Background: MurM utilizes aminoacyl-tRNAs and Lipid II for peptidoglycan biosynthesis in Streptococcus pneumoniae.
Results: MurM deacylates mischarged aminoacyl-tRNAs in the absence of Lipid II.
Conclusion: The ability of MurM to function in quality control can compensate for the absence of AlaXp proteins in S. pneumoniae.
Significance: MurM can function in translation as a lipid-independent trans editing factor.
Keywords: Aminoacyl-tRNA Synthesis, Aminoacyl-tRNA Synthetase, Peptidoglycan, Transfer RNA (tRNA), Translation
Abstract
Streptococcus pneumoniae is a causative agent of nosocomial infections such as pneumonia, meningitis, and septicemia. Penicillin resistance in S. pneumoniae depends in part upon MurM, an aminoacyl-tRNA ligase that attaches l-serine or l-alanine to the stem peptide lysine of Lipid II in cell wall peptidoglycan. To investigate the exact substrates the translation machinery provides MurM, quality control by alanyl-tRNA synthetase (AlaRS) was investigated. AlaRS mischarged serine and glycine to tRNAAla, as observed in other bacteria, and also transferred alanine, serine, and glycine to tRNAPhe. S. pneumoniae tRNAPhe has an unusual U4:C69 mismatch in its acceptor stem that prevents editing by phenylalanyl-tRNA synthetase (PheRS), leading to the accumulation of misaminoacylated tRNAs that could serve as substrates for translation or for MurM. Although the peptidoglycan layer of S. pneumoniae tolerates a combination of both branched and linear muropeptides, deletion of MurM results in a reversion to penicillin sensitivity in strains that were previously resistant. However, because MurM is not required for cell viability, the reason for its functional conservation across all strains of S. pneumoniae has remained elusive. We now show that MurM can directly function in translation quality control by acting as a broad specificity lipid-independent trans editing factor that deacylates tRNA. This activity of MurM does not require the presence of its second substrate, Lipid II, and can functionally substitute for the activity of widely conserved editing domain homologues of AlaRS, termed AlaXPs proteins, which are themselves absent from S. pneumoniae.
Introduction
Streptococcus pneumoniae is a Gram-positive diplococcus that is carried asymptomatically in the nasopharynx of 5–10% of healthy adults and 20–40% of healthy children. Clinically, S. pneumoniae is the common causative agent of several community and hospital acquired infections including pneumonia, otitis media, meningitis, and septicemia. According to the Centers for Disease Control (CDC), ∼5 million fatal cases of pneumococcal pneumonia in children under the age of five are reported globally each year (37). Pneumococci have an unusual lifestyle because they produce high levels of hydrogen peroxide that provides a competitive advantage for the organism during colonization of the nasopharynx (1, 2). In other organisms, it has been reported that exposure to increased levels of hydrogen peroxide can enhance cellular mistranslation rates both in vivo and in vitro (3, 4). For example, in mammalian cells, ∼1% of protein synthesis-directed methionine residues are aminoacylated onto noncognate tRNA molecules. This methionine misaminoacylation is increased as much as 10-fold in the presence of reactive oxygen species, such as hydrogen peroxide. Substitution of coded amino acids with methionine is believed to protect proteins against oxidative damage under stress conditions (3). In Escherichia coli, exposure to hydrogen peroxide causes reduction in the fidelity of translation. This effect has been directly attributed to oxidation of Cys-182 within threonyl-tRNA synthetase, which subsequently impairs the editing ability of the enzyme and results in the production of misaminoacylated Ser-tRNAThr (4). How translation quality control is maintained in pneumococci, which are routinely exposed to elevated hydrogen peroxide levels, is unknown.
Aminoacyl-tRNA synthetases are the first step in quality control of protein synthesis because they are responsible for amino acid activation and transfer to cognate tRNA (5). Following this process, the aminoacyl-tRNA is released from the synthetase and bound by elongation factor Tu (EF-Tu)2 for delivery to the ribosome and use in protein synthesis (6, 7). Aminoacyl-tRNA synthetases are usually highly selective for their cognate tRNA due to the availability of a large surface area for recognition, identity elements within the tRNA molecule itself, and also kinetic proofreading during the aminoacylation reaction (8–10). In contrast, some amino acids are difficult for aminoacyl-tRNA synthetases to distinguish with high accuracy as they can differ by as little as a single methyl group (11). For example, isoleucyl-tRNA synthetase has difficulty distinguishing between the isosteric amino acids isoleucine and valine, whereas the active site of alanyl-tRNA synthetase (AlaRS) is able to accommodate alanine, glycine, and serine (12–16). Misactivation of noncognate serine and glycine by AlaRS occurs at frequencies of 1/500 and 1/250, respectively. This is higher than the overall error rate for translation, which is typically from 1/3000 to 1/10,000 (10). Amino acid activation errors can be corrected both by the synthetase itself at a distinct editing site, as is the case for AlaRS, and also by free-standing editing domain homologues, as exemplified by the widely conserved AlaXPs proteins that edit Ser-tRNAAla (17, 18). S. pneumoniae encodes no known AlaXPs, suggesting that the genes encoding these AlaRS editing domain homologues may have been lost from the pneumococcal genome during gene shuffling, which occurs rapidly within the organism as a result of exposure to antibiotics (19). This loss would be feasible in the presence of another conserved protein able to perform the same function. One such candidate protein is MurM, an Ala/Ser-tRNA-dependent aminoacyl-tRNA ligase that is involved in the synthesis of branched structured muropeptides in pneumococcal peptidoglycan (20). MurM catalyzes the transfer of either alanine or serine to the stem peptide lysine of Lipid II and, in combination with MurN, generates the substrate for indirect cross-linking of peptidoglycan. Until recently, the only aminoacyl-tRNA substrates recognized by MurM were thought to be Ser-tRNASer, provided by seryl-tRNA synthetase, and Ala-tRNAAla, provided by AlaRS (38). However, MurM is also able to efficiently transfer serine to Lipid II from misaminoacylated Ser-tRNAAla, which is also produced by pneumococcal AlaRS (21). The observed preference for misaminoacylated Ser-tRNAAla suggests that MurM could function as a trans editing factor and influence translation quality control by channeling appropriate misaminoacylated tRNA species into the peptidoglycan biosynthesis pathway. Here we show that pneumococcal AlaRS also misaminoacylates an unusual tRNAPhe isoacceptor to generate substrates for MurM, which is able to catalyze deacylation in the absence of Lipid II.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and General Protein Expression and Purification
S. pneumoniae strain D39 chromosomal DNA for use as a template in the cloning of AlaRS, PheRS, EF-Tu, and MurM was a gift from B. Lazazzera (University of California, Los Angeles). An expression construct for producing His6-tagged E. coli AlaRS and pUC19 containing the E. coli tRNAAla gene for production by in vitro transcription were gifts from K. Musier-Forsyth (Ohio State University, Columbus, OH).
