Background: Binding to the ATP site results in poor selectivity; therefore, development of ATP-noncompetitive inhibitors is needed.
Results: A modified chrysin with anticancer activity targets Cdks and binds to a Cdk2 allosteric site, not the ATP pocket.
Conclusion: Modified chrysin is a novel ATP-noncompetitive inhibitor.
Significance: This pharmacophore model might provide insights for the development of new ATP-noncompetitive agents.
Keywords: ATP, CDK (Cyclin-dependent Kinase), Cell Cycle, Chemoprevention, Epidermal Growth Factor (EGF), ATP-noncompetitive, CDK Inhibitor, Chrysin Derivative, Natural Compound
Abstract
Chrysin (5,7-dihydroxyflavone), a natural flavonoid widely distributed in plants, reportedly has chemopreventive properties against various cancers. However, the anticancer activity of chrysin observed in in vivo studies has been disappointing. Here, we report that a chrysin derivative, referred to as compound 69407, more strongly inhibited EGF-induced neoplastic transformation of JB6 P+ cells compared with chrysin. It attenuated cell cycle progression of EGF-stimulated cells at the G1 phase and inhibited the G1/S transition. It caused loss of retinoblastoma phosphorylation at both Ser-795 and Ser-807/811, the preferred sites phosphorylated by Cdk4/6 and Cdk2, respectively. It also suppressed anchorage-dependent and -independent growth of A431 human epidermoid carcinoma cells. Compound 69407 reduced tumor growth in the A431 mouse xenograft model and retinoblastoma phosphorylation at Ser-795 and Ser-807/811. Immunoprecipitation kinase assay results showed that compound 69407 attenuated endogenous Cdk4 and Cdk2 kinase activities in EGF-stimulated JB6 P+ cells. Pulldown and in vitro kinase assay results indicated that compound 69407 directly binds with Cdk2 and Cdk4 in an ATP-independent manner and inhibited their kinase activities. A binding model between compound 69407 and a crystal structure of Cdk2 predicted that compound 69407 was located inside the Cdk2 allosteric binding site. The binding was further verified by a point mutation binding assay. Overall results indicated that compound 69407 is an ATP-noncompetitive cyclin-dependent kinase inhibitor with anti-tumor effects, which acts by binding inside the Cdk2 allosteric pocket. This study provides new insights for creating a general pharmacophore model to design and develop novel ATP-noncompetitive agents with chemopreventive or chemotherapeutic potency.
Introduction
Carcinogenesis involves at least three distinct stages, including initiation, promotion, and progression. Tumor promotion requires chronic exposure to tumor promoters, such as 12-O-tetradecanoylphorbol-13-acetate (1, 2) and epidermal growth factor (EGF) (3, 4), that complete the conversion of a cell to the neoplastic state. In contrast to initiation, the tumor promotion stage is prolonged and also reversible if tumor promoter treatment is ceased (5), making it a potential target for prevention strategies. One of the molecular mechanisms underlying EGF-triggered neoplastic cell transformation is EGF-induced aberrant cell cycle progression, which is one of the hallmarks of tumor formation and progression.
The cyclin-dependent kinase (Cdk)4 family of serine/threonine kinases was identified as a key regulator of cell cycle progression. At least nine Cdks (Cdk1–9) have been identified, and Cdk1–7 are involved in cell cycle control. Cell cycle progression is driven by the sequential and periodic activation of various Cdk-cyclin complexes (6). Mouse embryonic fibroblasts lacking Cdk4 are resistant to transformation by oncogenes (7). When co-transfected with activated H-Ras, Cdk4 displayed oncogenic potential by provoking foci formation in primary rat embryonic fibroblasts (8) and generating malignant human epidermal tumorigenesis (9). Mouse embryonic fibroblasts expressing disrupted Cdk2 exhibit reduced susceptibility to transformation by oncogenes (10, 11). Activation of Cdk2 is required for foci formation induced by cyclin E and SV40 small T antigen co-expression in human fibroblasts (12). Thus, Cdk2 and Cdk4 are closely associated with oncogenic transformation.
