Background: CD44 is cleaved, releasing a C-terminal intracellular domain with unknown function in the cytoplasm.
Results: Overexpression of this intracellular domain negatively affects the function of full-length CD44.
Conclusion: Determining the mechanism of interference highlighted the interaction of CD44 with ankyrin-3 adaptor proteins.
Significance: Released CD44 domains amplify the loss of function resulting from the initial cleavage.
Keywords: Cd44, Chondrocytes, Ezrin, Hyaluronate, Osteoarthritis, Ankyrin-3
Abstract
The hyaluronan receptor CD44 undergoes sequential proteolytic cleavage at the cell surface. The initial cleavage of the CD44 extracellular domain is followed by a second intramembranous cleavage of the residual CD44 fragment, liberating the C-terminal cytoplasmic tail of CD44. In this study conditions that promote CD44 cleavage resulted in a diminished capacity to assemble and retain pericellular matrices even though sufficient non-degraded full-length CD44 remained. Using stable and transient overexpression of the cytoplasmic domain of CD44, we determined that the intracellular domain interfered with anchoring of the full-length CD44 to the cytoskeleton and disrupted the ability of the cells to bind hyaluronan and assemble a pericellular matrix. Co-immunoprecipitation assays were used to determine whether the mechanism of this interference was due to competition with actin adaptor proteins. CD44 of control chondrocytes was found to interact and co-immunoprecipitate with both the 65- and 130-kDa isoforms of ankyrin-3. Moreover, this interaction with ankyrin-3 proteins was diminished in cells overexpressing the CD44 intracellular domain. Mutating the putative ankyrin binding site of the transiently transfected CD44 intracellular domain diminished the inhibitory effects of this protein on matrix retention. Although CD44 in other cells types has been shown to interact with members of the ezrin/radixin/moesin (ERM) family of adaptor proteins, only modest interactions between CD44 and moesin could be demonstrated in chondrocytes. The data suggest that release of the CD44 intracellular domain into the cytoplasm of cells such as chondrocytes exerts a competitive or dominant-negative effect on the function of full-length CD44.
Introduction
Many cells depend upon extracellular matrix receptors to sense changes in their external milieu and mount an appropriate response to maintain optimal cell homeostasis (1). This is especially important in tissues such as cartilage where chondrocytes are entirely dependent on their extracellular matrix for nutrient support and external signal recognition (2). In degenerative disease states such as osteoarthritis, there is proteolytic loss of extracellular matrix that not only affects overall tissue integrity but may also result in deterioration of exposed matrix receptors. Such is the case of the hyaluronan receptor CD44. We have recently shown proteolytic cleavage or shedding of CD44 in articular chondrocytes. Moreover, this degradation process is pronounced in cells from patients undergoing total knee replacement or in vitro models used to mimic osteoarthritis (3). The shedding of CD44 could be blocked by inhibitors of matrix metalloproteinases. This initial cleavage occurs by the action of membrane-type metalloproteases such as membrane type I (MT1-MMP) or ADAM17 or ADAM10 (4, 5). An interesting aspect of this cleavage is that the residual C-terminal fragment of CD44 (CD44-EXT)3 (6, 7) is also a substrate for intramembranous cleavage by γ-secretase, releasing an intracellular domain fragment of CD44 (CD44-ICD) into the cytoplasm (8) in a process similar to the generation of the Notch-ICD (9). Thus, this signature pattern of sequential proteolytic cleavage of CD44 generates at least three fragments, one of which is released inside the cell.
CD44 is a single-pass transmembrane glycoprotein receptor. CD44 appears to exhibit a capacity for cell signaling induced by alterations in CD44-hyaluronan interactions (10–12). However, the exact mechanisms responsible for CD44-mediated signaling remain unclear and appear to differ depending on the cell type (10, 13–15). The cytoplasmic domain of CD44 has no intrinsic kinase activity but has been shown to interact with members of the Src and Ras family of GTPases (16–18). CD44 has also been shown to act as a co-receptor influencing the activity of various receptor tyrosine or serine/threonine kinases including IGF1-Rβ, EGF-R, ErbB2, BMP-R, TGFβ-R, and PDGF-Rβ (19–27). Furthermore, in many of the cell lines that have been investigated, CD44 co-immunoprecipitates with these receptors as part of a larger complex. Thus, it is likely that CD44-mediated signal transduction will be impacted by proteolytic fragmentation if only due to the loss of the hyaluronan binding ectodomain.
In some cell types the cytoplasmic domain of CD44 has also been shown to interact with cytoskeletal adaptor proteins of the ankyrin (28) and ezrin/radixin/moesin (ERM) (29, 30) families. The ERM binding domain of CD44 includes amino acids 292–300 and is located between the transmembrane domain and the intracellular membrane-proximal domain. Distal to the ERM binding domain is the ankyrin binding motif including amino acids 304–318 (31). Studies have shown that disruption of the actin cytoskeleton using cytochalasin D reduced CD44-hyaluronan interactions and a loss of the pericellular matrix or “coat” surrounding the cells (32–34). In addition, overexpressing a competitor of the CD44/ankyrin binding motif blocked hyaluronan-mediated Ca2+ signaling in endothelial cells (35). Therefore, binding of the CD44 cytoplasmic tail to the cytoskeleton may be essential for the ability of cells to retain a pericellular matrix. Moreover, CD44-mediated signal transduction also occurs in part by interactions with the cytoskeleton via an ERM and/or ankyrin binding domains (36). To date, there is only indirect, cytochalasin-based evidence to suggest that CD44 interacts with cytoskeletal proteins in articular chondrocytes (32). A direct co-immunoprecipitation between CD44 and cytoskeleton adaptor proteins has never been shown in chondrocytes. Also, it remains unclear whether CD44 can interact with both ERM and ankyrin simultaneously forming a single complex or whether CD44/ERM and CD44/ankyrin are two separate mutually exclusive complexes that are utilized in a tissue-dependent manner.
Given that CD44 fragmentation occurs, we wanted to determine whether, in addition to the loss of the CD44 ectodomain, the released CD44 intracellular domain (CD44-ICD) also affected either cell signaling or matrix retention. Previous studies have suggested that the released CD44-ICD goes directly to the nucleus and activates gene transcription (37). However, it is unknown whether a fraction of the CD44-ICD remains in the cytoplasm and, if so, whether the CD44-ICD that resides in the cytoplasm exhibits any functional significance. This study addresses the biological consequences of cytoplasmic CD44-ICD and further investigates the mechanisms whereby CD44-ICD interferes with intracellular interactions of endogenous full-length CD44 in chondrocytes.
EXPERIMENTAL PROCEDURES
Materials
Primer design for cloning strategies and real time RT-PCR were developed using the Integrated DNA Technologies (Coralville, IA) website. Cloning vectors were purchased from Invitrogen with the exception of pEGFP-N2 vector (Clontech, Mountain View, CA). Also from Invitrogen were the Gene Tailor site-directed mutagenesis kit, NuPAGE gels, Calcein-AM, TRIzol, anti-GFP antibodies, enhanced chemiluminescence reagents, and magnetic beads for co-immunoprecipitation assays. DMEM was obtained from Mediatech (Herndon, VA), and FBS was purchased from Hyclone (South Logan, UT). Pronase and collagenase P used in dissociation of tissues were obtained from EMD Scientific (San Diego, CA) and Roche Applied Science (Indianapolis, IN), respectively. Protease inhibitor mixture was from Thermo Fisher (Waltham, MA), and Clear Blue x-ray film was from Genesee Scientific (San Diego, CA). Transient transfection was performed using AMAXA nucleofection technology (Lonza, Germany). Stable cell lines were generated using Flp-InTM system from Invitrogen. The qScriptTM cDNA synthesis kit was obtained from Quanta Biosciences (VWR), and RT2 Real TimeTM SYBR Green reagents were from SA Biosciences (Frederick, MD). IL-1β was from R&D Systems, Inc. (Minneapolis, MN). Cell lysis buffer, anti-ERM, and anti-phospho-ERM antibodies were from Cell Signaling Technologies (Danvers, MA). Anti-ankyrin-1 and ankyrin-3 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-moesin and anti-human CD44 antibodies were from EMD-Millipore (Billerica, MA), and anti-rat CD44 (OX49) was from BD Biosciences. All other enzymes and chemicals, either molecular biology or reagent grade materials, were purchased from Sigma. Human pigmented epithelial cells (ARPE-19) were obtained from ATCC (Manassas, VA); immortalized human mammary epithelial (HMLE) cells were a kind gift from Dr. Robert Weinberg, Whitehead Institute, Massachusetts Institute of Technology (Cambridge, MA).
Generation of CD44-ICD Constructs
Primers were designed to amplify the CD44-ICD domain from a human CD44H sequence, extending from Ala288 to the stop codon. Both untagged and C-terminal Myc-tagged CD44-ICD constructs were generated as previously described (3). An N-terminal GFP epitope tag was incorporated into the CD44-ICD sequence using a pcDNA3.1/NT-GFP-TOPO vector. To prepare a C-terminal enhanced GFP (EGFP)-tagged CD44-ICD construct, primers were designed to amplify the previously cloned untagged CD44-ICD including the start site and Kozak sequence from the pCMV/myc/cyto vector and subcloned into a pcDNA3.1/V5-His-TOPO vector. The upstream primer was engineered to include a HindIII site 5′-CAAGCTTTAACTAGAGAACCCGTGGCC-3′, and the downstream primer included an ApaI site, 5′-TTCGGGCCCCACCCCAATCTTCATGTCCACATTCTGCAGGTTG-3′. The PCR product was subcloned into pcDNA3.1/V5-His-TOPO vector and digested with HindIII and ApaI restriction enzymes. The resulting insert was then subcloned into a pEGFP-N2 vector (Clontech) to generate a C-terminal EGFP tag. Site-directed mutagenesis (to produce a control irrelevant 15-kDa peptide), and ERM and ankyrin mutants were generated using a Gene Tailor site-directed mutagenesis kit. The untagged CD44-ICD sequence was used as a template, and the desired mutations were incorporated in the upstream primers. The control 15-kDa irrelevant peptide was generated by adding a single nucleotide (extra dG shown in bold) to the upstream primer (5′-CCCGTGGCCACCATGGCCCAGGGTGCAGCTGCAG-3′) at the beginning of the coding sequence of the untagged CD44-ICD, generating a frameshift that translated into a unique and distinct sequence. This peptide was sequenced at the East Carolina University DNA sequencing facility to ensure no premature stop codons were generated. To destroy the ankyrin binding site point, mutations were developed to modify two positively charged amino acids in the putative CD44 binding site (Arg313–Lys314 to Ile313–Asn314). These point mutations were achieved using a single primer, 5′-AATGGAGCTGTGGAGGACACAAACCCAGTGGACTC-3′ (altered sequence shown in bold). All constructs were sequenced at the East Carolina University DNA sequencing facility.
Cell Culture
Bovine articular chondrocytes were isolated from metacarpophalangeal joints of 18–24-month-old steers, and human articular chondrocytes were from human knee cartilage after joint replacement surgery, both as described previously (3). All human cartilage was obtained within 24 h of death and with institutional approval. The rat chondrosarcoma cell line (RCS) was a continuous long-term culture line derived from the Swarm rat chondrosarcoma tumor (38). The RCS cell line in the Knudson laboratories was a gift of Dr. James H. Kimura (formerly of Rush University Medical Center) and represented an early clone of cells that eventually became known as long term culture RCS (39). Bovine and RCS chondrocytes were cultured as high density monolayers (2.0 × 106 cells/cm2) in DMEM containing 10% FBS and 1% l-glutamine and penicillin-streptomycin and incubated at 37 °C in a 5% CO2 environment. The RCS cells were passaged at confluence using 0.25% trypsin, 2.21 mm EDTA. To allow the bovine primary chondrocytes to undergo de-differentiation, at confluence the cells were passaged by treatment with 0.25% trypsin, 2.21 mm EDTA and used at passage 3. ARPE-19 and HMLE cells were grown as described previously in DME-F-12 medium containing 10% FBS (40) or DME-F-12 medium containing insulin/EGF/hydrocortisone and 5% bovine calf serum (41), respectively.
