Psmd11 | 26 | 10 |
| Mitochondria fission 1 protein | Fis1 | 25 | 7 |
| Mediator of RNA polymerase II transcription subunit 27 | Med27 | 23 | 0 |
| Segment polarity protein disheveled homolog DVL-2 | Dvl2 | 23 | 0 |
| Conserved oligomeric Golgi complex subunit 8 | Cog8 | 22 | 0 |
| Guanine nucleotide-binding protein, subunit β-1a | Gnb1 | 237 | 215 |
| Guanine nucleotide-binding protein, subunit α-1a | Gnat1 | 8 | 84 |
| Guanine nucleotide-binding protein, subunit γ-T1a | Gngt1 | 0 | 21 |
| Tubulin α-1Aa | Tuba1a | 441 | 560 |
| Tubulin β-5a | Tubb5 | 505 | 624 |
| Tubulin β-2Ca | Tubb2c | 102 | 124 |
| Tubulin β-2Aa | Tubb2a | 72 | 60 |
| Actin, cytoplasmic 1a | Actb | 85 | 67 |
a Selection criteria of spectral count >20 and Tg(+)/Tg(−) > 2 are not met.
Other less abundant protein partners of Δ1–83PhLP-FLAG were functionally categorized into several groups, including membrane biogenesis and dynamics (Palm, Osbp2, Dstn, Exoc1, and Fis1), protein biosynthesis and degradation (Pmm1, Psmd6, Psmd11, and Cog8), intracellular trafficking (Snx12, Snx1, and Cask), signal transduction (Itgb1, Klraq1, and Dvl2), nucleic acid biosynthesis and regulation (Polr1a, Wdr82, and Med27), metabolism (Aldoc), and cell death (Maged2). It is plausible that some of them may represent substrate proteins trapped inside the CCT folding chamber, whereas others interact with PhLP directly. Indeed, several bona fide clients of CCT from the actin and tubulin families were present in significant amounts in the pull-downs. However, all of them also appeared in the controls; therefore, the specificity of these interactions could not be reliably verified (Fig. 3 and Table 2).
PhLPs Forms a Complex with CCT Preloaded with Nascent Gβ
The Δ1–83PhLP-FLAG pull-downs from the retina contain significant amounts of Gβ1, a subunit of rod transducin and a known client of CCT. This Gβ1 was not associated with Gγ1, indicating that the Gβ1 is nascent protein (Table 2). Also, Δ1–83PhLP-FLAG lacks the first 83 amino acids, which include several points of interaction with G protein β subunit (14), and these proteins do not co-precipitate in the in vitro assay (31). Thus, this Gβ1 appears to be client protein trapped inside the folding chamber of the chaperonin co-precipitated with Δ1–83PhLP-FLAG. The abundance of Gβ1 in the pull-downs indicates that Δ1–83PhLP-FLAG forms a stable complex with those CCT that are preloaded with Gβ. To test whether Δ1–83PhLP-FLAG binding to CCT is indeed stimulated by Gβ, we utilized cell culture. In these experiments, Δ1–83PhLP-FLAG or PhLP-FLAG was transiently expressed in HEK 293 cells, with and without Gβ1-myc (Fig. 4). Both proteins were captured from cell extracts with an excess of anti-FLAG magnetic beads, and the resulting pull-downs were assayed by Western blotting. We found that the endogenous CCT of HEK 293 cells robustly co-precipitated with either PhLP-FLAG or Δ1–83PhLP-FLAG, and when Gβ1-myc was overexpressed in the cells, this protein was present in the pull-downs as well (Fig. 4B). Importantly, overexpressing Gβ1-myc markedly increased the amount of Δ1–83PhLP-FLAG and co-precipitated CCT while having no such effect on PhLP-FLAG (Fig. 4C). These data further support the notion that Δ1–83PhLP-FLAG forms a stable tertiary complex with the chaperonin CCT preloaded with nascent Gβ.
FIGURE 4.

Interaction of PhLP and PhLPs with CCT in cell culture. A, HEK 293 cells were co-transfected with the plasmids encoding the indicated epitope-tagged proteins and analyzed by pull-down with anti-FLAG magnetic beads after 48 h. B, representative blots showing captured PhLP-FLAG or Δ1–83PhLP-FLAG, co-precipitated with its subunits of CCT (TCP-1α, -1β, -1γ, and -1ϵ) and Gβ1-myc. C, protein bands in B were quantified, and their values in Gβ1-myc (+) cells were normalized to those in Gβ1-myc (−) cells. Error bars, S.E. with n = 6 for Δ1–83PhLP-FLAG and n = 3 for PhLP-FLAG and p < 0.1 (*), 0.01(**), and 0.001(***), as determined by paired t test.