The genes encoding S. pneumoniae AlaRS full-length protein, AlaRS residues 1–460 (catalytic domain only), EF-Tu, and MurM were cloned into pET21b (Novagen) by virtue of the NdeI and XhoI restriction sites. Subsequent expression constructs allowed for the production of recombinant proteins tagged at the C termini with a hexahistidine tag. The genes encoding S. pneumoniae PheRS α and β subunits were cloned into pET Duet-1 (Novagen) multiple cloning sites one and two, respectively, such that the protein was produced with an N-terminal hexahistidine tag. All proteins were overexpressed in E. coli strain BL21 (DE3) by the addition of a final concentration of 1 mm isopropyl-β-d-1-thigalactopyranoside at an A600 of 04–0.6 followed by a reduction in growth temperature from 37 °C to 28 °C for 3–5 h. Proteins were purified on BD TALON cobalt resin using equilibration/wash buffer (50 mm sodium phosphate, pH 7.2, 500 mm sodium chloride, and 20% glycerol) containing 250 mm imidazole. MurM was solubilized prior to purification as described (21, 38). S. pneumoniae tRNAAla and tRNAPhe (wild type and U4G) and the E. coli wild type equivalents were produced by in vitro T7 RNA polymerase runoff transcription as described (22).
Site-directed Mutagenesis
Site-directed mutagenesis of pneumococcal tRNAPhe was carried out by the polymerase chain reaction using PfuTurbo polymerase. Methodology was obtained from the Stratagene site-directed mutagenesis manual.
Aminoacylation
Aminoacylation time courses were carried out across a time period of 1 h at 37 °C in the presence of 0.1 m Na-HEPES, pH 7.2, 30 mm KCl, 10 mm MgCl2, 2 mm ATP, 7 μm tRNAAla/Phe transcript, 40 or 110 μm [3H]Ser/[14C]Ala/[14C]Gly (150–200 cpm/pmol), and 310 nm active AlaRS (as determined by active site titration). Reactions were repeated in the presence of 50 nm active S. pneumoniae PheRS, 300 nm S. pneumoniae MurM, or 3 μm activated S. pneumoniae EF-Tu. EF-Tu was activated in 50 mm Tris-HCl, 1 mm DTT, 68 mm KCl, 6.7 mm MgCl2, 2.5 mm phosphoenolpyruvate, 0.5 mm GTP, and 30 mg/ml pyruvate kinase as described (23). 10-μl samples were taken for each time point and spotted onto 3-mm Whatman filter paper discs, which were immediately dropped into 5% TCA. Discs were subjected to further washes with 5% TCA and ethanol prior to drying and scintillation counting.
Kinetics of Phenylalanylation of tRNAPhe Wild Type and U4G by PheRS
To determine Michaelis-Menten kinetics for pneumococcal PheRS with either wild type or mutant U4G tRNAPhe, phenylalanylation time courses were carried out at 37 °C at both the lowest (0.05 μm) and the highest (10.0 μm) tRNA concentration in the presence of 0.1 m Na-HEPES, pH 7.2, 30 mm KCl, 10 mm MgCl2, 2 mm ATP, 50 μm [14C]Phe (200 cpm/pmol), and 50 nm active PheRS (as determined by active site titration). Because the linear region was determined to be within the first 2 min, 10-μl samples were spotted onto 3-mm Whatman filter paper and dropped into 5% TCA at four time points (0.5, 1.0, 1.5, and 2 min) at each of the tRNA concentrations (0.05, 0.10, 0.20, 0.40, 0.60, 0.80, 1.00, 3.00, 5.00, 10.00 μm) for determination of gradients and key kinetic parameters from triplicate data sets.
Deacylation Assays
Aminoacylation reactions were set up in four 200-μl reactions each consisting of 30 mm HEPES, pH 7.6, 15 mm MgCl2, 10 mm DTT, 2 mm ATP, 110 μm [3H]serine or [3H]alanine (with a specific activity of ∼300 cpm/pmol), 10 μm S. pneumoniae tRNAAla transcript (prior to use, stock was resuspended in 4 mm MgCl2 and heated at 80 °C for 10 min followed by slow cooling to room temperature to allow refolding), 2 μmol min−1 ml−1 inorganic pyrophosphatase, and 3 μm alanyl-tRNA synthetase catalytic domain. The reactions were incubated at 37 °C for 2 h and then quenched by the addition of 20 μl of 3 m sodium acetate, pH 4.5. [3H]Aminoacyl-tRNA purification was achieved by the addition of 220 μl of phenol, pH 4.5, to each of the four reactions followed by mixing and centrifugation at 13,000 rpm for 5 min. After centrifugation, the aqueous phase was retained, and an equal volume of 24:1 chloroform isoamyl alcohol was added. 550 μl of −20 °C RNase-free ethanol was added to the aqueous phase, which was subsequently incubated at −80 °C for 1 h. The precipitated aminoacyl-tRNA was pelleted by centrifugation at 13,000 rpm for 30 min and washed with 1 ml of 70% ethanol. After a final centrifugation step at 13,000 rpm for 30 min, the pellet was dried at room temperature for 5 min and resuspended in 50 μl of 3 mm sodium acetate, pH 4.5. At this point, all four reactions were pooled, yielding 200 μl of [3H]Ser or [3H]Ala-tRNAAla, the concentration of which was determined by 5% TCA precipitation of 2 μl of the stock and scintillation counting. Deacylation assays were carried out by incubation of 50 pm [3H]Ser or Ala-tRNAAla in buffer composed of 0.1 m Na-HEPES, pH 7.2, 30 mm KCl, and 10 mm MgCl2. In addition, 0.5 μm AlaRS, 0.5 μm MurM, or an equal volume of protein storage buffer was added to the reaction, which was monitored by TCA precipitation and scintillation counting.