Chemoprevention is acknowledged as an important and practical strategy for the management of cancer. Accumulating research evidence suggests that many dietary phytochemicals have anticancer activity, low toxicity, and few adverse side effects making them safe for human use. These natural or dietary compounds could be a source of prototype molecules from which to synthetically develop new chemotherapeutic agents with potent anticancer properties. Chrysin (5,7-dihydroxyflavone), a natural flavonoid widely distributed in many plant extracts, and honey and propolis, reportedly has chemopreventive properties such as anti-proliferative and pro-apoptosis activities against various cancers (13, 14). However, the anticancer activity of chrysin observed in in vivo studies has been disappointing (15, 16). To address this issue, several chrysin derivatives have been synthesized in recent years (17–19), suggesting the feasibility of improving the biological activities of chrysin as an antitumor agent that is more potent, with lower toxicity and minimal side effects by modifying its structure.
The majority of protein kinase inhibitors are ATP-competitive (type I) agents, which typically bind to the ATP pocket that is highly conserved across most of the kinases of the human genome. The lack of selectivity is an issue with type I inhibitors, which can lead to so-called “off-target” effects (20). The relatively poor selectivity of type I inhibitors can be addressed by type II inhibitors, which bind not only the ATP pocket but, in addition, interact with a site adjacent to the pocket. Type III inhibitors bind to regions that are remote from the ATP pocket. These regions are typically not highly conserved across all the kinases, providing for better selectivity (21). Type IV inhibitors target protein kinases distal to the substrate binding pocket, and type V are bi-substrate and bivalent inhibitors (22). Types II–IV are allosteric (noncompetitive) inhibitors with distinct allosteric binding characteristics. To date, only a small number of noncompetitive inhibitors have been identified (21, 23). Most were identified serendipitously and were later determined to be ATP-noncompetitive agents through examination of x-ray co-structures (24). Although comparatively few agents remain in development, in particular phytochemicals, chemical strategies for converting known type I inhibitors into corresponding type II inhibitors with different kinase selectivity profiles and exceptionally potent cellular activity have been reported (24). This raises the possibility that natural phytochemicals could serve as core scaffolds that can be further designed and developed to obtain inhibitors with the desired spectrum of inhibitory activities.
Because of the important role of Cdks in carcinogenesis, these kinases have long been considered ideal targets for anticancer agents. As a result, many Cdk inhibitors have been developed, some of which have progressed to clinical trials. However, none are currently approved for clinical use because the numerous potential drug leads are ATP-competitive type I compounds, leading to a lack of target selectivity. An ever-increasing demand exists for the development of ATP-noncompetitive Cdk inhibitors, especially those from natural and dietary sources. Indeed, progress has been made in identifying Cdk inhibitors that act through novel mechanisms. A novel structural pocket present on Cdk2, which is conserved on Cdks 1, 4, and 6, has been identified. Small molecules, identified by a high throughput in silico screening of this pocket, exhibit cytostatic effects and act by decreasing the function of Cdks in cells by binding to this site (25). Recently, an allosteric ligand-binding site, away from the ATP site, in Cdk2 was also discovered. Binding of two 1-anilino-8-naphthalene sulfonate molecules is accompanied by substantial structural changes in Cdk2, resulting in a C-helix conformation that is incompatible for cyclin A association (26). A phytochemical Cdk inhibitor described as an ATP-noncompetitive inhibitor has also been reported. However, a mechanism of action that is distinct from that of ATP competitive inhibitors remains undisclosed (27).
Here, we report that a modified chrysin derivative, compound 69407, inhibits EGF-induced anchorage-independent growth of JB6 P+ cells and suppresses anchorage-dependent and -independent growth of A431 human epidermoid carcinoma cells. It also exhibited tumor suppression effects in an A431 mouse xenograft model. Compound 69407 was shown to be an ATP-noncompetitive inhibitor of Cdks. This study investigated a possible mechanism by which ATP did not compete with compound 69407 for binding to Cdk2. Our results provide new information for creating a general pharmacophore model through which the design and development of new ATP-noncompetitive agents (based on parental phytochemicals) with potent activity that target therapeutically relevant kinases with minimal or no side effects.