PhLP but Not PhLPs Stimulates the Assembly of Gβγ Subunit Complexes
As described above, Δ1–83PhLP-FLAG specifically targets chaperonin complexes engaged in the folding of G protein β subunits. Such a mode of action, together with the significant reduction in the level of Gγ1 observed in rods (Fig. 2B), was consistent with Δ1–83PhLP-FLAG disrupting the assembly of Gβ1 and Gγ1 subunits in these cells. To test this possibility directly, we studied the assembly of Gβγ dimers in HEK 293 cells from transiently expressed, epitope-tagged Gβ1-myc and Gγ1-HA (Fig. 5A). Specifically, we sought to determine how this process was affected by the overexpression of PhLP-FLAG or Δ1–83PhLP-FLAG in the same cells (Fig. 5B). Our assay was based on capturing the majority of expressed Gγ1-HA with an excessive amount of anti-HA-agarose and detecting the co-purified Gβ subunits by Western blotting (Fig. 5C). We found that exogenous Gγ1-HA was successfully processed by the endogenous Gβγ folding pathway, as evident from its forming indiscriminately a dimer with both Gβ1-myc and other Gβ subunits of the HEK 293 cells (Fig. 5C, lane 2). This process was suppressed by Δ1–83PhLP-FLAG (Fig. 5C, compare lanes 2 and 3). Conversely, PhLP-FLAG strongly stimulated the association between Gγ1 and Gβ (Fig. 5C, compare lanes 2 and 4). On average, overexpressing PhLP-FLAG in cells increased the amounts of Gγ1-HA in the pull-downs by 4.8 ± 2.6-fold and the co-precipitated Gβ1-myc and endogenous Gβ subunits by 7.1 ± 2.7- and 2.5 ± 0.9-fold, respectively (means ± S.E.; n = 5) (Fig. 5D). These data demonstrate that PhLP-FLAG markedly stimulates the assembly of Gβγ subunit complexes in the cells, whereas Δ1–83PhLP-FLAG shows a tendency to suppress this process. Hence, the observed decline of Gγ1 in rods was probably caused by a major disruption in Gβ1γ1 dimer assembly by Δ1–83PhLP-FLAG preventing the folded Gβ1 from being released from CCT.
FIGURE 5.
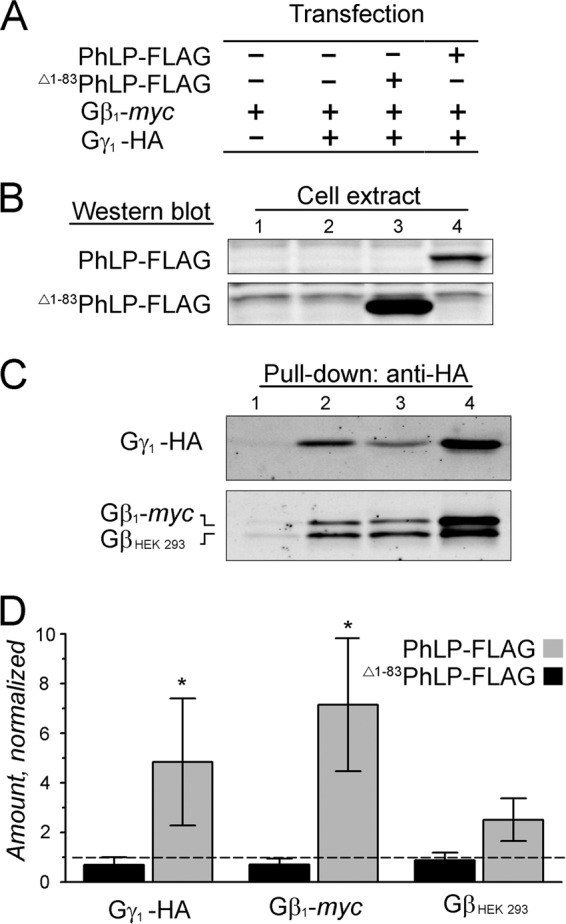
The effect of PhLP isoforms on the assembly of the Gβγ dimer in cell culture. A, HEK 293 cells were co-transfected with plasmids encoding the indicated epitope-tagged proteins and analyzed by pull-down with anti-HA-agarose after 48 h. B, representative Western blots showing the levels of PhLP-FLAG and Δ1–83PhLP-FLAG in the indicated cellular extracts. C, Western blots showing the amounts of Gγ1-HA and co-precipitated Gβ1-myc and endogenous GβHEK 293 subunits in the pull-downs. D, designated protein bands in C were quantified, and their values from lanes 3 and 4 were normalized to those in lane 2. Error bars, S.E. with n = 5 and p < 0.1 (*) as determined by t test.