Preparation of Stable Transfectants of RCS
RCS-CD44-ICD stable transfectant cells were produced using the Flp-In system from Invitrogen. First, to generate the flip recombination target (FRT) site into the RCS cell line, the pFRT/lacZeo plasmid was transfected into parental RCS cells using Amaxa nucleofection system to produce a stable RCS-FRT host cell line. After transfection, the cells were cultured in 10% FBS DMEM containing 100 μg/ml zeocin to select RCS-FRT cells. We then subcloned the CD44-ICD untagged into a pcDNA5 shuttle vector and co-transfected 4 μg of pcDNA5/FRT-CD44-ICD and 36 μg of pOG44 into the stable RCS-FRT cell line to produce a stable RCS-CD44-ICD cell line. Forty-eight hours after transfection, cells were cultured in hygromycin (200 μg/μl)-containing media for several weeks to select CD44-ICD stable transfectants.
Treatment of Cells
To induce proteolysis of either endogenous or exogenous CD44, confluent cultures of bovine chondrocytes or RCS were treated for varying time periods with 10 ng/ml IL-1β, 10 nm phorbol myristate acetate (PMA), or 250 μg/ml of hyaluronan oligosaccharides (a mixture of octa- and hexasaccharides derived from degraded rooster comb hyaluronic acid) prepared as described previously (3). Cells were first incubated overnight in serum free media followed by a 48-h incubation with either IL-1β, HA oligosaccharides, or PMA in serum-free media. Cell lysates were then analyzed for CD44 fragmentation by Western blotting using an antibody specific to the C-terminal cytoplasmic domain (anti-cytotail) (3). Cells were also treated with 200 turbidity reducing units/ml of bovine testicular hyaluronidase or 5 units/ml Streptomyces hyaluronidase in serum-free media for 90 min to disrupt CD44-HA interactions. To induce an epithelial-mesenchymal transition, ARPE-19 and HMLE cells were treated for 1 day in medium containing TNF-α (10 ng/ml) plus TGFβ2 (5 ng/ml) (40) or TGFβ1 (2.5 ng/ml) (41), respectively.
Western Blotting
Total protein was extracted using cell lysis buffer containing protease and phosphatase inhibitor cocktails. Equivalent protein concentrations were loaded into 4–12% NuPAGE® Novex® Tris acetate gradient mini gels. After electrophoresis, proteins within the acrylamide gel were transferred to a nitrocellulose membrane using a Criterion blotter apparatus (Bio-Rad), and the nitrocellulose membrane was then blocked in 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T). CD44-specific antibody to the ectodomain (BU52; Calbiochem) and CD44 cytoplasmic tail-specific antibody (anti-cytotail) (3) were used to detect endogenous CD44 and CD44 fusion proteins. A GFP antibody was used to detect both GFP- and EGFP-tagged fusion proteins. After incubation with corresponding secondary antibody, detection was performed using enhanced chemiluminescence reagents.
Co-immunoprecipitation
Cell monolayers were extracted using 10 mm Tris, pH 7.5, with 2 mm EDTA, 1% Triton X-100, and protease and phosphatase inhibitors. Magnetic protein G Dynabeads® were used after the product protocol that for most experiments included incubation of a preformed antibody-Dynabead complex with cell lysates for 60 min. In other experiments the antibody-Dynabead complexes were incubated overnight with cell lysates. Typically, between 100 and 500 μg of protein lysate was incubated with antibody-Dynabead complex (10 μg of a primary antibody diluted in 200 μl of PBS-Tween 20). The non-binding protein in the immunoprecipitate (termed the flow-through (or “F” fraction) was collected for further analysis. The immunoprecipitated proteins were eluted from the beads-antibody-antigen complex using 20 μl of elution buffer (50 mm Glycine, pH 2.8). The eluted protein fraction is termed the bound or “B” fraction. The recovered protein fractions were mixed with 10 μl of NuPAGE® LDS sample buffer, 4 μl of reducing agent, and 6 μl of deionized water and incubated 10 min at 70 °C and loaded into a 4–12% NuPAGE® Novex® Tris acetate gradient mini gel and analyzed by Western blot analysis. Other lysis buffers of different detergent conditions and concentrations were used to ensure that the lack of protein-protein interactions was not caused by harsh buffer conditions. Lysis buffer used contained 50 mm Tris-HCl, 150 mm NaCl, and 5 mm EDTA and varying concentrations of Igepal (previously known as Nonidet P-40, 0.1, 0.25, 0.5, and 1%), Triton X-100 (0.5%), or CHAPS (0.5 and 1%). Several antibodies were conjugated with HRP following the protocol for the Lightning-Link HRP conjugation kit (Innova Biosciences) to avoid cross-reactivity with immunoglobins of other species in co-immunoprecipitation assays by avoiding the use of secondary reagents. The HRP-conjugated antibodies included anti-CD44-cytotail-HRP, anti-ERM-HRP, anti-pERM-HRP, anti-ankyrin1-HRP, and anti-ankyrin3-HRP. Other antibodies used as non-HRP-conjugated included anti-moesin.
Particle Exclusion Assay
Bovine or RCS chondrocytes were cultured overnight in 35-mm wells. The medium was replaced with a suspension of formalin-fixed erythrocytes in PBS, 0.1% BSA (3). Cells were photographed using a Nikon TE2000 inverted phase-contrast microscope, and images were captured digitally in real time using a Retiga 2000R digital camera (QImaging, Surrey, British Columbia, Canada). The presence of cell-bound extracellular matrix is seen as the particle-excluded zone surrounding the chondrocytes. In some experiments, cells were treated with calcein-AM to provide contrast and delineate the extent of the plasma membrane.
Differential Extraction
Cells were extracted first in 200 μl of a light-detergent buffer containing 50 mm Tris-HCl, 150 mm NaCl, 5 mm EDTA, 0.1% Igepal and 1X protease inhibitor mixture and incubated on ice for 10 min. Cells were then centrifuged for 10 min at 1,300 × g, and supernatant was collected as the “lower” Nonidet P-40 extraction conditions. The pellet was re-suspended in 200 μl of a harsher-detergent buffer containing 50 mm Tris-HCl, 150 mm NaCl, 5 mm EDTA, 0.5% Igepal (Nonidet P-40), 1% Empigen and 1X protease inhibitor mixture and incubated on ice for an additional 10 min. Following centrifugation for 10 min at 13,600 × g, the supernatant was collected as the “higher” Igepal extraction conditions. Lysates were run side by side on a gel for Western blotting, loading equal volume rather than equal protein.
Fluorescence Microscopy and Flow Cytometry
For immunofluorescence studies, RCS and RCS-CD44-ICD stable transfectants were plated into 4-well chamber slides at 50% confluence and cultured for 24 h. The cells were fixed in 2% paraformaldehyde for 30 min at 4 °C, quenched with 0.2 m glycine in PBS, and then blocked in 1% BSA in PBS for 30 min at room temperature. Biotinylated hyaluronan-binding protein (Seikagaku USA, Ijamsville, MD) at a 2 μg/ml concentration was added to the cells and incubated for 1 h at 4 °C. The cells were rinsed with PBS and mounted using a medium containing DAPI nuclear stain. Cells were visualized using a Nikon Eclipse E600 microscope equipped with Y-Fl Epi-fluorescence (Melville, NY), a 60× 1.4 n.a. oil-immersion objective, and FITC (green) and DAPI (blue) filters. Images were captured digitally in real time using a Retiga 2000R digital camera and processed using NIS Elements BR imaging software (Nikon, Lewisville, TX).
For flow cytometry studies, RCS chondrocytes were pretreated with 100 turbidity reducing units/ml of testicular hyaluronidase in serum-containing DMEM for 60 min to remove all cell surface glycosaminoglycans and then released from the monolayer culture using enzyme-free cell dissociation solution (Millipore). Live cells were next preincubated for 30 min ± unlabeled hyaluronan followed by incubation ± 60 μg/ml FITC-conjugated hyaluronan for 1 h at room temperature. FITC-hyaluronan was prepared as described previously (42). After washing and centrifugation, cell surface immunofluorescence was then quantified using a FACScanTM cytometer (BD Biosciences) with CELL Quest software. For 10,000 cells, log fluorescence channel versus cells per channel was plotted.
Real Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from the bovine and RCS cultures according to the manufacturer's instructions for the use of TRIzol® reagent. The RNA was reverse-transcribed with Q-Script cDNA supermix reagents (random and oligo-dT primers) and amplified using the PTC-100TM Programmable Thermal Controller (MJ Research, Inc., Watertown, MA) at 42 °C for 30 min. For real time RT-PCR, the PCR products were detected by RT2 Real TimeTM SYBR Green reagents. Primer-specific amplification was at 60 °C for 30 s. However, fluorescence quantification was performed at a higher temperature, 72 °C. These quantification temperatures were set below the individual melting peak of each PCR product. The primer rat-specific sequences are: GAPDH, forward (5′-TTCTGGCAAAGTGGACATCGTCG3-′) and reverse (5′-TGGCCTTTCCATTGATGACGAGC-′; CD44, forward (5′-TCTGCAAGGCCTTTAATAGCACGC-3′) and reverse (5′-TGTTCGCAGCACAGATGGAATTGG-3′), and hyaluronan synthase-2, forward (5′-TCACTGACTTCCTAGACAATGG-3′ and reverse (5′-ACTGGCGGTCAGCGTCGTAGT-3′. Thermal cycling and fluorescence detection were done using a Smart Cycler system (Cepheid, Sunnyvale, CA). Real time RT-PCR efficiency (E) was calculated as E = 10[−1/slope] (3, 15). The -fold increase in copy numbers of mRNA was calculated as a relative ratio of target gene to GAPDH following the mathematical model introduced by Pfaffl (43) as described previously (3, 15).
RESULTS
Experimental Conditions That Enhance CD44 Cleavage Are Associated with a Loss of Pericellular Matrix
One of the typical phenotypic characteristics of bovine chondrocytes in culture is the retention of large pericellular matrices surrounding rounded or polygonal cells (Fig. 1, A and B). The pericellular matrices are composed at a minimum of hyaluronan-aggrecan aggregates anchored to the cell surface by hyaluronan binding to CD44 (44–46). When bovine chondrocytes were grown under conditions that affect CD44 cleavage (3), namely treatment with IL-1β (Fig. 1, C and D) or PMA (Fig. 1, E and F), the cells maintained their rounded morphology but lost their capacity to retain a pericellular matrix. These results may be co-incidental and reflect pleiotropic changes in many cell products. We have previously observed a similar loss of matrices on cytochalasin-treated chondrocytes (32). Therefore, we postulated that release of the CD44-ICD into the cytoplasm could also disrupt interactions between full-length CD44 (CD44FL) and cytoskeletal adaptor protein interactions that are required to stabilize retention of a pericellular matrix. To test this possibility, bovine chondrocytes were transiently co-transfected with an expression plasmid containing CD44-ICD and a GFP plasmid. As shown in Fig. 1, G and H, all successfully transfected CD44-ICD cells (GFP-positive cells) did not exhibit pericellular matrices, whereas most, if not all of the non-transfected chondrocytes within the same field (GFP-negative cells) retained their pericellular matrices. Thus, the presence of intracellular CD44-ICD appears to exert a negative influence on matrix retention.