Due to our inability to successfully isolate sufficient yields of misaminoacylated tRNAPhe, an aminoacylation-coupled deacylation reaction was developed. 100-μl aminoacylation reactions were incubated at 37 °C in the presence of 0.1 m Na-HEPES, pH 7.2, 30 mm KCl, 10 mm MgCl2, 2 mm ATP, 7 μm tRNAAla/Phe transcript, 100 μm [3H]Ser (PerkinElmer)/[3H]Ala (Moravek Biochemicals) at a specific activity of 200–300 cpm/pmol, and 0.5–1.0 μm active AlaRS catalytic domain (or full-length AlaRS in the case of cognate Ala-tRNAAla). After a 1-h incubation, sodium chloride (final concentration 100 mm) and adenosine 5′-triphosphatase from porcine cerebral cortex (final concentration of 0.5 units, Sigma-Aldrich) were added to the reaction. Incubation at 37 °C was continued for an additional 30 min to the plateau of aminoacylation prior to the addition of the potential deacylation factor (enzyme storage buffer in the case of the control, full-length AlaRS, PheRS, or MurM) in a 34-μl volume where 4 μl was composed entirely of 100 mm unlabeled serine or alanine as appropriate. After equilibration for 2 min, a 10-μl volume was taken from the reaction and spotted onto 3-mm Whatman filter paper, which was immediately dropped into 5% TCA. This was used as the zero time point, and the deacylation reaction was monitored for an additional 10 min with samples taken at 1, 2, 4, 8, and 10 min. After all time points had been taken, filter papers were washed a further two times in 5% TCA and then once in 100% ethanol prior to drying and liquid scintillation counting. All samples were repeated in triplicate. Full-length AlaRS, PheRS, and MurM were at final concentrations of 0.6, 1, and 1 μm, respectively.
RESULTS
Pneumococcal AlaRS Displays Relaxed Specificity for Amino Acid and tRNA Recognition
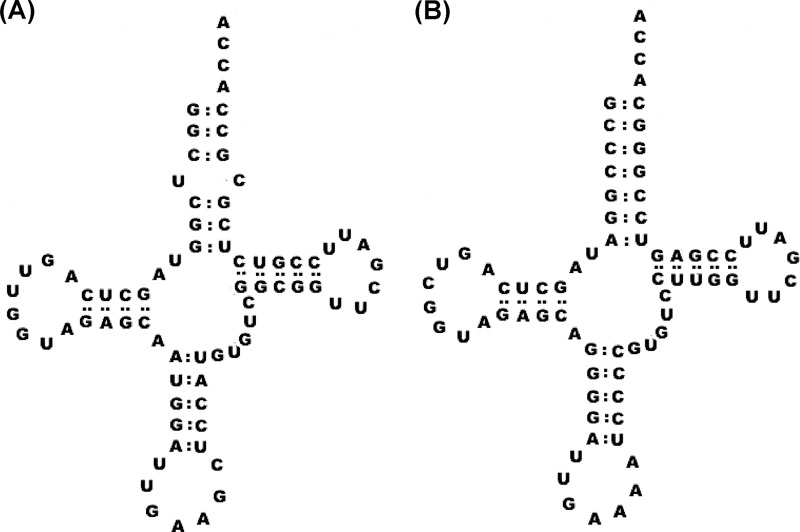
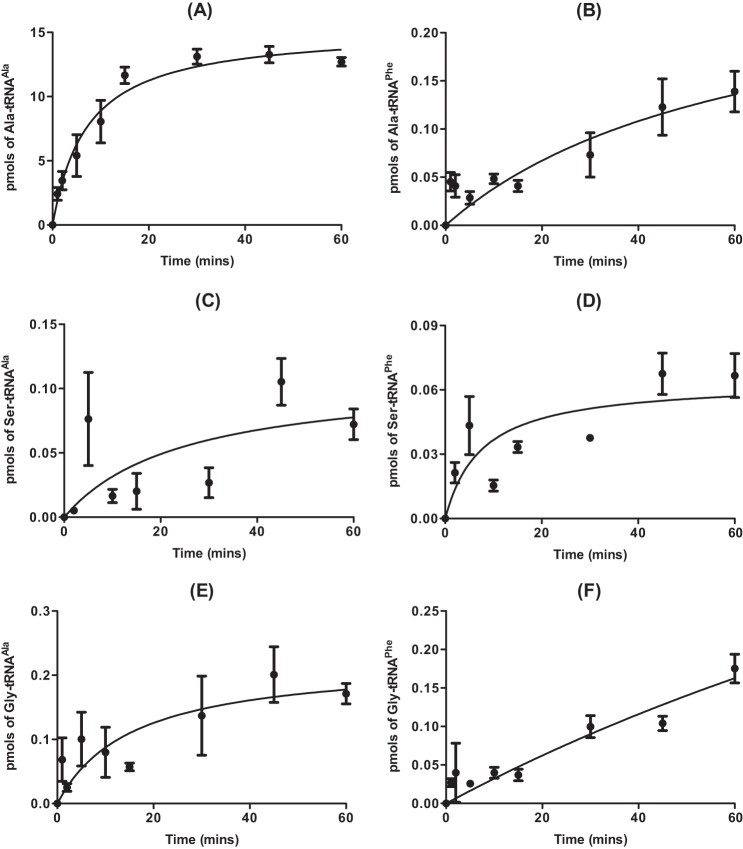
The ability of AlaRS to misactivate both serine and glycine is well documented, as is its high specificity for cognate tRNAAla, which results from recognition of the conserved G3:U70 base pair in the acceptor stem (14, 24). The potential ability of pneumococcal AlaRS to utilize noncognate tRNAPhe as a substrate was investigated here due to the presence of an unusual U4:C69 mismatch within the acceptor stem of the molecule, which may affect the ability of AlaRS to accurately discriminate against it (Fig. 1). In vitro assays showed that pneumococcal AlaRS was able to aminoacylate both tRNAAla and tRNAPhe with any of Ala, Gly, or Ser (Fig. 2). As expected, an equivalent active concentration of the isolated catalytic domain of AlaRS, which has no editing function, was more efficient in tRNA misaminoacylation than the full-length editing-proficient enzyme (Fig. 3).