Binding of the γ Subunit Triggers the Release of PhLP from Gβ
Having observed the robust stimulatory effect of PhLP on Gβγ dimer formation, we next tested whether PhLP and Gβγ are engaged in direct interaction. In these experiments, HEK 293 cells were also co-transfected with phosducin, a close PhLP homolog abundantly expressed in rod photoreceptors (Fig. 6A). When Gγ1-HA was captured from the cells, we again observed that it was assembled with Gβ1-myc as well as endogenous Gβ subunits, and the amount of these complexes in the cells increased in the presence of PhLP-FLAG (Fig. 6B, compare lanes 2 and 3). On average, the amounts of Gγ1-HA in the pull-downs increased by 3.6 ± 0.8-fold (p < 0.05), and the amounts of co-precipitated Gβ1-myc and endogenous Gβ subunits increased by 3.3 ± 0.5-fold (p < 0.05) and 1.4 ± 0.6-fold, respectively (means ± S.E.; n = 3). Surprisingly, no detectable amounts of PhLP-FLAG were retained by these newly assembled Gβ·γ1-HA complexes, which instead contained substantial amounts of bound phosducin (Fig. 6B). Moreover, when PhLP-FLAG itself was pulled down from the same cells with anti-FLAG-agarose, it co-precipitated with Gβ, which was essentially free from Gγ1-HA (Fig. 6C). These data support a model according to which PhLP interacts exclusively with nascent Gβ that has already been folded by CCT, perhaps to keep this protein in a conformation optimal for assembly with Gγ. The ensuing association of Gβ with Gγ triggers PhLP release, and from now on, the protection of the newly assembled Gβγ dimer becomes a prerogative of a different molecular chaperone, phosducin. As our model predicts, the amount of Gβ co-precipitated with PhLP-FLAG decreased when the cells were co-transfected with Gγ1-HA (Fig. 6D). Among two types of Gβ-containing complexes captured in this assay, Gβ·CCT·PhLP-FLAG and Gβ·PhLP-FLAG, only the second type is expected to dissociate in the presence of increasing amounts of Gγ1-HA. The degree of the observed Gβ reduction indicates that Gβ·CCT·PhLP-FLAG is significantly more abundant in the cell than a transient Gβ·PhLP-FLAG.
FIGURE 6.
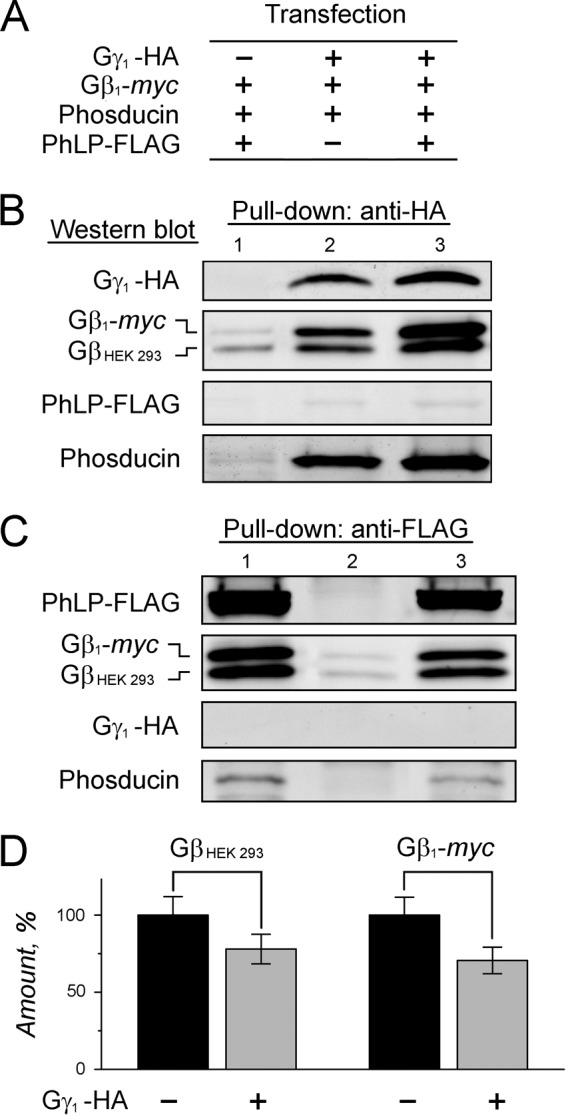
Analysis of the interaction of PhLP with Gβ and Gβγ subunits in cell culture. A, HEK 293 cells were co-transfected with the plasmids encoding the indicated epitope-tagged proteins and analyzed by pull-down with anti-HA or anti-FLAG-agarose after 48 h. B, representative blots showing captured Gγ1-HA and its co-precipitated endogenous GβHEK 293, Gβ1-myc, PhLP-FLAG, and phosducin. C, representative blots showing captured PhLP-FLAG and its co-precipitated GβHEK 293, Gβ1-myc, Gγ1-HA, and phosducin. D, the Gβ1-myc and GβHEK 293 protein bands in C were quantified, and their values were normalized to that of PhLP-FLAG and expressed as a percentage fraction in lane 3 compared with lane 1; GβHEK 293 and Gβ1-myc were reduced by 22 ± 10% (p = 0.094) and 29 ± 9% (p = 0.023). Error bars, S.E. with n = 5. Paired t test was applied.