FIGURE 1.
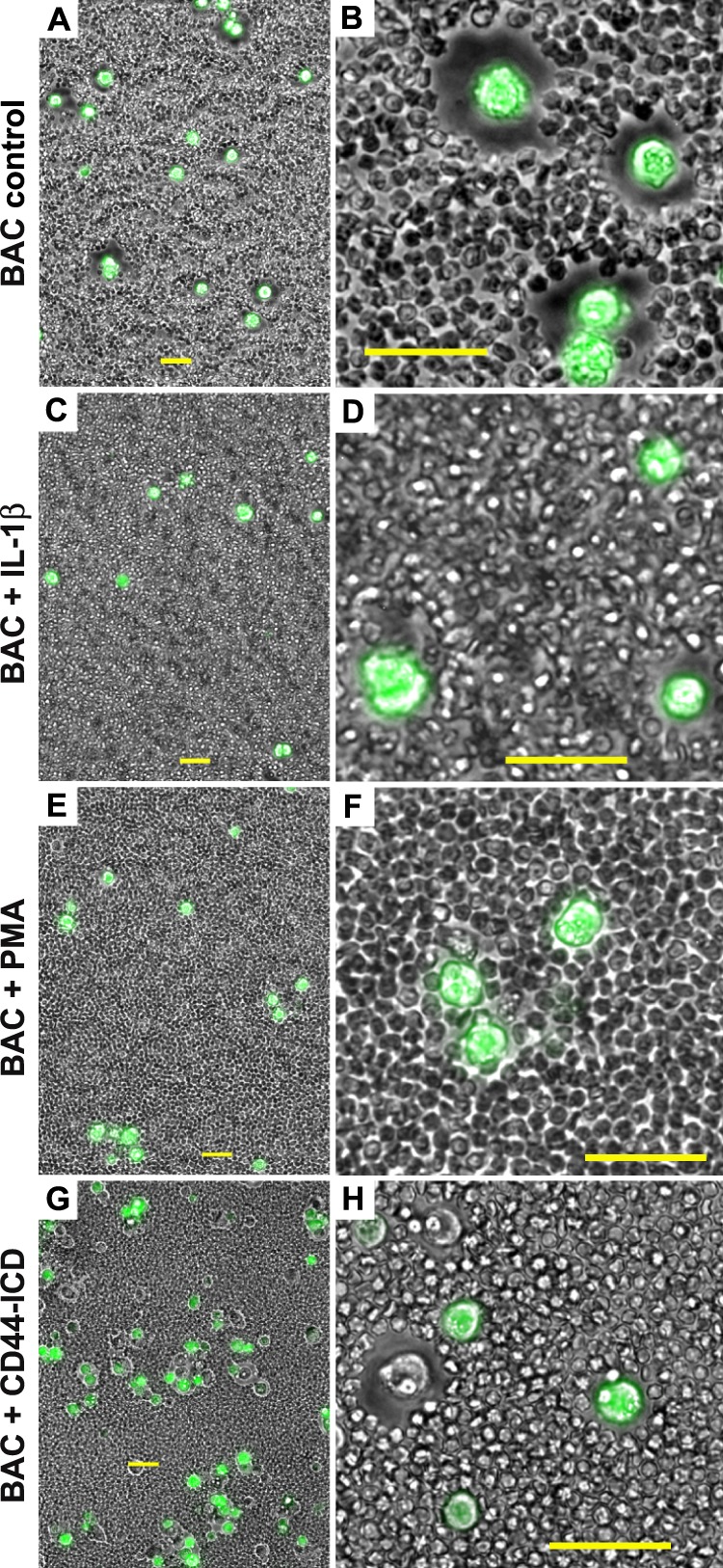
Conditions that promote CD44 cleavage alter the chondrocyte pericellular matrix. Pericellular matrices were visualized by the exclusion of formalin-fixed erythrocyte particles from the cells surface. Bovine articular chondrocytes (BAC) were incubated in the absence (A and B) or presence of 10 ng/ml IL-1β (panels C and D) or 10 mm PMA (E and F). Images shown in panels A--F represent two-color overlays of phase contrast and green fluorescence images, green fluorescence due to calcein-AM that stains live cells and was used to provide contrast. In panels G and H, bovine articular chondrocytes were co-transfected with CD44-ICD-myc and empty pEGFP-N2 plasmid. Images shown in G and H represent two-color overlays of phase contrast and green fluorescence due to EGFP. As such, successfully transfected bovine articular chondrocytes are GFP-positive and express the CD44-ICD; non-transfected cells within the same field are GFP-negative and serve as controls. Panels A, C, E, and G are lower magnification views to allow visualization of multiple cells; panels B, D, F, and H are higher magnification views to highlight details. All bars are 50 μm.
Differential Extraction of CD44FL from the Plasma Membrane
We and others have used a differential extraction approach to assay CD44FL association with the actin cytoskeleton, interactions that are likely mediated via adaptor proteins (32). In most cell types two pools of CD44 can be distinguished. As shown in Fig. 2A, considerable CD44FL protein was released from monolayer cultures of bovine chondrocytes by a 10-min treatment with a detergent containing a lower 0.1% concentration of Igepal (lanes labeled as L). After this treatment the plasma membranes remained visually intact. Subsequent treatment with a detergent containing a higher 0.5% concentration of Igepal and 1% Empigen effected complete cell lysis, and a second pool of CD44FL was recovered (Fig. 2A, lanes labeled H). The first pool of CD44 that can be released without complete dissolution of the membrane structure is a fraction that likely is not anchored to a well organized membrane or cytoskeletal complex. In control bovine chondrocytes, the lower detergent extracted pool was clearly a minor proportion of the total CD44. However, when the bovine chondrocytes were first pretreated with IL-1β, hyaluronan oligosaccharides, or PMA, a larger proportion of the CD44FL was recovered in the lower detergent pool, isolated at the same time as control cells (Fig. 2A). This suggests that these conditions have effected a shift in CD44 anchorage within the plasma membrane. In a second series of experiments, a larger percentage of the CD44FL was also recovered in the lower detergent pool of bovine chondrocytes transfected with CD44-, CD44-ICD/GFP, or CD44-ICD/myc expression plasmids (Fig. 2B) as compared with chondrocytes expressing a transfection control plasmid (t-Ctr) or non-transfected control cells (nt-Ctr).
FIGURE 2.
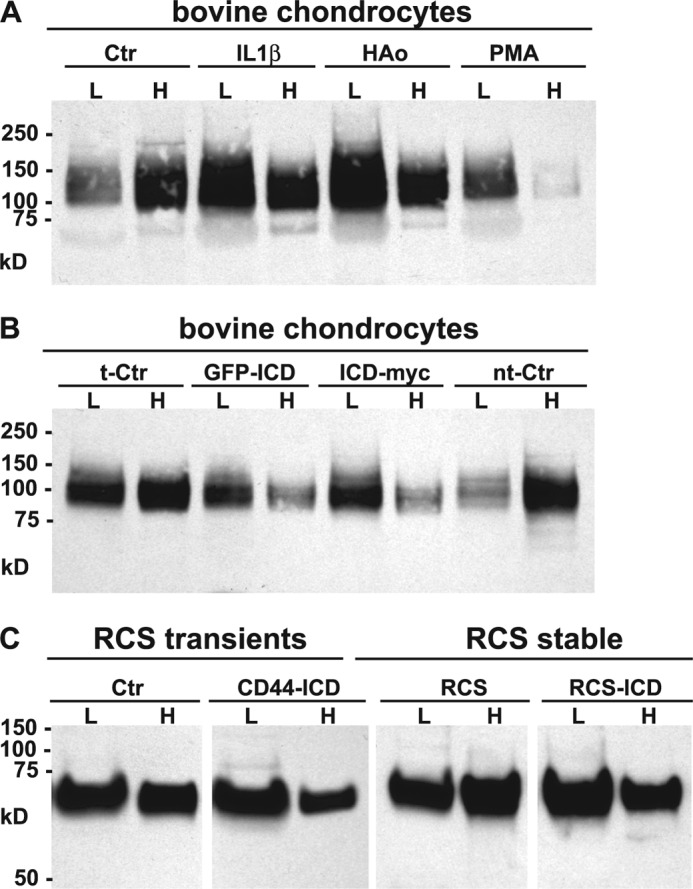
Differential extraction of full-length CD44 from chondrocytes. High density monolayers of bovine articular or RCS chondrocytes were treated under a variety of conditions and then detergent extracted using the sequential differential extraction approach (0.1% Igepal, lanes labeled as L; 0.5% Igepal and 1% Empigen, lanes labeled H). Equal-volume aliquots were analyzed by Western blotting using the CD44 anti-cytotail antibody for detection. Panel A is a representative experiment of bovine chondrocytes treated without or with IL-1β, HA oligosaccharides (HAo) or PMA. Panel B is a representative experiment of bovine chondrocytes that were transfected with a control plasmid (t-Ctr), N-terminal GFP-tagged CD44-ICD (GFP-ICD), C-terminal myc-tagged CD44-ICD (ICD-myc), or non-transfected control chondrocytes (nt-Ctr). On these Western blots, full-length CD44 of bovine chondrocytes is represented by a diffuse band between 95 and 150 kDa. Panel C is a representative experiment depicting control RCS cells or RCS cells transiently or stably transfected with CD44-ICD. Full-length CD44 of RCS chondrocytes is represented by a diffuse band between 65–85 kDa.
Stable Transfection and Use of Rat Chondrosarcoma Cells
In an effort to establish stable transfectants over-expressing CD44-ICD, a chondrocyte cell line was needed, but one that met several requirements necessary for our study. We previously reported (47) that the RCS cell line expresses CD44 and the CD44 of RCS binds hyaluronan and aggrecan and uses this tripartite complex to assemble large pericellular matrices such as those shown in Fig. 3, A and B. We have also shown that RCS cells can bind and internalize hyaluronan via CD44 (47). Like the bovine chondrocytes, transient transfection of RCS cells with CD44-ICD-containing plasmid resulted in a loss of pericellular matrix retention. For example, within the same field of view shown in Fig. 3, C and D, successfully transfected cells (GFP-positive) exhibited no pericellular matrices, whereas neighboring GFP-negative cells continued to display large matrices. CD44-ICD transiently transfected RCS also exhibited a higher proportion of the total CD44FL recovered in the lower detergent extraction pool (Fig. 2C). However, unlike the bovine chondrocytes, the control RCS cells naturally exhibited a substantial proportion of CD44FL in the lower detergent pool, making the differences due to CD44-ICD transfection more subtle.
FIGURE 3.