EXPERIMENTAL PROCEDURES
Reagents
Compound 69407 (97% purity) was purchased from InterBioScreen (Moscow, Russia). Chrysin (>97% purity) and other chemical reagents, including Tris, NaCl, and SDS, for molecular biology and buffer preparation were purchased from Sigma. (R)-Roscovitine was from Selleck Chemicals (Houston, TX). Restriction enzymes and a DNA ligation kit (version 2.0) were purchased from Takara Bio, Inc. (Shiga, Japan). CNBr-Sepharose 4B beads were obtained from Amersham Biosciences, and the protein assay kit was from Bio-Rad. Active Cdk2/cyclin E was purchased from Millipore (Dundee, UK). Active Cdk4/cyclin D1, active Cdk6/cyclin D1, active Cdk7/cyclin H1/MNAT1, recombinant human Rb protein (773–928 amino acids; substrate for Cdk4/cyclin D1 and Cdk6/cyclin D1), and myelin basic protein (MBP) (substrate for Cdk7/cyclin H1/MNAT1) were purchased from SignalChem (Richmond, British Columbia, Canada). Recombinant human histone H1 (substrate for Cdk2/cyclin E) was purchased from New England Biolabs (Ipswich, MA). Antibodies for immunoprecipitation (IP) or immunoblotting were purchased from Cell Signaling Technology (Beverly, MA), Santa Cruz Biotechnology (Santa Cruz, CA), Millipore (Dundee, UK), or Upstate Biotechnology, Inc. (Charlottesville, VA). For immunoprecipitation, protein extracts were immunoprecipitated by using antibodies against Cdk2 (sc-163, Santa Cruz Biotechnology), Cdk4 (sc-260, Santa Cruz Biotechnology), cyclin A (sc-751, Santa Cruz Biotechnology), cyclin D1 (sc-753, Santa Cruz Biotechnology), or normal rabbit IgG (NI01, Calbiochem). Antibodies to detect Cdk1, cyclin E, and cyclin B1 were purchased from Cell Signaling Technology. Antibodies against GST, Cdk6, Cdk7, Rb, and phosphorylated Rb (Ser-807/811) were purchased from Santa Cruz Biotechnology. Antibodies against phosphorylated Rb (Ser-795) and β-actin were purchased from Sigma. Antibodies for Western blotting analysis of other EGF-stimulated signaling pathways were from Cell Signaling Technology, Santa Cruz Biotechnology, or Upstate Biotechnology.
Cell Culture
JB6 P+ mouse epidermal cells were cultured in monolayers in minimum essential medium (MEM) Eagle's containing 5% fetal bovine serum (FBS), 2 mm l-glutamine, and 25 μg/ml gentamicin at 37 °C under a 5% CO2 atmosphere. Human skin epidermoid carcinoma A431 cells were cultured in DMEM supplemented with 10% FBS, 1% l-glutamate, penicillin, and streptomycin at 37 °C under a 5% CO2 atmosphere.
Cell Viability Assay
Cell viability was measured using the 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate (WST-1) assay kit (ITSBIO, Seoul, Korea) according to the manufacturer's instructions. The assay is based on the cleavage of WST-1 to formazan dye by cellular mitochondrial dehydrogenases. Because cleavage of WST-1 to formazan dye occurs only in viable cells, the amount of dye produced, measured in OD values, directly corresponds with the number of viable cells present in culture (17, 28). Briefly, for the cytotoxicity assay, JB6 P+ cells were seeded (1 × 104) into 96-well plates in 100 μl of 5% FBS/MEM and incubated in a 37 °C, 5% CO2 incubator. After culturing for 12 h, different concentrations of compound 69407 were added to each well. After culturing for another 24 or 48 h, 10 μl of WST-1 were added to each well, and cells were incubated for 2 h at 37 °C. For the proliferation assay, JB6 P+ cells (1 × 103) or A431 cells (2 × 103) were seeded into 96-well plates in 100 μl of medium and incubated in a 37 °C, 5% CO2 incubator. After culturing for 12 h, different concentrations of compound 69407 were added to each well. After culturing for another 24, 48, 72, or 96 h, 10 μl of WST-1 was added to each well, and cells were incubated for 2 h at 37 °C. The cellular reduction of WST-1 to formazan and its absorbance were measured at 450 nm.