DISCUSSION
These studies reveal a mechanism whereby the cellular levels of heterotrimeric G proteins are controlled by two splice isoforms of phosducin-like protein (Fig. 7). We demonstrate that a short transcript produced by the alternative splicing of the phosducin-like protein gene, PhLPs, previously found in rat brain (28), human retina (29), and chromaffin cells of bovine adrenal gland (36), is widely expressed in normal human tissues at levels typically lower than those of PhLP. Seeking to understand the biological significance of PhLPs, we expressed this protein in the rod photoreceptors of transgenic mice and found that this manipulation ablated the expression of the rod major heterotrimeric G protein, transducin (Gαt1β1γ1). All transducin subunits were affected on either transcriptional or posttranslational levels. The decrease of Gαt1 was primarily caused by a dramatic and rather specific down-regulation of its transcript, although a mutual posttranslational destabilization due to the Gβ1γ1 deficiency in these cells, similar to that observed in Gγ1-null mice (37, 38), may also contribute to the reduction. The molecular mechanism of this negative transcriptional feedback remains unknown and will be the subject of future studies. The decline of two other transducin subunits, Gβ1 and Gγ1, and a remarkable propensity of PhLPs toward the chaperonin CCT responsible for the posttranslational stabilization of these proteins, suggests that PhLPs acts as a CCT suppressor. This notion gains additional support from our cell culture studies, demonstrating that, in sharp contrast to PhLP, which markedly stimulates the dimerization of Gβ and Gγ subunits, PhLPs suppresses this process. This observation, thus, further corroborates the notion that PhLP acts as a co-factor of CCT, facilitating the assembly of the Gβγ subunit complex (2), whereas PhLPs competitively inhibits this function. According to the currently accepted model, a permanent association of the G protein β and γ subunits, via a high affinity coiled-coil interaction (39), is required for their posttranslational stabilization in cells (4) and occurs subsequent to the folding of Gβ by CCT (2). PhLP may potentially facilitate this interaction by rendering the newly folded Gβ competent to bind Gγ. As evident from the lack of any significant co-precipitation of PhLP and Gγ1 observed in all of our experiments, binding of Gγ1 apparently triggers the release of PhLP from Gβ1 and also serves as a signal precluding the assembled Gβ1γ1 dimer from re-entering CCT. All of these latter steps in the posttranslational processing of Gβ are inhibited by PhLPs, which binds CCT preloaded with nascent Gβ with a high affinity and, being unable to productively assist with the release of Gβ, traps this protein in the folding chamber of the chaperonin. Such a mode of action is consistent with a broad suppression of Gβ, which was indeed observed in rods unable to maintain a normal level of Gβ5L.
FIGURE 7.