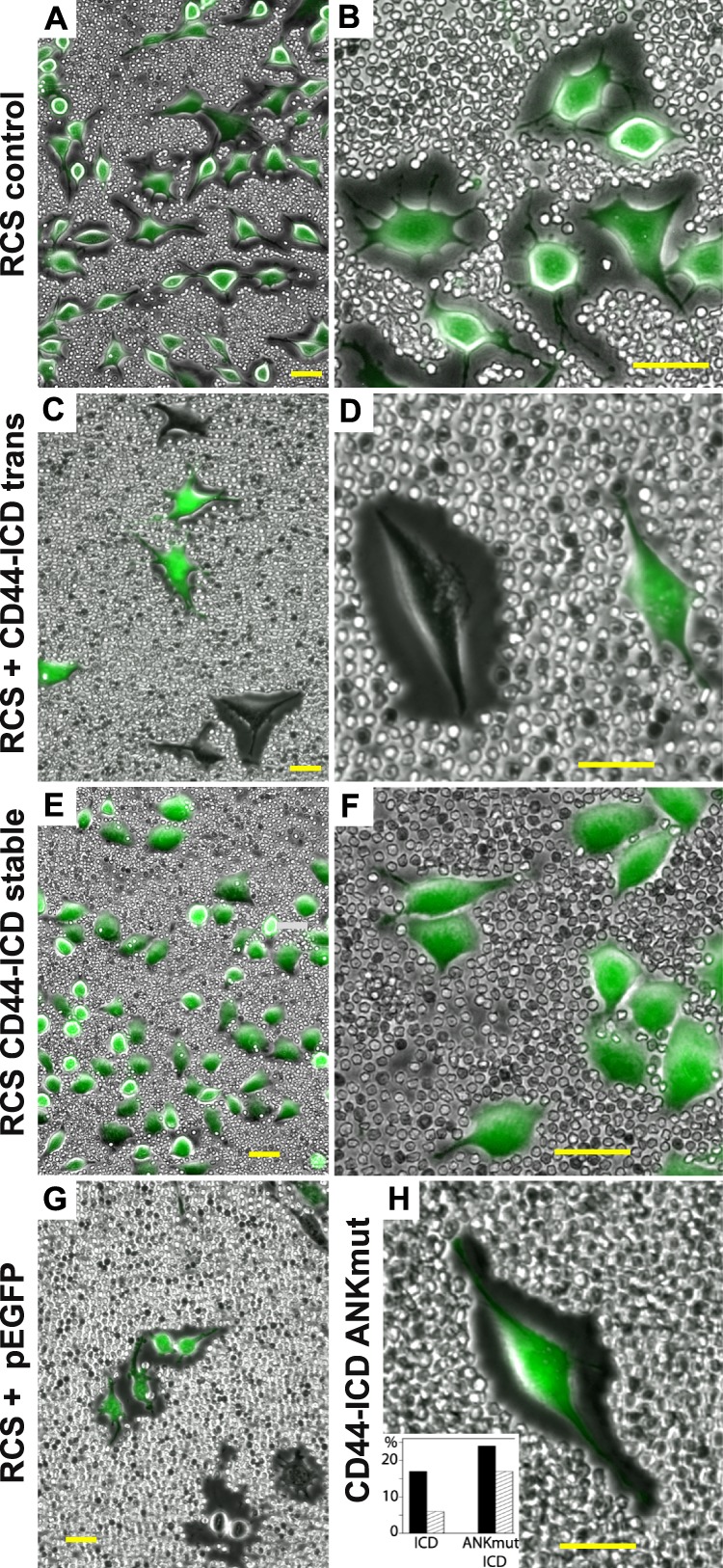
Effect of CD44-ICD on pericellular matrix retention in RCS chondrocytes. Pericellular matrices were visualized by the exclusion of formalin-fixed erythrocyte particles from the cells surface. Control, untreated RCS cells (panels A and B), and CD44-ICD stable transfectants of RCS (RCS CD44-ICD stable, panels E and F) are shown as two-color overlays of phase contrast and green fluorescence due to calcein-AM to provide contrast. RCS cells transiently transfected with C-terminal-EGFP-CD44-ICD (C and D), empty pEGFP-N2 plasmid (panel G), or C-terminal-EGFP-CD44-ICD-R313I,K314N (panel H) are shown as two-color overlays of phase contrast and green fluorescence due to EGFP. The inset in panel H summarizes the percentage of RCS cells successfully transfected (dark bars) with CD44-ICD (ICD) or C-terminal-EGFP-CD44-ICD-R313I,K314N (ANKmut ICD). The light hatched bars represent the percentage of these transfected cells that retained a coat of >1.15 coat/cell diameters. Panels A, C, E, and G are lower magnification views to allow visualization of multiple cells, and panels B, D, F, and H are higher magnification views to highlight details. All bars are 50 μm.
To establish stable transfectants in RCS cells, a Flp recombinase site was first inserted into the genome followed by cell selection using zeocin. The zeocin-positive RCS clones were then transfected with a Flp-In CD44-ICD-containing plasmid and re-selected using hygromycin. As shown in Fig. 4B, the 15-kDa CD44-ICD protein fragment was clearly seen in the resultant stable transfectants (arrows and ICD labels), a band conspicuously absent in the parental RCS cells (Fig. 4A). Moreover, the relative abundance of transgene CD44-ICD product in the stable transfectants (Fig. 4B) was similar to the level of endogenous CD44-ICD that can be observed in lysates of some human osteoarthritic patient chondrocytes (Fig. 4C, lane 1). Interestingly, the same human osteoarthritic chondrocytes (shown in Fig. 4C, lane 1) when grown as three-dimensional cultures in alginate beads, exhibited diminished generation of CD44-EXT and CD44-ICD bands (Fig. 4C, lane 2). All of the RCS cells exhibit the CD44FL at ∼75 kDa. As was observed in bovine or human chondrocytes (3), both RCS (Fig. 4A) and RCS-CD44-ICD (Fig. 4B) cells responded to IL-1β (Fig. 4, A and B, lanes 2) or PMA (lanes 5), resulting in an increase in CD44-EXT bands. When the cells were preincubated with the γ-secretase inhibitor DAPT together with IL-1β, the CD44-EXT band accumulation was enhanced (Fig. 4, A and B, lanes 3), whereas pre-incubation with the matrix metalloproteinase inhibitor GM6001 blocked the generation of CD44-EXT bands (Fig. 4, A and B, lanes 4). Preincubation with proteasome inhibitor MG132 to block CD44-ICD intracellular breakdown (together with IL-1β) increased both CD44-EXT and CD44-ICD accumulation (Fig. 4, lanes 6). Thus, the generation of endogenous CD44-ICD occurs in RCS cells as it does in bovine or human chondrocytes, a process that can be blocked by the appropriate chemical inhibitors. Another important observation from these data was that the overexpression of recombinant CD44-ICD did not inhibit or promote the proteolytic fragmentation of the endogenous CD44FL (Fig. 4B). Nonetheless, all of the CD44-ICD stable RCS cells lost the capacity to assemble pericellular matrices (Fig. 3, E and F). In addition, like the transient transfectants, a larger percentage of the CD44FL was also recovered in the lower detergent pool of the stable CD44-ICD cells as compared with the parental RCS cells (Fig. 2C).
FIGURE 4.
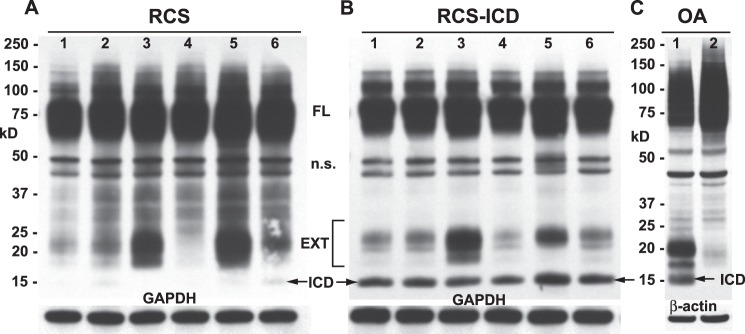
Changes in CD44 protein after stable transfection of RCS cells with CD44-ICD. High density monolayers of parental rat chondrosarcoma chondrocytes (panel A) or RCS cells stably transfected with CD44-ICD (panel B) were treated under a variety of conditions. After treatment, the cells were lysed, and aliquots of equivalent protein were analyzed by Western blotting using the anti-CD44 cytotail antibody for detection. Depicted are untreated control cells (lanes 1) or cells treated with IL-1β (lanes 2), IL-1β + DAPT (lanes 3), IL-1β + GM6001 (lanes 4), PMA (lanes 5), or IL-1β + MG132 (lanes 6). Bands represent full-length CD44 (FL), nonspecific bands (n.s.), CD44-EXT fragments (EXT), and CD44-ICD fragments (ICD). After detection of CD44, the Western blots were reprobed for GAPDH. The CD44 profile of a human osteoarthritic (OA) chondrocyte lysate is shown for comparison (panel C). Lysates derived from cells grown in monolayer (lane 1) or alginate beads (lane 2) were immunoblotted using anti-CD44 cytotail antibody. Arrows in all panels are used to further highlight CD44-ICD bands.
Stable RCS-CD44-ICD Cells Exhibit Reduced Capacity to Bind Hyaluronan
When examined by real time RT-PCR, no significant differences were observed in the base-line expression of CD44 or hyaluronan synthase 2 mRNA between RCS and CD44-ICD cells (data not shown). Thus, the lack of matrix assembly observed in RCS-CD44-ICD cells was not due to an indirect, metabolic effect on the expression of these two genes critical to matrix assembly. Using a biotinylated hyaluronan-binding protein probe, the parental RCS cells exhibited surface retention of endogenous hyaluronan typical of chondrocytes, namely a uniform but fuzzy or cloud-like appearance (Fig. 5, A and B). The staining intensity for hyaluronan on RCS-CD44-ICD cells was similar but appeared non-uniform in appearance (Fig. 5, C, D, and E). Some groups of RCS-CD44-ICD cells exhibited a discontinuous punctuate pattern of hyaluronan staining at the cell surface (Fig. 5, C and D), whereas other groups of RCS-CD44-ICD cells exhibited disrupted surface staining and the appearance of thread-like cables between cells (Fig. 5E). The staining specificity of the hyaluronan binding probe was demonstrated using hyaluronidase pretreatment of cells (Fig. 5F). One interpretation of these results is that CD44 binding activity is reduced in the RCS-CD44-ICD cells, resulting in retention of hyaluronan at the plasma membrane by way of the hyaluronan synthase alone as occurs in GFP-hyaluronan synthase 3-transfected MCF-7 cells (48). To explore the possibility that CD44-mediated hyaluronan binding was in fact reduced in RCS-CD44-ICD (as compared with RCS control cells), the binding of fluorescein-conjugated hyaluronan was compared using flow cytometry. As shown in Fig. 5, G and H, mean channel fluorescence for fluorescein-conjugated hyaluronan binding to RCS-CD44-ICD cells was reduced by ∼22% as compared with RCS cells; peak channel fluorescence was reduced by 35%.
FIGURE 5.