Anchorage-independent Cell Growth Assay
Cells (8 × 103 per well) were suspended in basal medium Eagle (1 ml with 10% FBS and 0.33% agar) in the absence or presence of EGF (10 ng/ml) with various concentrations of compound 69407 or chrysin and plated over a layer of solidified BME, 10% FBS, 0.5% agar (3 ml) with various concentrations of compound 69407 or chrysin. The cultures were maintained at 37 °C in a 5% CO2 incubator for 2 or 3 weeks, and colonies were counted under a microscope as described by Colburn et al. (29).
Cell Cycle Analysis
JB6 P+ cells were serum-starved in 0.1% FBS/MEM for 36 h to synchronize the cells at G0 phase and treated for 1 h with DMSO or the indicated concentrations of compound 69407, chrysin, or (R)-roscovitine, and then exposed to EGF (10 ng/ml) for 24 h. Trypsinized cells were stained with propidium iodide with the CycleTESTTM PLUS DNA reagent kit (BD Biosciences) according to the manufacturer's recommendations, and the cell cycle phases were analyzed in a FACSCalibur flow cytometer (BD Biosciences) using the CellQuest Pro (BD Biosciences) software package.
Preparation of Sepharose 4B Beads
Sepharose 4B beads (0.3 g) were washed with 300 ml of 1 mm HCl three times for 5 min each by gentle inversion and then incubated with 3 mg of compound 69407 or DMSO in coupling buffer (0.1 m NaHCO3 and 0.5 m NaCl, pH 8.3) at 4 °C overnight. The samples were washed five times with coupling buffer and incubated with blocking buffer (0.1 m Tris-HCl, pH 8.0) at 4 °C overnight. The samples were alternatively washed with 0.1 m acetic acid buffer, pH 4.0, and with 0.1 m Tris-HCl and 0.5 m NaCl, pH 8.0, three times and then resuspended in 1 ml of PBS for use.
Pulldown Assays
For pulldown assays, compound 69407-conjugated Sepharose 4B beads (100 μl, 50% slurry) were incubated with 100 ng of recombinant Cdk2-cyclin E (Millipore), Cdk4-cyclin D1, Cdk6-cyclin D1, or Cdk7-cyclin H1-MNAT1 protein complexes (SignalChem), or cellular supernatant fractions of JB6 P+ cells (500 μg) in immunoprecipitation reaction buffer (50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 150 mm NaCl, 1 mm dithiothreitol, 0.01% Nonidet P-40, and 0.02 mm phenylmethylsulfonyl fluoride) containing 2 μg/ml bovine serum albumin and 1× protease inhibitor mixture at 4 °C with gentle rocking overnight. The beads were then washed five times with immunoprecipitation reaction buffer, and the proteins bound to the beads were analyzed by immunoblotting (30).
In Vitro Kinase Assay
Cdk in vitro kinase assays were performed as described previously (31) with the following modifications. Different concentrations of compound 69407 were incubated with the active recombinant Cdk2-cyclin E (Millipore), Cdk4-cyclin D1, Cdk6-cyclin D1, or Cdk7-cyclin H1-MNAT1 protein complexes (SignalChem) at 30 °C for 10 min. Then, 1 μg of purified histone H1 (substrate for Cdk2-cyclin E, New England Biolabs), 1 μg of Rb protein (773–928 amino acids, substrate for Cdk4-cyclin D1 and Cdk6-cyclin D1, SignalChem) or 1 μg of MBP (substrate for Cdk7-cyclin H1-MNAT1, SignalChem) was added, and reactions were carried out in 1× kinase buffer (25 mm Tris-Cl, pH 7.5, 5 mm β-glycerophosphate, 0.1 mm Na3VO4, 10 mm MgCl2, and 2 mm dithiothreitol) containing 50 μm unlabeled ATP with or without 10 μCi of [γ-32P]ATP at 30 °C for 30 min. Reactions were terminated and then proteins resolved by 12% SDS-PAGE and visualized by autoradiography.