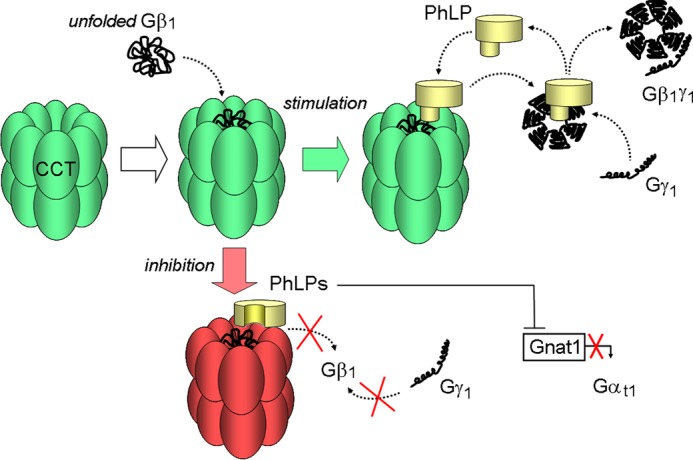
Model illustrating the opposite effects of PhLP and PhLPs on the biosynthesis of rod heterotrimeric G protein. Unfolded Gβ1 enters the folding chamber of CCT (white arrow). PhLP stimulates the subsequent steps of this process by facilitating the assembly of processed Gβ1 with Gγ1 (green arrow). PhLPs inhibits these steps by blocking the release of Gβ1 from CCT and also suppresses the transcription of Gαt1 subunit (red arrow).
The only physiological function of PhLPs demonstrated thus far was its ability to inhibit Ca2+-induced exocytosis in chromaffin cells (36). Our data provide the first evidence that PhLPs can also effectively regulate the expression of heterotrimeric G proteins in vivo. What could be the physiological role of PhLPs in photoreceptors? At this point, we can only speculate that the effects of PhLPs would become significant only when its protein level is up-regulated through alternative mRNA splicing. If such regulation does occur in the photoreceptors under a stress condition (e.g. excessive light stimulation), the ensuing temporal blockage of heterotrimeric G protein-mediated visual signaling and perhaps Ca2+-mediated membrane trafficking may play a protective role, increasing the chance of photoreceptors to survive under this condition. Determining whether such a stress response, encompassing global suppression of extracellular signaling, is widespread among eukaryotes will remain the subject of future studies.
This work was supported, in whole or in part, by National Institutes of Health Grants EY019665 (to M. S.) and EY017035 (to V. R.) and an unrestricted Research to Prevent Blindness Grant awarded to the West Virginia University Eye Institute. The Transgenic Animal Core Facility at West Virginia University was supported by Centers of Biomedical Excellence (CoBRE) Grants RR031155 and RR016440.

This article contains supplemental Table 1.
- CCT
- chaperonin containing t-complex protein 1
- PhLP
- phosducin-like protein
- PhLPs
- phosducin-like protein short isoform
- TCP-1
- t-complex protein 1
- qPCR
- quantitative PCR.
REFERENCES
- 1. Lagerström M. C., Schiöth H. B. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 2. Willardson B. M., Howlett A. C. (2007) Function of phosducin-like proteins in G protein signaling and chaperone-assisted protein folding. Cell. Signal. 19, 2417–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dupre D. J., Robitaille M., Rebois R. V., Hébert T. E. (2009) The role of Gβγ subunits in the organization, assembly, and function of GPCR signaling complexes. Annu. Rev. Pharmacol. Toxicol. 49, 31–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mende U., Schmidt C. J., Yi F., Spring D. J., Neer E. J. (1995) The G protein γ subunit. Requirements for dimerization with β subunits. J. Biol. Chem. 270, 15892–15898 [DOI] [PubMed] [Google Scholar]
- 5. Muñoz I. G., Yébenes H., Zhou M., Mesa P., Serna M., Park A. Y., Bragado-Nilsson E., Beloso A., de Cárcer G., Malumbres M., Robinson C. V., Valpuesta J. M., Montoya G. (2011) Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat. Struct. Mol. Biol. 18, 14–19 [DOI] [PubMed] [Google Scholar]
- 6. Dekker C., Roe S. M., McCormack E. A., Beuron F., Pearl L. H., Willison K. R. (2011) The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 30, 3078–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thulasiraman V., Yang C. F., Frydman J. (1999) In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 18, 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lambright D. G., Sondek J., Bohm A., Skiba N. P., Hamm H. E., Sigler P. B. (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379, 311–319 [DOI] [PubMed] [Google Scholar]
- 9. Camasses A., Bogdanova A., Shevchenko A., Zachariae W. (2003) The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12, 87–100 [DOI] [PubMed] [Google Scholar]