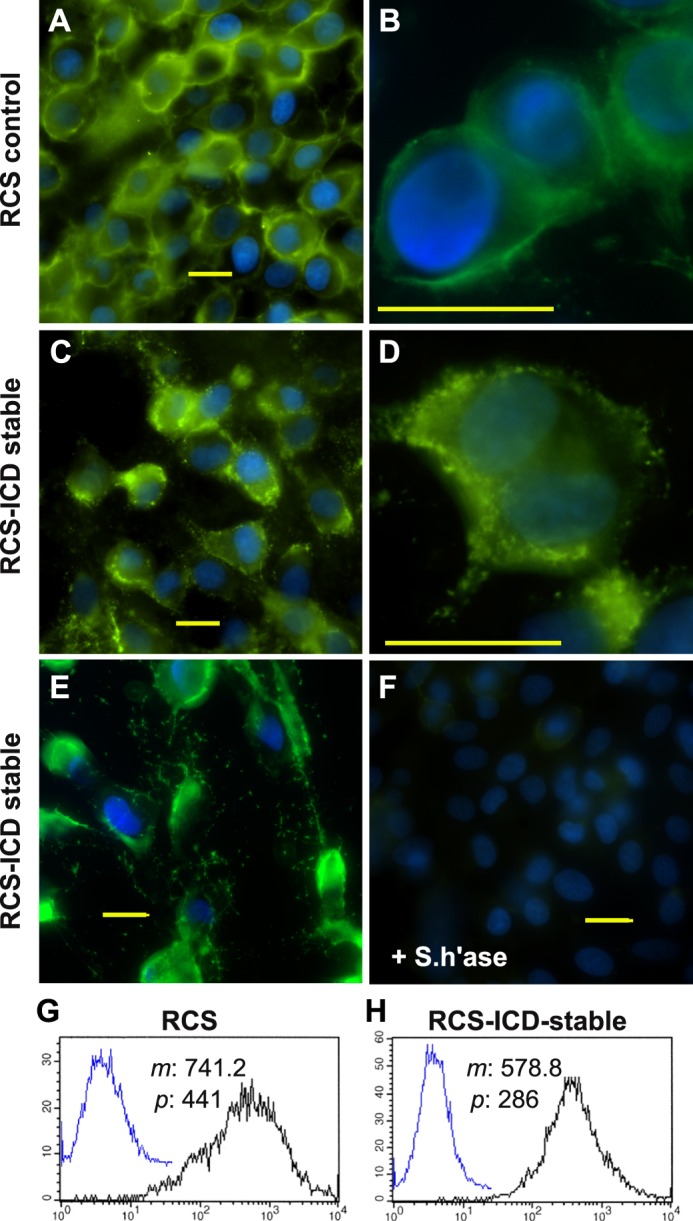
Cell surface retention of endogenous and exogenous hyaluronan on RCS cells. Control, parental RCS cells (panels A and B) were compared with CD44-ICD stable transfectants of RCS (RCS-ICD stable, panels C–E) for retention of hyaluronan at the cell surface. Cells grown on coverslips were untreated (panels A–E) or pretreated with 5 units/ml Streptomyces hyaluronidase (+S.h'ase) for 3 h at 37 °C (panel F). The washed cells were then fixed and stained for hyaluronan using biotinylated hyaluronan-binding protein followed by neutravidin-FITC, mounted in medium containing DAPI, and visualized by fluorescence microscopy. Shown are digital overlay images of green (hyaluronan) and blue (nucleus) fluorescence channels. Bars indicate 20 μm. Flow cytometric analysis was used to compare the binding of exogenous hyaluronan to control RCS cells (panel G) and CD44-ICD stable transfectants of RCS (panel H). High density monolayers of each cell type were pretreated with testicular hyaluronidase to remove endogenous matrix. The cells were next brought into suspension using non-enzymatic dissociation solution and then washed. One group of washed cells was left untreated to serve as controls (panels G and H, overlay blue line tracing). Another group of cells was incubated with 60 μg/ml fluorescein-conjugated hyaluronan (panels G and H, black line). Cell surface immunofluorescence was quantified using a FACScanTM cytometer; mean channel fluorescence (m) and peak channel fluorescence (p) values are included. Cells preincubated with unlabeled hyaluronan before the addition of fluorescein-conjugated hyaluronan exhibited substantially reduced mean channel fluorescence (data not shown).
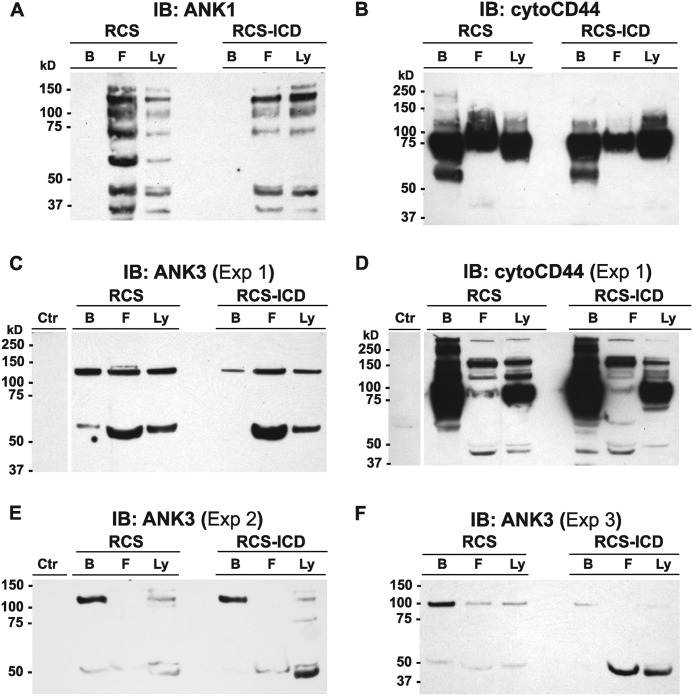
Chondrocyte CD44 Interacts with Ankyrin-3 Adaptor Proteins
One family of actin adaptor proteins that has been shown to link CD44 to actin are members of the ankyrin family. Ankyrin-1 proteins were detected in lysates of RCS and RCS-CD44-ICD cells as a series of bands ranging from 25 to 150 kDa in size (Fig. 6A, lanes Ly). Upon immunoprecipitation of CD44 from these lysates using a polyclonal anti-cytotail CD44 antibody followed by retention on protein-G magnetic microbeads, all of the ankyrin-1 proteins observed in the whole lysates were recovered in the flow-through fractions (Fig. 6A, lanes F). In this set of experiments re-blots for CD44 detected CD44 in both the bound (lanes B) and flow-through fractions (Fig. 6B). Ankryin-3 proteins were observed as 65- and 130-kDa proteins in total lysates (Fig. 6C). Upon immunoprecipitation of CD44 from these lysates of RCS or RCS-CD44-ICD cells, both 65- and 130-kDa ankyrin-3 proteins were recovered in the flow-through fractions (Fig. 6C). However, ankyrin-3 proteins were also observed in the bound fractions, indicating co-immunoprecipitation with CD44 (Fig. 6C). Moreover, the RCS-CD44-ICD cells exhibited less 130-kDa and no 65-kDa ankyrin-3 in the CD44-retained bound fraction as compared with the parental RCS cells even though both RCS and RCS-CD44-ICD cells exhibited equivalent levels of CD44FL retained on the protein-G microbeads (Fig. 6D). These experiments were repeated several times. In each separate experiment, the percent distribution of the 130- and 65-kDa ankyrin-3 proteins in the bound and flow-through fractions varied. Nonetheless, the RCS-CD44-ICD cells consistently exhibited lower levels of 130-kDa and no 65-kDa ankyrin-3 protein co-immunoprecipitation with CD44 as compared with parental, control RCS cells grown and harvested under identical conditions (Fig. 6, E and F). These results suggest that the CD44-ICD domain alters the interaction between CD44FL and the actin adaptor protein, ankyrin-3. As a preliminary confirmation of this result, Arg313 and Lys314 residues within the putative ankyrin binding domain (31) within the CD44-ICD were mutated to Ile313 and Asn314. Transient transfection of RCS chondrocytes with CD44-ICD-R313I,K314N no longer affected a loss of pericellular matrix (Fig. 3H); rather, matrix retention was similar to control transfected cells (Fig. 3G). The effects on matrix loss or matrix retention were quantified in replicates of this experiment by analysis of the diameter of the coat/cell ratio with greater than 1.15 being considered positive (Fig. 3H, inset). Of 265 cells total, 17% were successfully transfected with CD44-ICD (as in Fig. 3, C and D), and of these 17%, only ⅓, continued to exhibit detectable coats (6% of the total cells). However, in an analysis of another 200 cells, 24% were successfully transfected with the CD44-ICD-R313I,K314N (CD44-ICD ANKmut), and of these 24%, 71% continued to exhibit positive expression of coats (17% of total cells).
FIGURE 6.
Effect of CD44-ICD on the interaction of chondrocyte full-length CD44 with ankyrin adaptor proteins. High density monolayers of RCS or CD44-ICD stable transfectants of RCS (RCS-ICD) chondrocytes were detergent-extracted, and lysates were analyzed by Western blotting directly (lanes labeled Ly) or immunoprecipitated using the anti-CD44-cytotail antibody. Antibody-antigen complexes were captured on protein-G magnetic microbeads, and non-bound flow-through fractions were collected (lanes labeled F) followed by the elution of bound proteins (lanes labeled B). Equivalent volume aliquots of all fractions were processed for Western blot analysis with immunoblotting (IB) performed using HRP-conjugated anti-ankyrin-1 (ANK1; panel A) or anti-ankyrin-3 antibodies (ANK3; panel C, E, and F). After detection, blots were re-probed using HRP-conjugated anti-CD44-cytotail antibody (panels B and D). As a control, the protein-G microbeads were incubated with the anti-CD44-cytotail antibody but without the addition of a cell lysate. The eluted fraction from this control condition (labeled Ctr in panels C and D) was then immunoblotted using HRP-conjugated anti-ANK3 or anti-CD44-cytotail antibodies.
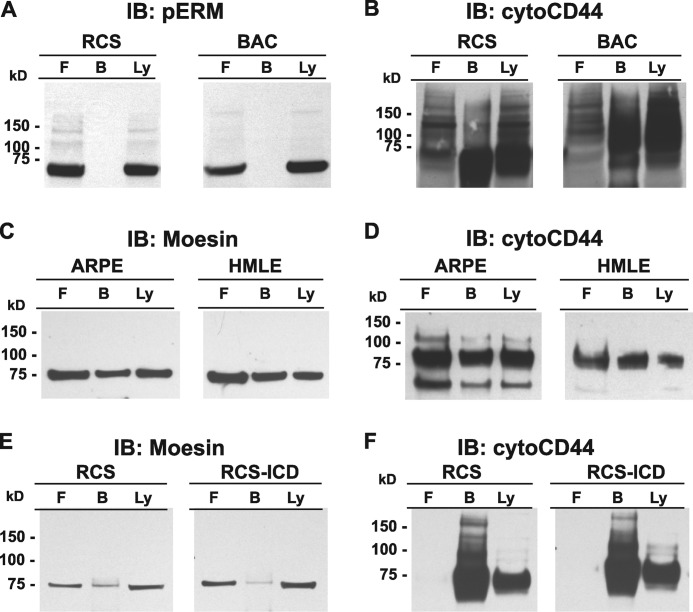
Chondrocyte CD44 Interacts Poorly with ERM Adaptor Proteins
Several studies suggest that the intracellular domain of CD44 is tethered to the actin cytoskeleton by way of adaptor proteins ezrin, radixin and moesin (ERM) or more specifically, phosphorylated ERM (pERM) (49). To determine whether CD44-ICD interferes with CD44FL binding to pERM, additional co-immunoprecipitation studies were performed. Analyses of lysates of primary bovine chondrocytes as well as parental RCS cells clearly show the accumulation of pERM (Fig. 7A). However, after immunoprecipitation, no pERM was recovered in the bound fraction of the column of either cell type (Fig. 7A, lanes B); instead, all of the pERM was recovered in the flow-through fraction (Fig. 7A, lanes F). Nonetheless, when the same blots were stripped and re-probed for CD44FL, the majority of the CD44 was recovered in the bound fraction as expected (Fig. 7B).
FIGURE 7.
Interaction of chondrocyte full-length CD44 with Ezrin, Radixin, Moesin adaptor proteins. High density monolayers of RCS or bovine articular (BAC) chondrocytes, ARPE-19 (ARPE), and HMLE human epithelial cells were detergent-extracted using 10 mm Tris, pH 7.5, with 2 mm EDTA, 1% Triton X-100, and protease and phosphatase inhibitors. The lysates were then either analyzed by Western blotting directly (lanes labeled Ly) or immunoprecipitated using the CD44 anti-cytotail antibody (panels A and B), anti-human CD44 (panels C and D), or OX49 anti-rat CD44 (panels E and F). Antibody-antigen complexes were captured on protein-G magnetic microbeads, and non-bound flow-through fractions were collected (lanes labeled F) followed by the elution of bound proteins (lanes labeled B). Equivalent volume aliquots of all fractions were processed for Western blot analysis with immunoblotting (IB) performed using HRP-conjugated anti-phospho-ERM (pERM) (panel A) or anti-moesin followed by anti-rabbit-HRP secondary antibody (panels C and E). After detection, blots were reprobed using HRP-conjugated anti-CD44-cytotail antibody (right panels of B, D, and F).