FIGURE 1.
Cloverleaf structure of pneumococcal tRNAPhe. A and B, cloverleaf structures of pneumococcal (A) and E. coli (B) tRNAPhe (anticodon GAA). The distorted region in the acceptor stem of pneumococcal tRNAPhe was removed in the mutant species used in this study (termed tRNAPhe U4G) by replacement of the uracil at position 4 with a guanine.
FIGURE 2.
Error-prone aminoacylation of tRNAAla and tRNAPhe by full-length pneumococcal AlaRS. A–F, aminoacylation time courses in the presence of 40 μm [14C]alanine (A and B), [14C]serine (C and D), or [14C]glycine (E and F) for 310 nm active full-length pneumococcal AlaRS. Wild type pneumococcal tRNAAla (A, C, and E) or tRNAPhe (B, D, and F) were used at a concentration of 7 μm. Data sets are the average of three independent experiments. Error bars indicate S.E.
FIGURE 3.
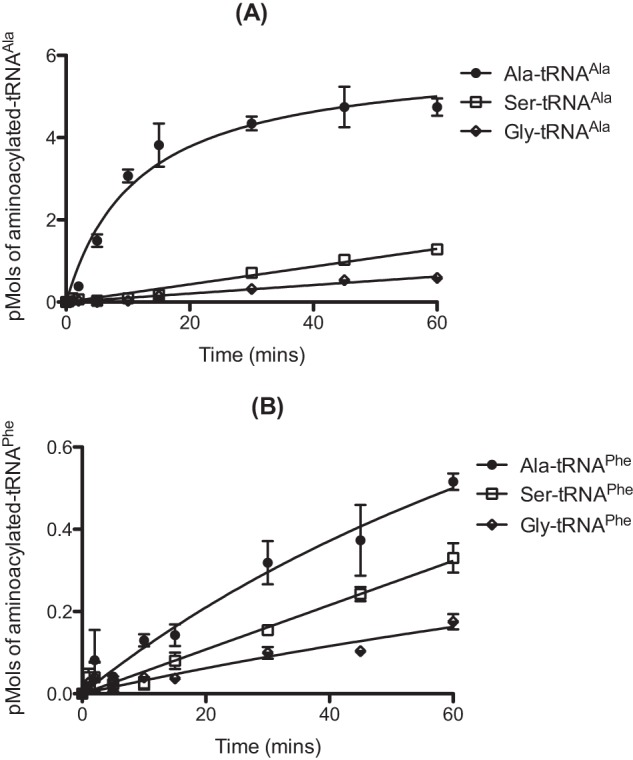
Error-prone aminoacylation of tRNAAla and tRNAPhe by catalytic domain of pneumococcal AlaRS. Aminoacylation time courses in the presence of 40 μm [14C]alanine, [14C]serine, or [14C]glycine for 310 nm pneumococcal AlaRS catalytic domain. A and B, wild type pneumococcal tRNAAla (A) or tRNAPhe (B) was used at a concentration of 7 μm. Data sets are the average of three independent experiments. Error bars indicate S.E.
To determine whether aminoacylation of tRNAPhe by AlaRS is a unique feature of the pneumococcal system, alanylation time courses were carried out with an equivalent concentration of full-length E. coli AlaRS and either the cognate E. coli tRNAAla or the noncognate E. coli tRNAPhe (data not shown). E. coli AlaRS does not use tRNAPhe as a substrate, consistent with the absence of either the G3:U70 alanylation identity element or other atypical structures such as the U4:C69 mismatch present in S. pneumoniae tRNAPhe (Fig. 1). Previous studies have indicated that mutation of E. coli tRNAPhe to contain the G3:U70 identity element is required for aminoacylation by E. coli AlaRS (24).
Pneumococcal PheRS Is Unable to Edit Misaminoacylated tRNAPhe
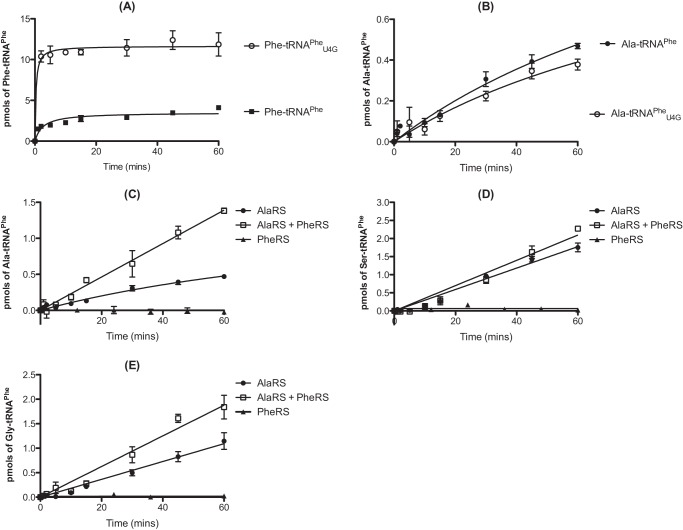
The ability of pneumococcal PheRS to aminoacylate both wild type pneumococcal tRNAPhe and mutant tRNAPheU4G was tested. In the later species, the distorted region of the acceptor stem was replaced by a Watson-Crick base pair conformation, making the structure more closely resemble that of E. coli tRNAPhe (Fig. 1). The tRNAPheU4G mutant was aminoacylated ∼5-fold more efficiently than the wild type species by PheRS (Fig. 4A), whereas full-length AlaRS was able to alanylate both tRNAs equally efficiently (Fig. 4B). Kinetic characterization of PheRS with both wild type and mutant tRNAPhe indicated that this effect was not caused by a change in Km but rather by an approximate 2.5-fold increase in kcat for tRNAPheU4G (Table 1).