- 10. Spiess C., Meyer A. S., Reissmann S., Frydman J. (2004) Mechanism of the eukaryotic chaperonin. Protein folding in the chamber of secrets. Trends Cell Biol. 14, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kubota S., Kubota H., Nagata K. (2006) Cytosolic chaperonin protects folding intermediates of Gβ from aggregation by recognizing hydrophobic β-strands. Proc. Natl. Acad. Sci. U.S.A. 103, 8360–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valpuesta J. M., Martín-Benito J., Gómez-Puertas P., Carrascosa J. L., Willison K. R. (2002) Structure and function of a protein folding machine. The eukaryotic cytosolic chaperonin CCT. FEBS Lett. 529, 11–16 [DOI] [PubMed] [Google Scholar]
- 13. Blaauw M., Knol J. C., Kortholt A., Roelofs J., Ruchira, Postma M., Visser A. J., van Haastert P. J. (2003) Phosducin-like proteins in Dictyostelium discoideum. Implications for the phosducin family of proteins. EMBO J. 22, 5047–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaudet R., Bohm A., Sigler P. B. (1996) Crystal structure at 2.4 ångstroms resolution of the complex of transducin βγ and its regulator, phosducin. Cell 87, 577–588 [DOI] [PubMed] [Google Scholar]
- 15. Schröder S., Lohse M. J. (2000) Quantification of the tissue levels and function of the G-protein regulator phosducin-like protein (PhlP). Naunyn Schmiedebergs Arch. Pharmacol. 362, 435–439 [DOI] [PubMed] [Google Scholar]
- 16. McLaughlin J. N., Thulin C. D., Hart S. J., Resing K. A., Ahn N. G., Willardson B. M. (2002) Regulatory interaction of phosducin-like protein with the cytosolic chaperonin complex. Proc. Natl. Acad. Sci. U.S.A. 99, 7962–7967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Humrich J., Bermel C., Bünemann M., Härmark L., Frost R., Quitterer U., Lohse M. J. (2005) Phosducin-like protein regulates G-protein βγ folding by interaction with tailless complex polypeptide-1α. Dephosphorylation or splicing of PhLP turns the switch toward regulation of Gβγ folding. J. Biol. Chem. 280, 20042–20050 [DOI] [PubMed] [Google Scholar]
- 18. Martín-Benito J., Bertrand S., Hu T., Ludtke P. J., McLaughlin J. N., Willardson B. M., Carrascosa J. L., Valpuesta J. M. (2004) Structure of the complex between the cytosolic chaperonin CCT and phosducin-like protein. Proc. Natl. Acad. Sci. U.S.A. 101, 17410–17415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lukov G. L., Baker C. M., Ludtke P. J., Hu T., Carter M. D., Hackett R. A., Thulin C. D., Willardson B. M. (2006) Mechanism of assembly of G protein βγ subunits by protein kinase CK2-phosphorylated phosducin-like protein and the cytosolic chaperonin complex. J. Biol. Chem. 281, 22261–22274 [DOI] [PubMed] [Google Scholar]
- 20. Wells C. A., Dingus J., Hildebrandt J. D. (2006) Role of the chaperonin CCT/TRiC complex in G protein βγ-dimer assembly. J. Biol. Chem. 281, 20221–20232 [DOI] [PubMed] [Google Scholar]
- 21. Sokolov M., Strissel K. J., Leskov I. B., Michaud N. A., Govardovskii V. I., Arshavsky V. Y. (2004) Phosducin facilitates light-driven transducin translocation in rod photoreceptors. Evidence from the phosducin knockout mouse. J. Biol. Chem. 279, 19149–19156 [DOI] [PubMed] [Google Scholar]
- 22. Krispel C. M., Sokolov M., Chen Y. M., Song H., Herrmann R., Arshavsky V. Y., Burns M. E. (2007) Phosducin regulates the expression of transducin βγ subunits in rod photoreceptors and does not contribute to phototransduction adaptation. J. Gen. Physiol. 130, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belcastro M., Song H., Sinha S., Song C., Mathers P. H., Sokolov M. (2012) Phosphorylation of phosducin accelerates rod recovery from transducin translocation. Invest. Ophthalmol. Vis. Sci. 53, 3084–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flanary P. L., DiBello P. R., Estrada P., Dohlman H. G. (2000) Functional analysis of Plp1 and Plp2, two homologues of phosducin in yeast. J. Biol. Chem. 275, 18462–18469 [DOI] [PubMed] [Google Scholar]