These experiments were repeated using different detergent concentrations and compositions of the lysis buffer. Varying concentrations of Triton-X100, CHAPS, or Nonidet P-40 were used for solubilization of RCS cells or bovine chondrocytes, and still no pERM or total ERM was recovered in the bound fraction; instead, all the pERM was recovered in the flow-through fractions (data not shown). Reverse approaches such as using anti-ERM, anti-moesin, or pERM antibodies for immunoprecipitation and immunoblotting with anti-CD44 also failed to detect co-immunoprecipitation of CD44 and ERM proteins. Various treatments before detergent solubilization of the cells were also investigated including (i) removing the hyaluronan-rich pericellular matrices with Streptomyces or testicular hyaluronidase or (ii) passage of cells in culture at low density to allow the chondrocytes to “de-differentiate” and take on a more fibroblastic-like morphology with prominent actin cables or bundles (3). These treatments did not promote the detectable interaction between CD44 and ERM proteins (data not shown).
To determine whether the failure to observe CD44/ERM complexes in chondrocytes was a technical issue, CD44 was immunoprecipitated from lysates of HMLE (41) or human retinal pigmented epithelial cells (ARPE-19) (40) after the cells were induced to undergo an epithelial-mesenchymal transition, a transition known to promote CD44/ERM interactions (40). The immunoprecipitation protocol outlined by Takahashi et al. (40) was utilized including using a rat anti-human CD44 antibody reacting with the extracellular of CD44 for immunoprecipitation and anti-moesin for immunoblotting. As shown in Fig. 7C, a substantial proportion of the moesin present in the lysates of both cell lines was now detected as retained in the bound fraction. The proportion of moesin in the bound fraction was also roughly equivalent to the proportion of CD44 retained on the microbeads, as shown when the membranes were reprobed for CD44 (detected using the anti-cytotail CD44) (Fig. 7D). Next, RCS chondrocyte lysates were immunoprecipitated with a mouse anti-rat CD44 (extracellular domain) antibody (OX49) and immunoblotted with anti-moesin. As shown in Fig. 7E, a faint band for moesin was now detected in the bound fraction. Moreover, less bound moesin was found in extracts from the RCS-CD44-ICD stable cells than from the parental RCS cells, although this is difficult to conclusively quantify at this low level of detection. The faint detection of bound moesin was striking considering that the majority of the chondrocyte CD44 was retained by the OX49 microbeads as seen upon reprobing the nitrocellulose blots with anti-CD44 antibodies (Fig. 7F). In comparison to the human epithelial cells (Fig. 7, C and D), it would appear that in chondrocytes there is a smaller pool of CD44 interacting with ERM proteins.
DISCUSSION
Proteolytic cleavage of CD44 occurs in a variety of tissues and results in the shedding of the CD44 ectodomain into the extracellular space and release of a free intracellular fragment (CD44-ICD) into the cytoplasm. Enhanced cleavage of CD44 is most often associated with tumor invasion and metastasis (8, 37, 50–57). Recently, however, we demonstrated that CD44 fragmentation was also a prominent feature exhibited by the chondrocytes of osteoarthritic cartilage (3). Interestingly, the steady-state level of CD44-ICD present in some osteoarthritic chondrocyte lysates is similar to the levels observed due to overexpression of CD44-ICD used in this study (Fig. 4C). Although CD44 fragmentation is known to occur, little is understood about the effects of this event on cell physiology. As for the effects due to the CD44-ICD, three explanations appeared possible. The CD44-ICD could be an inert peptide destined for degradation. The CD44-ICD could translocate to the nucleus and function as a transcription factor like Notch-ICD as had been suggested in early studies (37, 56). Last, CD44-ICD could remain within the cytoplasm and interfere with complex formation between CD44FL and assorted binding partners; that is, partners required to maintain normal cell functions dependent on CD44.
One of the clearest functions of CD44FL in chondrocytes is the anchoring of hyaluronan and aggrecan proteoglycan to the chondrocyte cell surface (44). The addition of agents known to induce cleavage of endogenous CD44FL, namely IL-1β, hyaluronan oligosaccharides, and PMA, resulted in the loss of hyaluronan-dependent pericellular matrices on bovine articular chondrocytes (Fig. 1). The fact that all three conditions induce proteolytic cleavage CD44FL is not surprising. Saya and co-workers suggested that proteolytic cleavage is regulated by several signal pathways, including the extracellular influx of Ca2+ followed by activation of protein kinase C, Ras oncoprotein, and Rho family GTPases (58, 59). Multiple events certainly occur in these IL-1β- or PMA-treated chondrocytes that might explain the loss of pericellular matrices. However, given that the overexpression of CD44-ICD alone also affected a loss of pericellular matrices (Fig. 1, G and H) suggests that the generation of this peptide, such as through cytokine stimulation, may have the potential to interfere with CD44FL-dependent function.
A study by Duterme et al. (49) provides support for our hypothesis that the CD44-ICD might act as a dominant negative and interfere with critical intracellular complex formation events of CD44FL. They demonstrated that overexpression of hyaluronidase-2 in v-src-transformed rat BB16 fibroblasts altered the function of CD44FL including the loss of pericellular matrices and reduced capacity of these cells to bind exogenous fluorescein-conjugated hyaluronan. Moreover, they further demonstrated that hyaluronidase-2 overexpression induced the proteolytic cleavage of CD44FL and that the loss of function caused by hyaluronidase-2 overexpression was due to a loss of CD44FL interaction with pERM actin adaptor proteins. Interestingly, the authors suggested that hyaluronidase-2 overexpression mimicked the effects of hyaluronan oligosaccharides. In this study, the displacement of pericellular matrix via hyaluronan oligosaccharides (outside-in effects) mimicked the effects of IL-1β, PMA, and overexpression of CD44-ICD (Fig. 2, inside-out effects).
CD44FL tethering to actin filaments is required for matrix retention, even in chondrocytes with a more delicate cortical actin cytoskeleton. We demonstrated this previously using cytochalasin and latrunculin disruption of actin filaments (32). Moreover, direct disruption of the actin cytoskeleton by cytochalasin or latrunculin resulted in more CD44 that could be extracted from chondrocytes under lower detergent conditions similar to conditions used in this study in Fig. 2 (32). Thus, when IL-1β, hyaluronan oligosaccharides, PMA, and CD44-ICD overexpression all affected enhanced CD44FL extraction in a lower detergent (Fig. 2), these results provided a clue that the mechanism of action was linked to changes in the interaction between CD44FL and the actin cytoskeleton. However, the exact mechanism for why and how there is reduced CD44-based retention of matrix when CD44/cytoskeleton linkages are broken remains to be determined. One start is to identify the principal actin adaptor proteins that might be involved.
Several studies suggest that the cytoplasmic domain of CD44FL couples to the actin cytoskeleton by way of adaptor proteins of the ezrin/radixin/moesin or ERM family (29, 30). A crystal structure analysis of the FERM/CD44 complex revealed that the CD44/ERM binding region is contained in a cluster of basic amino acids, Lys-Lys-Lys, followed by a nonpolar region that binds to the groove formed between helix α1C and the β-strand 5 of subdomain C (β5C) of the FERM domain (60). A functional analysis of the ezrin binding site in CD44 found that deletion of residues 289–295 or 296–300 completely abolished association with ezrin but that phosphorylation of CD44 at Ser325 does not regulate ezrin binding. Interestingly, the CD44 ERM binding domain mutants did not affect hyaluronan binding, suggesting that in these cells hyaluronan binding to CD44 was not dependent upon CD44 binding to these adaptor proteins (61). Given the work of Duterme et al. (49), a change in the interaction between CD44FL and pERM was expected after the overexpression of CD44-ICD. However, co-immunoprecipitation of CD44 and pERM could not be demonstrated in lysates from bovine or rat chondrocytes. Co-immunoprecipitations are known to be sensitive to a variety of conditions, especially the strength of the lysis detergent. However, no interactions between CD44 and pERM could be observed using the same lysis buffer as Duterme et al. (49) or the use of alternative detergents, varying detergent concentrations, or cell growth conditions predicted to affect the cortical cytoskeleton. Thus, even under the least stringent detergent conditions, no interaction between CD44 and pERM could be demonstrated in chondrocytes. However, if pERM is a limited subfraction of the total ERM proteins and only minor levels of moesin could be detected, co-immunoprecipitating with chondrocyte CD44 (Fig. 7E), pERM proteins are likely well below our detection limits. Thus, in chondrocytes, pERM binding to CD44FL may represent a weaker interaction, interaction as part of a large, dynamic complex, or be restricted to a select subpool of CD44 (Fig. 8). Interestingly, ERM phosphorylation levels in total lysates were consistently lower in total cell lysates from CD44-ICD stable transfectants as compared with parental RCS cells (data not shown), suggesting that CD44-ICD overexpression does affect ERM phosphorylation at some level.
FIGURE 8.
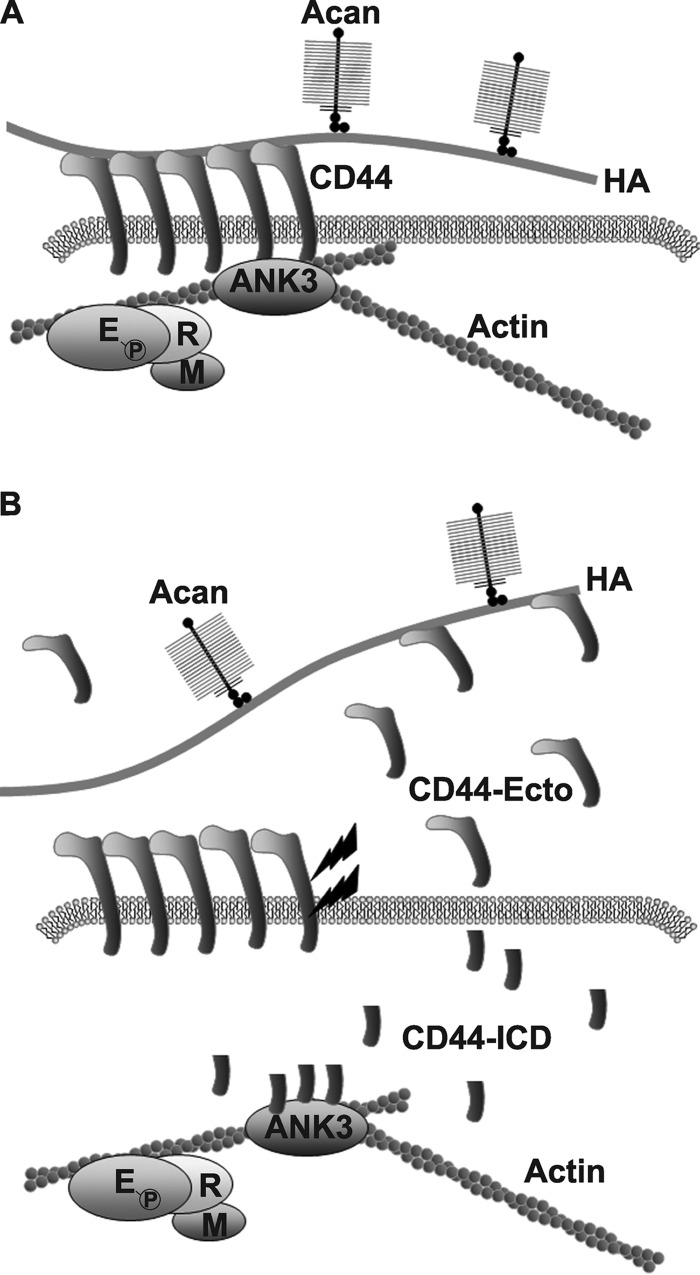
A proposed model for CD44-ICD mediated interference with the interaction of full-length CD44 with actin adaptor proteins in chondrocytes. The chondrocyte pericellular matrix is anchored to the cell surface in large part by the binding of aggrecan (Acan)/link protein/hyaluronan (HA) complexes to CD44 (panel A). CD44 in turn is anchored to the intracellular cortical actin cytoskeleton via interaction with the adaptor protein, ankyrin 3 (ANK3). Members of the ERM adaptor proteins are also present within the complex. When CD44 undergoes proteolytic cleavage (lightning bolts), fragmented products are released including a remnant CD44 extracellular domain (CD44-Ecto) and an intracellular CD44 domain (CD44-ICD) (panel B). Whether by natural cleavage or overexpression of a recombinant transgene, CD44-ICD competes with the binding of full-length CD44 with ANK3. This interference by CD44-ICD results in blockage of binding of the remaining full-length CD44 (receptors that would be otherwise functional) with the underlying actin cytoskeleton. Under these conditions, the capacity of CD44FL to bind extracellular HA is reduced, and the CD44 is more readily solubilized from the membranes with lower Igepal detergent conditions. As such, the CD44-ICD could be considered a dominant-negative agent, producing an inside-out effect on coat retention.