FIGURE 4.
The effect of the U4:C69 wobble on aminoacylation of tRNAPhe by PheRS and AlaRS. A, comparison of the ability of pneumococcal PheRS to aminoacylate wild type and mutant U4G pneumococcal tRNAPhe. Time courses were carried out in the presence of 40 μm [14C]phenylalanine with 500 nm active full-length pneumococcal PheRS and either 7 μm wild type tRNAPhe or 7 μm mutant tRNAPheU4G. B, comparison of the ability of pneumococcal full-length AlaRS to aminoacylate wild type and mutant U4G pneumococcal tRNAPhe. Time courses were carried out in the presence of 110 μm [14C]alanine and either 7 μm wild type tRNAPhe or 7 μm mutant tRNAPheU4G. C–E, misaminoacylation time courses for 310 nm active full-length pneumococcal AlaRS in the presence of 50 nm active pneumococcal PheRS and 7 μm wild type pneumococcal tRNAPhe. The concentration of [14C]alanine (C), [3H]serine (D), and [14C]glycine (E) used was 110 μm. Data sets are the average of three independent experiments. Error bars indicate S.E.
TABLE 1.
Kinetic parameters for phenylalanylation of tRNAPhe wild type of U4G by PheRS
| Km | Vmax | kcat | kcat/Km | |
|---|---|---|---|---|
| μm | μm/min/mg | s−1 | ||
| Wild type tRNAPhe | 1.05 ± 0.19 | 0.15 ± 0.04 | 0.32 ± 0.09 | 0.30 |
| Mutant tRNAPheU4G | 1.09 ± 0.30 | 0.38 ± 0.06 | 0.78 ± 0.10 | 0.72 |
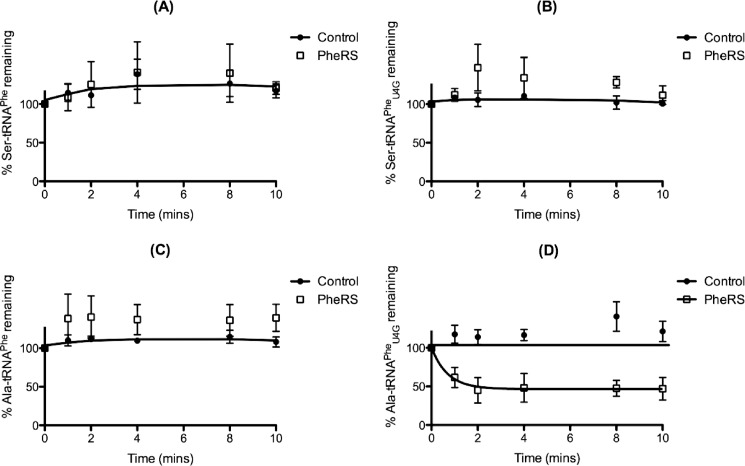
To test whether pneumococcal PheRS could hydrolyze misaminoacylated Ala-, Ser-, and Gly-tRNAPhe, AlaRS-catalyzed misaminoacylation reactions were repeated in the presence of PheRS (Fig. 4, C–E). PheRS addition caused a marked enhancement of Ala-tRNAPhe production by pneumococcal AlaRS, suggesting that PheRS binds and protects this misaminoacylated tRNA from spontaneous deacylation (Fig. 4C). Some protection was also demonstrated for Gly-tRNAPhe (Fig. 4E) but not for Ser-tRNAPhe (Fig. 4D). In the absence of a successful procedure for isolating adequate yields of misaminoacylated tRNAPhe, a coupled aminoacylation/deacylation assay was developed to assess the ability of PheRS to edit both wild type and mutant Ala- and Ser-tRNAPhe. PheRS addition had no effect on the deacylation of either Ser-tRNAPhe species (Fig. 5, A and B, respectively). For Ala-tRNAPhe, PheRS addition led to deacylation of Ala-tRNAPheU4G but not wild type Ala-tRNAPhe (Fig. 5, D and C, respectively). This suggests that the U4:C69 mismatch in wild type pneumococcal tRNAPhe is an anti-determinant for PheRS-mediated editing of Ala-tRNAPhe.
FIGURE 5.
The U4:C69 mismatch is an anti-determinant for PheRS editing of Ala-tRNAPhe. A–D, deacylation time courses for AlaRS-generated Ser-tRNAPhe wild type (A), Ser-tRNAPheU4G (B), Ala-tRNAPhe wild type (C), and Ala-tRNAPheU4G (D) in the presence of 0.6 μm active PheRS. Aminoacylation reactions were carried out using 0.5 μm AlaRS catalytic domain, 7 μm tRNA transcript, and 100 μm [3H]serine or [3H]alanine. Data sets are the average of three independent experiments. Error bars indicate S.E.
The Effect of Pneumococcal EF-Tu on tRNA Misaminoacylation
The sequence of pneumococcal EF-Tu is diverged from that of other bacteria at four conserved positions: P129K, M140L, T230S, and E234D. Of particular interest is the substitution of Thr-230 with Ser in the third β strand of the second domain of the protein, which comprises the aminoacyl-tRNA binding pocket (25). The effects of pneumococcal EF-Tu addition on the generation of misaminoacylated tRNAAla and tRNAPhe by AlaRS were investigated. EF-Tu addition resulted in modest enhancement of the production of Gly-tRNAAla by pneumococcal AlaRS (Fig. 6) but had no significant effect on other AlaRS-catalyzed reactions (data not shown).