- 25. Stirling P. C., Srayko M., Takhar K. S., Pozniakovsky A., Hyman A. A., Leroux M. R. (2007) Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression. Mol. Biol. Cell 18, 2336–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCormack E. A., Altschuler G. M., Dekker C., Filmore H., Willison K. R. (2009) Yeast phosducin-like protein 2 acts as a stimulatory co-factor for the folding of actin by the chaperonin CCT via a ternary complex. J. Mol. Biol. 391, 192–206 [DOI] [PubMed] [Google Scholar]
- 27. Stirling P. C., Cuéllar J., Alfaro G. A., El Khadali F., Beh C. T., Valpuesta J. M., Melki R., Leroux M. R. (2006) PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J. Biol. Chem. 281, 7012–7021 [DOI] [PubMed] [Google Scholar]
- 28. Miles M. F., Barhite S., Sganga M., Elliott M. (1993) Phosducin-like protein. An ethanol-responsive potential modulator of guanine nucleotide-binding protein function. Proc. Natl. Acad. Sci. U.S.A. 90, 10831–10835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Craft C. M., Xu J., Slepak V. Z., Zhan-Poe X., Zhu X., Brown B., Lolley R. N. (1998) PhLPs and PhLOPs in the phosducin family of Gβγ binding proteins. Biochemistry 37, 15758–15772 [DOI] [PubMed] [Google Scholar]
- 30. Humrich J., Bermel C., Grubel T., Quitterer U., Lohse M. J. (2003) Regulation of phosducin-like protein by casein kinase 2 and N-terminal splicing. J. Biol. Chem. 278, 4474–4481 [DOI] [PubMed] [Google Scholar]
- 31. Lukov G. L., Hu T., McLaughlin J. N., Hamm H. E., Willardson B. M. (2005) Phosducin-like protein acts as a molecular chaperone for G protein βγ dimer assembly. EMBO J. 24, 1965–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Posokhova E., Song H., Belcastro M., Higgins L., Bigley L. R., Michaud N. A., Martemyanov K. A., Sokolov M. (2011) Disruption of the chaperonin containing TCP-1 function affects protein networks essential for rod outer segment morphogenesis and survival. Mol. Cell Proteomics 10, M110.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xing Y., Kapur K., Wong W. H. (2006) Probe selection and expression index computation of affymetrix exon arrays. PloS One 1, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kapur K., Xing Y., Ouyang Z., Wong W. H. (2007) Exon arrays provide accurate assessments of gene expression. Genome Biol. 8, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chittum H. S., Lane W. S., Carlson B. A., Roller P. P., Lung F. D., Lee B. J., Hatfield D. L. (1998) Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37, 10866–10870 [DOI] [PubMed] [Google Scholar]
- 36. Gensse M., Vitale N., Chasserot-Golaz S., Bader M. F. (2000) Regulation of exocytosis in chromaffin cells by phosducin-like protein, a protein interacting with G protein βγ subunits. FEBS Lett. 480, 184–188 [DOI] [PubMed] [Google Scholar]
- 37. Lobanova E. S., Finkelstein S., Herrmann R., Chen Y. M., Kessler C., Michaud N. A., Trieu L. H., Strissel K. J., Burns M. E., Arshavsky V. Y. (2008) Transducin γ-subunit sets expression levels of α- and β-subunits and is crucial for rod viability. J. Neurosci. 28, 3510–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kolesnikov A. V., Rikimaru L., Hennig A. K., Lukasiewicz P. D., Fliesler S. J., Govardovskii V. I., Kefalov V. J., Kisselev O. G. (2011) G-protein βγ-complex is crucial for efficient signal amplification in vision. J. Neurosci. 31, 8067–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garritsen A., van Galen P. J., Simonds W. F. (1993) The N-terminal coiled-coil domain of β is essential for γ association. A model for G-protein βγ subunit interaction. Proc. Natl. Acad. Sci. U.S.A. 90, 7706–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eng J. K., McCormack A. L., Yates J. R., 3rd (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]