Other studies have suggested that CD44 links to the actin cytoskeleton via interactions with members of the ankyrin family of adaptor proteins (28, 35, 62, 63). Moreover, some of these reports have demonstrated that the binding affinity between CD44 and ankyrin is much higher than that of CD44 and ERM (28, 64) and that CD44/ERM interactions require an activation of the Rho or protein kinase C pathways (65, 66). Zhu and Bourguignon (62) documented that CD44 and ankyrin-1 co-immunoprecipitated with CD44 in SKOV3 human ovarian carcinoma cells. However in chondrocytes, we observed no ankyrin-1 co-immunoprecipitation with CD44FL in the cell lysates of parental RCS or stable RCS-CD44-ICD transfectants (Fig. 6A). Nonetheless, the ankyrin family includes three homologous genes (ANK1, ANK2, ANK3) with each gene producing different isoforms through alternative splicing. Ankyrin-3 is highly expressed in many tissues (67) but to our knowledge has never been examined for expression in cartilage or chondrocytes. Two isoforms of ankyrin-3 (65 and 130 kDa) were present in direct lysates of chondrocytes (Fig. 6, C, E, and F). In parental RCS cell lysates, both ankyrin-3 isoforms were detected in both the flow-through fraction and the fraction of proteins that co-immunoprecipitate with CD44FL (Fig. 6, C, E, and F). However, the 130-kDa isoform appeared as the predominant species in the bound fraction and the 65-kDa predominant in the non-bound fraction. More importantly, in lysates of the RCS-CD44-ICD stable transfectants, less 130-kDa ankyrin-3 was observed in the CD44-immunoprecipitate fraction, and all of the 65-kDa isoform was in the flow-through fraction (Fig. 6, C, E, and F). Interestingly, we also observed ankyrin-3 isoforms in CD44FL immunoprecipitates of SKOV3 human ovarian carcinoma cell lysates (data not shown). Altogether the results suggest that CD44-ICD competes with CD44FL binding to ankyrin-3. To further demonstrate this binding, mutation of the putative ankyrin binding site in CD44-ICD was generated to limit the capacity of this domain to compete for ankyrin-3. Initial experiments confirm this hypothesis, as the transient transfection of CD44-ICD-R313I,K314N (CD44-ICD ankyrin mutant; Fig. 3H) no longer results in the loss of pericellular matrices in RCS cells. This suggests that the actin-ankyrin-3-CD44-hyaluronan axis is important in chondrocyte pericellular matrix retention (Fig. 8A) and that CD44-ICD can disrupt this linkage (Fig. 8B).
We have proposed that release of the CD44-ICD into the cytoplasm of cells such as chondrocytes would exert a competitive or dominant-negative effect on the CD44FL function (Fig. 8B). This study has provided support for this hypothesis as there is little change in the overall content of CD44FL during conditions in which CD44-ICD is overexpressed. Thus, there should be sufficient, non-degraded CD44FL available to bind hyaluronan (Fig. 8A), but in the presence of CD44-ICD, the CD44-FL does not support proper pericellular matrix or fluorescein-conjugated hyaluronan retention (Fig. 8B). Also, in CD44-ICD containing cells, the CD44FL is no longer strongly anchored to the cell cytoskeleton and exhibits lower binding to both ankyrin-3 isoforms. The clinical significance of these data relies on the fact that several pathological conditions result in the proteolytic cleavage of CD44FL. In preosteoarthritic chondrocytes an initial signal or trauma could result in an initial disruption of pericellular matrix in cartilage, triggering enhanced production of proteases and, as a consequence, enhanced intracellular accumulation of CD44-ICD as we have observed in chondrocytes of human osteoarthritis patients (3). In turn, the presence of CD44-ICD would limit the capacity of newly synthesized CD44FL to participate in matrix repair and retention of new pericellular matrix. A delay in reforming a pericellular matrix would induce further protease synthesis, thus perpetuating matrix loss. Osteoarthritis is often considered a result of attempted-but-ultimately failed repair. As such, CD44-ICD formation could represent a contributing factor in the failed repair, perpetuating a downward spiral that results in progressive osteoarthritis.
Acknowledgments
We thank Christy Holland, Nadege Etienne, and Kayla Britton for technical expertise and assistance with this project.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AR43384 (to W. K.) and R01-AR39507 (to C. B. K.) and RO1-AR043384–15S1 (NIAMS; Research Supplemental to Promote Diversity in Health-related Research; to L. M.).
- CD44-EXT
- CD44 domain remaining after extracellular proteolysis
- CD44-ICD
- CD44 intracellular domain
- ARPE-19
- spontaneously arising retinal pigment epithelial cells
- CD44FL
- full-length CD44
- ERM
- ezrin/radixin/moesin
- pERM
- phosphorylated ERM
- EGFP
- enhanced GFP
- FRT
- flip recombination target
- HMLE
- human mammary epithelial cells
- PMA
- phorbol myristate acetate
- RCS
- rat chondrosarcoma chondrocyte
- RCS-ICD
- CD44-ICD stable transfectants of RCS
- HMLE
- human mammary epithelial.
REFERENCES
- 1. Knudson C. B., Knudson W. (1993) Hyaluronan-binding proteins in development, tissue homeostasis, and disease. FASEB J. 7, 1233–1241 [PubMed] [Google Scholar]
- 2. Knudson W., Loeser R. F. (2002) CD44 and integrin matrix receptors participate in cartilage homeostasis. Cell. Mol. Life Sci. 59, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takahashi N., Knudson C. B., Thankamony S., Ariyoshi W., Mellor L., Im H. J., Knudson W. (2010) Induction of CD44 cleavage in articular chondrocytes. Arthritis Rheum. 62, 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nagano O., Murakami D., Hartmann D., De Strooper B., Saftig P., Iwatsubo T., Nakajima M., Shinohara M., Saya H. (2004) Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca2+ influx and PKC activation. J. Cell Biol. 165, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakamura H., Suenaga N., Taniwaki K., Matsuki H., Yonezawa K., Fujii M., Okada Y., Seiki M. (2004) Constitutive and induced CD44 shedding by ADAM-like proteases and membrane-type 1 matrix metalloproteinase. Cancer Res. 64, 876–882 [DOI] [PubMed] [Google Scholar]
- 6. Nagano O., Saya H. (2004) Mechanism and biological significance of CD44 cleavage. Cancer Sci. 95, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thorne R. F., Legg J. W., Isacke C. M. (2004) The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J. Cell Sci. 117, 373–380 [DOI] [PubMed] [Google Scholar]
- 8. Lammich S., Okochi M., Takeda M., Kaether C., Capell A., Zimmer A. K., Edbauer D., Walter J., Steiner H., Haass C. (2002) Presenilin-dependent intramembrane proteolysis of CD44 leads to the liberation of its intracellular domain and the secretion of an Aβ-like peptide. J. Biol. Chem. 277, 44754–44759 [DOI] [PubMed] [Google Scholar]
- 9. De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J. S., Schroeter E. H., Schrijvers V., Wolfe M. S., Ray W. J., Goate A., Kopan R. (1999) A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 398, 518–522 [DOI] [PubMed] [Google Scholar]
- 10. Ghatak S., Misra S., Toole B. P. (2002) Hyaluronan oligosaccharides inhibit anchorage-independent growth of tumor cells by suppressing the phosphoinositide 3-kinase/Akt cell survival pathway. J. Biol. Chem. 277, 38013–38020 [DOI] [PubMed] [Google Scholar]
- 11. Toole B. P. (2009) Hyaluronan-CD44 interactions in cancer. Paradoxes and possibilities. Clin. Cancer Res. 15, 7462–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohno S., Im H. J., Knudson C. B., Knudson W. (2005) Hyaluronan oligosaccharide-induced activation of transcription factors in bovine articular chondrocytes. Arthritis Rheum. 52, 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hosono K., Nishida Y., Knudson W., Knudson C. B., Naruse T., Suzuki Y., Ishiguro N. (2007) Hyaluronan oligosaccharides inhibit tumorigenicity of osteosarcoma cell lines MG-63 and LM-8 in vitro and in vivo via perturbation of hyaluronan-rich pericellular matrix of the cells. Am. J. Pathol. 171, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmitz I., Ariyoshi W., Takahashi N., Knudson C. B., Knudson W. (2010) Hyaluronan oligosaccharide treatment of chondrocytes stimulates expression of both HAS-2 and MMP-3, but by different signaling pathways. Osteoarthritis Cartilage 18, 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohno S., Im H. J., Knudson C. B., Knudson W. (2006) Hyaluronan oligosaccharides induce matrix metalloproteinase 13 via transcriptional activation of NFκB and p38 MAP kinase in articular chondrocytes. J. Biol. Chem. 281, 17952–17960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fitzgerald K. A., Bowie A. G., Skeffington B. S., O'Neill L. A. (2000) Ras, protein kinase Cζ, and IκB kinases 1 and 2 are downstream effectors of CD44 during the activation of NF-κB by hyaluronic acid fragments in T-24 carcinoma cells. J. Immunol. 164, 2053–2063 [DOI] [PubMed] [Google Scholar]
- 17. Sohara Y., Ishiguro N., Machida K., Kurata H., Thant A. A., Senga T., Matsuda S., Kimata K., Iwata H., Hamaguchi M. (2001) Hyaluronan activates cell motility of v-Src-transformed cells via Ras-mitogen-activated protein kinase and phosphoinositide 3-kinase-Akt in a tumor-specific manner. Mol. Biol. Cell 12, 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turley E. A., Noble P. W., Bourguignon L. Y. (2002) Signaling properties of hyaluronan receptors. J. Biol. Chem. 277, 4589–4592 [DOI] [PubMed] [Google Scholar]
- 19. Yu W. H., Woessner J. F., Jr., McNeish J. D., Stamenkovic I. (2002) CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev. 16, 307–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bourguignon L. Y., Singleton P. A., Zhu H., Zhou B. (2002) Hyaluronan promotes signaling interaction between CD44 and the transforming growth factor β receptor I in metastatic breast tumor cells. J. Biol. Chem. 277, 39703–39712 [DOI] [PubMed] [Google Scholar]
- 21. Wobus M., Rangwala R., Sheyn I., Hennigan R., Coila B., Lower E. E., Yassin R. S., Sherman L. S. (2002) CD44 associates with EGFR and erbB2 in metastasizing mammary carcinoma cells. Appl. Immunohistochem. Mol. Morphol. 10, 34–39 [DOI] [PubMed] [Google Scholar]