FIGURE 6.
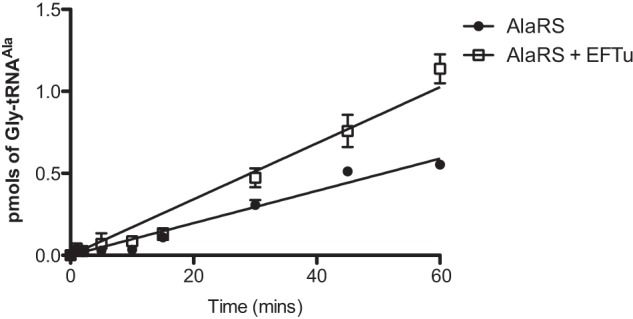
The effect of EF-Tu on Gly-tRNAAla misaminoacylation by AlaRS. Misaminoacylation time courses for generation of Gly-tRNAAla by 310 nm full-length pneumococcal AlaRS in the presence or absence of 3 μm activated pneumococcal EF-Tu are shown. The concentration of wild type pneumococcal tRNAAla used was 7 μm. The concentration of [14C]glycine used was 110 μm. Data sets are the average of three independent experiments. Error bars indicate S.E.
MurM Functions as a trans Editing Factor in Pneumococci
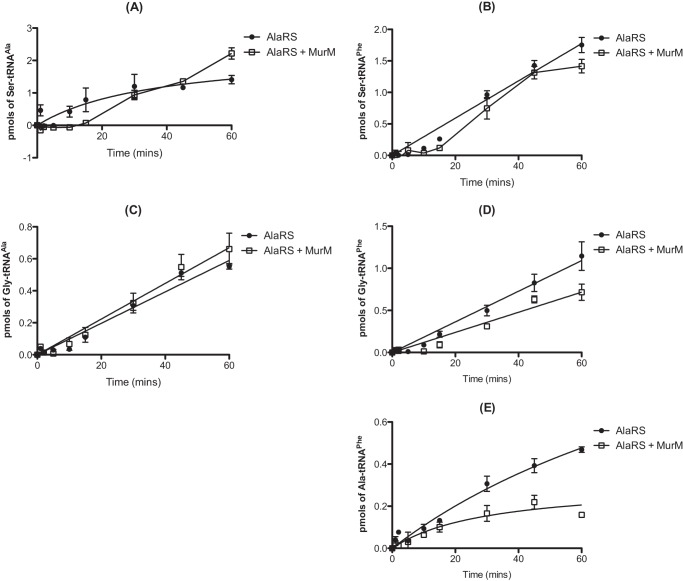
Pneumococcal peptidoglycan is unusual in that it typically consists of a combination of both branched and linear muropeptides. Within the structure, branched muropeptides consist of either a serine-alanine or an alanine-alanine dipeptide bridge attached to the stem peptide lysine of Lipid II. This dipeptide bridge is synthesized by the action of the MurM and MurN proteins (26–28). The substrates for MurM include pneumococcal Ser-tRNASer and Ala-tRNAAla provided by seryl-tRNA synthetase and AlaRS, respectively. Previous work also demonstrated that the catalytic efficiency of MurM is greater when Ser-tRNAAla is provided as a substrate as opposed to Ala-tRNAAla (21). The ability of MurM to utilize Ser-tRNAAla and the absence of any genome-encoded AlaXPs proteins in S. pneumoniae prompted us to investigate the ability of MurM to trans edit mischarged tRNAs. In the case of AlaRS synthesis of Ser-tRNAAla, the addition of MurM suppressed mischarging for the first 15 min of the reaction (Fig. 7A). After this time, product formation increased, possibly due to a loss of MurM stability during incubation for prolonged time periods at 37 °C. The initial suppression of mischarging suggests that MurM may act as a trans editing factor capable of hydrolyzing Ser-tRNAAla in the absence of its second substrate, Lipid II. AlaRS generation of Ala-tRNAPhe was also reduced by the addition of MurM to the mischarging time course, providing additional support for a trans editing function (Fig. 7E). The ability of MurM to hydrolyze Ser-tRNAAla and Ala-tRNAAla was investigated in the absence of Lipid II (Fig. 8, A and B, respectively). The addition of MurM led to rapid deacylation of both Ala-tRNAAla and Ser-tRNAAla, consistent with trans editing activity. Direct deacylation assays could not be performed with MurM and Ala-tRNAPhe due to the comparative instability of this mischarged tRNA species (see above).
FIGURE 7.
MurM decreases production of serylated tRNAAla/Phe and alanylated tRNAPhe by AlaRS. A–E, misaminoacylation time courses for generation of Ser-tRNAAla (A), Ser-tRNAPhe (B), Gly-tRNAAla (C), Gly-tRNAPhe (D), or Ala-tRNAPhe (E) by 310 nm active full-length pneumococcal AlaRS in the presence of 300 nm MurM. The concentration of wild type pneumococcal tRNAAla or tRNAPhe used was 7 μm. The concentration of [14C]alanine, [3H]serine, and [14C]glycine used was 110 μm. Data sets are the average of three independent experiments. Error bars indicate S.E.
FIGURE 8.
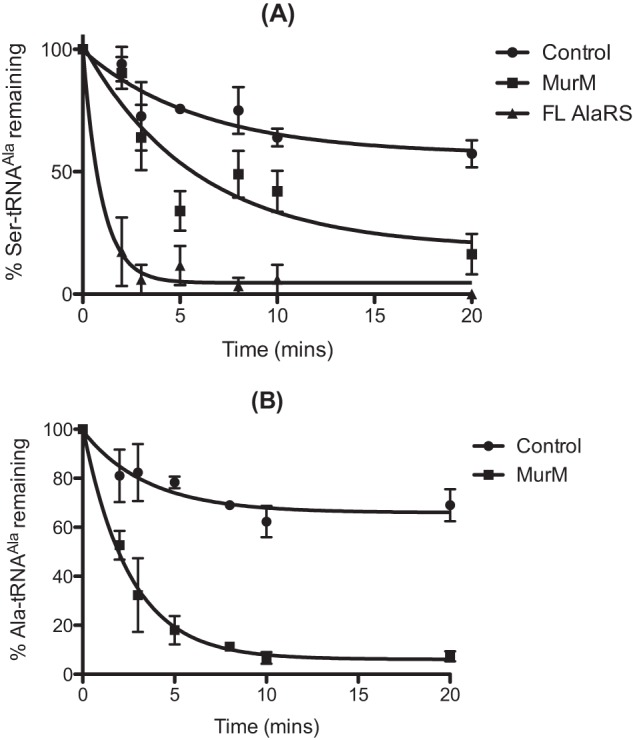
Deacylation of mischarged pneumococcal tRNAAla by MurM. A and B, deacylation time courses were carried out as described by incubation of 50 pm [3H]Ser-tRNAAla (A) or Ala-tRNAAla (B) with 0.5 μm pneumococcal AlaRS or 0.5 μm MurM. In the control reaction, an equal volume of protein storage buffer was added. Error bars indicate S.E. FL, full-length.
DISCUSSION
Pneumococcal AlaRS Is Error-prone during Aminoacylation
Extensive studies on E. coli AlaRS have indicated that the G3:U70 wobble base pair in the acceptor stem of tRNAAla is a critical identity element for alanylation by AlaRS (24, 29–32). In addition to this, it has been demonstrated that E. coli tRNAPhe can only be alanylated by E. coli AlaRS if the acceptor stem is mutated so that it contains the G3:U70 base pair (24, 29). The predicted cloverleaf structure of pneumococcal tRNAPhe indicates the presence of an unusual wobble base pair in its acceptor stem, U4:C69, which is not found in E. coli tRNAPhe (Fig. 1) (33, 34). In addition to misaminoacylating tRNAAla, pneumococcal AlaRS is also able to charge tRNAPhe with alanine, serine, and glycine. Production of high levels of hydrogen peroxide during the life cycle of pneumococcus could potentially accelerate misaminoacylation of tRNAAla and tRNAPhe by AlaRS. In support of this hypothesis, it has been shown that exposure of E. coli threonyl-tRNA synthetase to hydrogen peroxide results in exacerbated production of misaminoacylated Ser-tRNAThr. This is due to oxidation of an editing site cysteine residue and subsequent loss of zinc ion coordination (4). Therefore, in the absence of free-standing homologues of the editing domain of AlaRS (AlaXPs proteins), pneumococcus may require MurM to maintain translation quality control, particularly if tRNA mischarging is elevated under oxidative stress as has been previously observed in other organisms (3, 4).
Evolution of Pneumococcal tRNA for Dual Functions in Protein and Peptidoglycan Biosynthesis
The peptidoglycan structure of pneumococcus is particularly unusual in that it contains a combination of both branched and linear muropeptides. The MurMN proteins are responsible for the synthesis of branched muropeptides and are one of the requirements for high level penicillin resistance within this bacterium (27, 28). Therefore, the mechanisms used by pneumococcus to ensure sufficient division of aminoacylated tRNA species between peptidoglycan biosynthesis and protein synthesis are of great interest.
In Staphylococcus aureus, pentaglycine bridge formation is essential for cell viability and is catalyzed by the Gly-tRNAGly requiring FemXAB proteins (35). In S. aureus, four annotated tRNAGly isoacceptors have been identified in addition to a pseudogene encoding an unusual fifth tRNAGly isoacceptor. Although all five tRNAGly isoacceptors have been shown to be substrates for the S. aureus glycyl-tRNA synthetase, the pseudogene and two of the other tRNAGly isoacceptors with the same anticodon were shown to possess sequence identity elements favoring weak binding interaction with EF-Tu. This allows S. aureus to ensure proper division of Gly-tRNAGly between protein synthesis and peptidoglycan biosynthesis (36).
In pneumococcus, no unique tRNAAla or tRNASer isoacceptors have been identified that would enable the bacterium to achieve division of aminoacylated tRNA between protein synthesis and peptidoglycan biosynthesis in the same way S. aureus does. The inability of pneumococcal PheRS to edit misaminoacylated Ala- and Ser-tRNAPhe may ensure that these species are potential cellular substrates for the peptidoglycan biosynthesis pathway. This is supported by the finding that the mismatch within the acceptor stem of pneumococcal tRNAPhe may allow AlaRS to compete with PheRS for this substrate by compromising the aminoacylation activity of the latter protein. Our data suggest that, once released from AlaRS, misaminoacylated tRNA species can be hydrolytically cleaved by MurM, but it remains unclear whether they are specifically diverted away from the translation machinery as is the case for some forms of Gly-tRNAGly in S. aureus. Further characterization of these and other adaptations pneumococci have made to ensure that the fidelity of protein synthesis and peptidoglycan biosynthesis are maintained during antibiotic and oxidative stress may enable new drug targets to be identified in the future. This has the potential to result in subsequent restoration of the potency of penicillin in the treatment of infections by this bacterium.
Acknowledgments
We thank Drs. Karin Musier-Forsyth and Beth Lazazzera for gifts of materials.
This work was supported by National Science Foundation Grant MCB-1052344.
- EF-Tu
- elongation factor Tu
- AlaRS
- alanyl-tRNA synthetase
- PheRS
- phenylalanyl-tRNA synthetase.