- 22. Wakahara K., Kobayashi H., Yagyu T., Matsuzaki H., Kondo T., Kurita N., Sekino H., Inagaki K., Suzuki M., Kanayama N., Terao T. (2005) Bikunin down-regulates heterodimerization between CD44 and growth factor receptors and subsequently suppresses agonist-mediated signaling. J. Cell. Biochem. 94, 995–1009 [DOI] [PubMed] [Google Scholar]
- 23. Misra S., Toole B. P., Ghatak S. (2006) Hyaluronan constitutively regulates activation of multiple receptor tyrosine kinases in epithelial and carcinoma cells. J. Biol. Chem. 281, 34936–34941 [DOI] [PubMed] [Google Scholar]
- 24. Pályi-Krekk Z., Barok M., Kovács T., Saya H., Nagano O., Szöllosi J., Nagy P. (2008) EGFR and ErbB2 are functionally coupled to CD44 and regulate shedding, internalization and motogenic effect of CD44. Cancer Lett. 263, 231–242 [DOI] [PubMed] [Google Scholar]
- 25. Meran S., Luo D. D., Simpson R., Martin J., Wells A., Steadman R., Phillips A. O. (2011) Hyaluronan facilitates transforming growth factor-β1-dependent proliferation via CD44 and epidermal growth factor receptor interaction. J. Biol. Chem. 286, 17618–17630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peterson R. S., Andhare R. A., Rousche K. T., Knudson W., Wang W., Grossfield J. B., Thomas R. O., Hollingsworth R. E., Knudson C. B. (2004) CD44 modulates Smad1 activation in the BMP-7 signaling pathway. J. Cell Biol. 166, 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andhare R. A., Takahashi N., Knudson W., Knudson C. B. (2009) Hyaluronan promotes the chondrocyte response to BMP-7. Osteoarthritis Cartilage 17, 906–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lokeshwar V. B., Fregien N., Bourguignon L. Y. (1994) Ankyrin-binding domain of CD44(GP85) is required for the expression of hyaluronic acid-mediated adhesion function. J. Cell Biol. 126, 1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsukita S., Oishi K., Sato N., Sagara J., Kawai A. (1994) ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J. Cell Biol. 126, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morrison H., Sherman L. S., Legg J., Banine F., Isacke C., Haipek C. A., Gutmann D. H., Ponta H., Herrlich P. (2001) The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 15, 968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marhaba R., Zöller M. (2004) CD44 in cancer progression. Adhesion, migration, and growth regulation. J. Mol. Histol. 35, 211–231 [DOI] [PubMed] [Google Scholar]
- 32. Nofal G. A., Knudson C. B. (2002) Latrunculin and cytochalasin decrease chondrocyte matrix retention. J. Histochem. Cytochem. 50, 1313–1324 [DOI] [PubMed] [Google Scholar]
- 33. Lesley J., He Q., Miyake K., Hamann A., Hyman R., Kincade P. W. (1992) Requirements for hyaluronic acid binding by CD44. A role for the cytoplasmic domain and activation by antibody. J. Exp. Med. 175, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu D., Zhang D., Mori H., Sy M.-S. (1996) Binding of CD44 to hyaluronic acid can be induced by multiple signals and requires the CD44 cytoplasmic domain. Cell. Immunol. 174, 73–83 [DOI] [PubMed] [Google Scholar]
- 35. Singleton P. A., Bourguignon L. Y. (2004) CD44 interaction with ankyrin and IP3 receptor in lipid rafts promotes hyaluronan-mediated Ca2+ signaling leading to nitric oxide production and endothelial cell adhesion and proliferation. Exp. Cell Res. 295, 102–118 [DOI] [PubMed] [Google Scholar]
- 36. Knudson W., Peterson R. S. (2004) in Chemistry and Biology of Hyaluronan (Garg H. G., Hales C. A., eds) pp. 83–123, Elsevier, Boston [Google Scholar]
- 37. Okamoto I., Kawano Y., Murakami D., Sasayama T., Araki N., Miki T., Wong A. J., Saya H. (2001) Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J. Cell Biol. 155, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi H. U., Meyer K., Swarm R. (1971) Mucopolysaccharide and protein. Polysaccharide of a transplantable rat chondrosarcoma. Proc. Natl. Acad. Sci. U.S.A. 68, 877–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kucharska A. M., Kuettner K. E., Kimura J. H. (1990) Biochemical characterization of long-term culture of the Swarm rat chondrosarcoma chondrocytes in agarose. J. Orthop. Res. 8, 781–792 [DOI] [PubMed] [Google Scholar]
- 40. Takahashi E., Nagano O., Ishimoto T., Yae T., Suzuki Y., Shinoda T., Nakamura S., Niwa S., Ikeda S., Koga H., Tanihara H., Saya H. (2010) Tumor necrosis factor-α regulates transforming growth factor-β-dependent epithelial-mesenchymal transition by promoting hyaluronan-CD44-moesin interaction. J. Biol. Chem. 285, 4060–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Embry J. J., Knudson W. (2003) G1 domain of aggrecan cointernalizes with hyaluronan via a CD44-mediated mechanism in bovine articular chondrocytes. Arthritis Rheum. 48, 3431–3441 [DOI] [PubMed] [Google Scholar]
- 43. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knudson C. B. (1993) Hyaluronan receptor-directed assembly of chondrocyte pericellular matrix. J. Cell Biol. 120, 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Knudson W., Aguiar D. J., Hua Q., Knudson C. B. (1996) CD44-anchored hyaluronan-rich pericellular matrices. An ultrastructural and biochemical analysis. Exp. Cell Res. 228, 216–228 [DOI] [PubMed] [Google Scholar]
- 46. Knudson W., Bartnik E., Knudson C. B. (1993) Assembly of pericellular matrices by COS-7 cells transfected with CD44 lymphocyte-homing receptor genes. Proc. Natl. Acad. Sci. U.S.A. 90, 4003–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hua Q., Knudson C. B., Knudson W. (1993) Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J. Cell Sci. 106, 365–375 [DOI] [PubMed] [Google Scholar]
- 48. Rilla K., Tiihonen R., Kultti A., Tammi M., Tammi R. (2008) Pericellular hyaluronan coat visualized in live cells with a fluorescent probe is scaffolded by plasma membrane protrusions. J Histochem Cytochem. 56, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Duterme C., Mertens-Strijthagen J., Tammi M., Flamion B. (2009) Two novel functions of hyaluronidase-2 (Hyal2) are formation of the glycocalyx and control of CD44-ERM interactions. J. Biol. Chem. 284, 33495–33508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cichy J., Bals R., Potempa J., Mani A., Puré E. (2002) Proteinase-mediated release of epithelial cell-associated CD44. Extracellular CD44 complexes with components of cellular matrices. J. Biol. Chem. 277, 44440–44447 [DOI] [PubMed] [Google Scholar]
- 51. Guo Y. J., Liu G., Wang X., Jin D., Wu M., Ma J., Sy M. S. (1994) Potential use of soluble CD44 in serum as indicator of tumor burden and metastasis in patients with gastric or colon cancer. Cancer Res. 54, 422–426 [PubMed] [Google Scholar]
- 52. Masson D., Denis M. G., Denis M., Blanchard D., Loirat M. J., Cassagnau E., Lustenberger P. (1999) Soluble CD44. Quantification and molecular repartition in plasma of patients with colorectal cancer. Br. J. Cancer. 80, 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamane N., Tsujitani S., Makino M., Maeta M., Kaibara N. (1999) Soluble CD44 variant 6 as a prognostic indicator in patients with colorectal cancer. Oncology 56, 232–238 [DOI] [PubMed] [Google Scholar]
- 54. Okamoto I., Kawano Y., Matsumoto M., Suga M., Kaibuchi K., Ando M., Saya H. (1999) Regulated CD44 cleavage under the control of protein kinase C, calcium influx, and the Rho family of small G proteins. J. Biol. Chem. 274, 25525–25534 [DOI] [PubMed] [Google Scholar]
- 55. Pelletier L., Guillaumot P., Frêche B., Luquain C., Christiansen D., Brugière S., Garin J., Manié S. N. (2006) γ-Secretase-dependent proteolysis of CD44 promotes neoplastic transformation of rat fibroblastic cells. Cancer Res. 66, 3681–3687 [DOI] [PubMed] [Google Scholar]
- 56. Murakami D., Okamoto I., Nagano O., Kawano Y., Tomita T., Iwatsubo T., De Strooper B., Yumoto E., Saya H. (2003) Presenilin-dependent γ-secretase activity mediates the intramembranous cleavage of CD44. Oncogene 22, 1511–1516 [DOI] [PubMed] [Google Scholar]
- 57. Cichy J., Puré E. (2003) The liberation of CD44. J. Cell Biol. 161, 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Okamoto I., Kawano Y., Tsuiki H., Sasaki J., Nakao M., Matsumoto M., Suga M., Ando M., Nakajima M., Saya H. (1999) CD44 cleavage induced by a membrane-associated metalloprotease plays a critical role in tumor cell migration. Oncogene. 18, 1435–1446 [DOI] [PubMed] [Google Scholar]
- 59. Kawano Y., Okamoto I., Murakami D., Itoh H., Yoshida M., Ueda S., Saya H. (2000) Ras oncoprotein induces CD44 cleavage through phosphoinositide 3-OH kinase and the rho family of small G proteins. J. Biol. Chem. 275, 29628–29635 [DOI] [PubMed] [Google Scholar]
- 60. Mori T., Kitano K., Terawaki S., Maesaki R., Fukami Y., Hakoshima T. (2008) Structural basis for CD44 recognition by ERM proteins. J. Biol. Chem. 283, 29602–29612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Legg J. W., Isacke C. M. (1998) Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr. Biol. 8, 705–708 [DOI] [PubMed] [Google Scholar]
- 62. Zhu D., Bourguignon L. Y. (2000) Interaction between CD44 and the repeat domain of ankyrin promotes hyaluronic acid-mediated ovarian tumor cell migration. J. Cell. Physiol. 183, 182–195 [DOI] [PubMed] [Google Scholar]
- 63. Bourguignon L. Y. W., Zhu D., Zhu H. (1998) CD44 isoform-cytoskeleton interaction in oncogenic signaling and tumor progression. Front. Biosci. 3, d637–d649 [DOI] [PubMed] [Google Scholar]
- 64. Tsukita S., Yonemura S. (1997) ERM (ezrin/radixin/moesin) family. From cytoskeleton to signal transduction. Curr. Opin. Cell Biol. 9, 70–75 [DOI] [PubMed] [Google Scholar]
- 65. Hirao M., Sato N., Kondo T., Yonemura S., Monden M., Sasaki T., Takai Y., Tsukita S., Tsukita S. (1996) Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association. Possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol. 135, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S. (1998) Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol. 140, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Peters L. L., John K. M., Lu F. M., Eicher E. M., Higgins A., Yialamas M., Turtzo L. C., Otsuka A. J., Lux S. E. (1995) Ank3 (epithelial ankyrin), a widely distributed new member of the ankyrin gene family and the major ankyrin in kidney, is expressed in alternatively spliced forms, including forms that lack the repeat domain. J. Cell Biol. 130, 313–330 [DOI] [PMC free article] [PubMed] [Google Scholar]