REFERENCES
- 1. Pesakhov S., Benisty R., Sikron N., Cohen Z., Gomelsky P., Khozin-Goldberg I., Dagan R., Porat N. (2007) Effect of hydrogen peroxide production and the Fenton reaction on membrane composition of Streptococcus pneumoniae. Biochim. Biophys. Acta 1768, 590–597 [DOI] [PubMed] [Google Scholar]
- 2. Regev-Yochay G., Trzcinski K., Thompson C. M., Lipsitch M., Malley R. (2007) SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 189, 6532–6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Netzer N., Goodenbour J. M., David A., Dittmar K. A., Jones R. B., Schneider J. R., Boone D., Eves E. M., Rosner M. R., Gibbs J. S., Embry A., Dolan B., Das S., Hickman H. D., Berglund P., Bennink J. R., Yewdell J. W., Pan T. (2009) Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ling J., Söll D. (2010) Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. U.S.A. 107, 4028–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reynolds N. M., Lazazzera B. A., Ibba M. (2010) Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 8, 849–856 [DOI] [PubMed] [Google Scholar]
- 6. Ibba M., Söll D. (1999) Quality control mechanisms during translation. Science 286, 1893–1897 [DOI] [PubMed] [Google Scholar]
- 7. Ling J., So B. R., Yadavalli S. S., Roy H., Shoji S., Fredrick K., Musier-Forsyth K., Ibba M. (2009) Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell 33, 654–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McClain W. H. (1993) Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 234, 257–280 [DOI] [PubMed] [Google Scholar]
- 9. Guth E. C., Francklyn C. S. (2007) Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol. Cell 25, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ling J., Reynolds N., Ibba M. (2009) Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78 [DOI] [PubMed] [Google Scholar]
- 11. Fersht A. R., Schimmel P. R., Söll D., Abelson J. N. (1979) Editing mechanisms in the aminoacylation of tRNA. in Transfer RNA: Structure, Properties and Recognition, pp. 247–254, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 12. Eldred E. W., Schimmel P. R. (1972) Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 247, 2961–2964 [PubMed] [Google Scholar]
- 13. Fersht A. R. (1977) Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry 16, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 14. Tsui W. C., Fersht A. R. (1981) Probing the principles of amino acid selection using the alanyl-tRNA synthetase from Escherichia coli. Nucleic Acids Res. 9, 4627–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nureki O., Vassylyev D. G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T. L., Schimmel P., Yokoyama S. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280, 578–582 [DOI] [PubMed] [Google Scholar]
- 16. Beebe K., Ribas De Pouplana L., Schimmel P. (2003) Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 22, 668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo M., Chong Y. E., Beebe K., Shapiro R., Yang X. L., Schimmel P. (2009) The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science 325, 744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo M., Chong Y. E., Shapiro R., Beebe K., Yang X. L., Schimmel P. (2009) Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature 462, 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Croucher N. J., Harris S. R., Fraser C., Quail M. A., Burton J., van der Linden M., McGee L., von Gottberg A., Song J. H., Ko K. S., Pichon B., Baker S., Parry C. M., Lambertsen L. M., Shahinas D., Pillai D. R., Mitchell T. J., Dougan G., Tomasz A., Klugman K. P., Parkhill J., Hanage W. P., Bentley S. D. (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filipe S. R., Severina E., Tomasz A. (2000) Distribution of the mosaic structured murM genes among natural populations of Streptococcus pneumoniae. J. Bacteriol. 182, 6798–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shepherd J. (2011) Characterisation of Pneumococcal Peptidoglycan Cross-linking Enzymology. Ph.D. thesis, University of Warwick, Coventry, United Kingdom [Google Scholar]
- 22. Roy H., Ling J., Irnov M., Ibba M. (2004) Post-transfer editing in vitro and in vivo by the β subunit of phenylalanyl-tRNA synthetase. EMBO J. 23, 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ling J., Yadavalli S. S., Ibba M. (2007) Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA 13, 1881–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou Y. M., Schimmel P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333, 140–145 [DOI] [PubMed] [Google Scholar]
- 25. Ke D., Boissinot M., Huletsky A., Picard F. J., Frenette J., Ouellette M., Roy P. H., Bergeron M. G. (2000) Evidence for horizontal gene transfer in evolution of elongation factor Tu in enterococci. J. Bacteriol. 182, 6913–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fiser A., Filipe S. R., Tomasz A. (2003) Cell wall branches, penicillin resistance, and the secrets of the MurM protein. Trends Microbiol. 11, 547–553 [DOI] [PubMed] [Google Scholar]
- 27. Filipe S. R., Tomasz A. (2000) Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc. Natl. Acad. Sci. U.S.A. 97, 4891–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Filipe S. R., Severina E., Tomasz A. (2002) The murMN operon: a functional link between antibiotic resistance and antibiotic tolerance in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 99, 1550–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McClain W. H., Foss K. (1988) Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket”. Science 241, 1804–1807 [DOI] [PubMed] [Google Scholar]
- 30. Gabriel K., Schneider J., McClain W. H. (1996) Functional evidence for indirect recognition of G·U in tRNAAla by alanyl-tRNA synthetase. Science 271, 195–197 [DOI] [PubMed] [Google Scholar]
- 31. Beuning P. J., Yang F., Schimmel P., Musier-Forsyth K. (1997) Specific atomic groups and RNA helix geometry in acceptor stem recognition by a tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 94, 10150–10154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McClain W. H., Chen Y. M., Foss K., Schneider J. (1988) Association of transfer RNA acceptor identity with a helical irregularity. Science 242, 1681–1684 [DOI] [PubMed] [Google Scholar]
- 33. Lowe T. M., Eddy S. R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schattner P., Brooks A. N., Lowe T. M. (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ling B., Berger-Bächi B. (1998) Increased overall antibiotic susceptibility in Staphylococcus aureus femAB null mutants. Antimicrob. Agents Chemother. 42, 936–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Giannouli S., Kyritsis A., Malissovas N., Becker H. D., Stathopoulos C. (2009) On the role of an unusual tRNAGly isoacceptor in Staphylococcus aureus. Biochimie 91, 344–351 [DOI] [PubMed] [Google Scholar]
- 37. Centers for Disease Control and Prevention (2013) http://www.cdc.gov/pneumococcal/surveillance.html
- 38. Lloyd A. J., Gilbey A. M., Blewett A. M., De Pascale G., El Zoeiby A., Levesque R. C., Catherwood A. C., Tomasz A., Bugg T. D., Roper D. I., Dowson C. G. (2008) Characterization of tRNA-dependent peptide bond formation by MurM in the synthesis of Streptococcus pneumoniae peptidoglycan. J. Biol. Chem. 283, 6402–6417 [DOI] [PubMed] [Google Scholar]