Western Blotting Assays
Cell lysates were prepared with RIPA buffer (50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 1 mm EDTA, 1× protease inhibitor tablet). Protein concentrations were determined using a dye-binding protein assay kit (Bio-Rad) as described by the manufacturer, and equal amounts of proteins were used for each experiment. Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked with 5% nonfat dry milk for 1 h at room temperature and incubated with appropriate primary antibodies overnight at 4 °C. After washing with PBS containing 0.1% Tween 20, the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody at a 1:2,000 dilution, and the signal was detected with a SuperSignal West Pico Chemiluminescent kit (Thermo Scientific, Rockford, IL).
IP-Western Blotting Assays and IP Kinase Assays
Immunoprecipitation and IP kinase assays for Cdks were performed as described previously (31) with the following modifications. Quiescent JB6 P+ cells were serum-starved in 0.1% FBS/MEM for 36 h followed by treatment for 1 h with DMSO or the indicated concentrations of compound 69407, chrysin, or (R)-roscovitine and then exposed to EGF (10 ng/ml) for 24 h. Cells were harvested and disrupted in Cdk lysis buffer (50 mm Tris-HCl, pH 7.4, 0.25 m sodium chloride, 0.1% v/v Nonidet P-40, 5 mm EDTA, 50 mm sodium fluoride, 1 mm sodium orthovanadate, 1 mm sodium pyrophosphate, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture (Roche Applied Science) prepared fresh each time and stored on ice. The samples were sonicated on ice (output 10, 10 s on, 5 s off, total 50 s; Sonicator-4000, Misonix) and incubated on ice for 30 min. The lysates were clarified by centrifugation at 13,000 rpm at 4 °C for 10 min. The protein concentration was determined using a dye-binding protein assay kit (Bio-Rad) as described by the manufacturer. To pre-clear cell lysates, 40 μl of protein A/G-agarose beads (sc-2003, Santa Cruz Biotechnology) were added to 250 μg of cell lysate to a final volume of 750 μl using Cdk lysis buffer and rotating at 4 °C for 1 h. The pre-cleared cell lysates were immunoprecipitated with 2 μg of anti-Cdk2, anti-Cdk4, anti-cyclin A, or anti-cyclin D1 at 4 °C overnight, followed by 2 h of incubation with 40 μl of protein A/G-agarose beads. For IP-Western blotting assays, immunocomplexes were resolved by SDS-PAGE, and co-immunoprecipitated proteins were detected by Western blotting using antibodies as indicated in the specific experiments. For IP kinase assays, immunocomplexes were preincubated with DMSO and different concentrations of compound 69407, chrysin, or (R)-roscovitine at 30 °C for 10 min and then incubated in the presence of appropriate substrates in Cdk kinase buffer (50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 1 mm dithiothreitol) supplemented with 50 μm ATP and 10 μCi of [γ-32P]ATP at 30 °C for 30 min. Reactions were terminated and resolved by SDS-PAGE. To verify equal loading, the gel was stained with Coomassie Brilliant Blue to visualize substrate proteins before it was dried and subjected to autoradiography.
RNA Interference
A431 cells were grown in 6-well plates and transfected with a Cdk2-specific small interfering RNA oligonucleotide (si-Cdk2; catalog no. 8618; 100 pmol; Cell Signaling), a Cdk4-specific small interfering RNA oligonucleotide (si-Cdk4; catalog no. s2822; 100 pmol; Applied Biosystems), or a scrambled oligonucleotide (si-scrambled; catalog no. 6568; 100 pmol; Cell Signaling) using HiPerFect transfection reagent (catalog no. 301705, Qiagen) for 72 h according to the manufacturer's instructions. To confirm Cdk2 or Cdk4 knockdown, cells transfected with si-Cdk2, si-Cdk4, or scrambled oligonucleotide were harvested for protein extraction and immunoblotting.
Construction of Expression Vectors
pCMV-HA-Cdk2 (Addgene plasmid 1884) containing human full-length Cdk2 cDNA was constructed by van den Heuvel and Harlow (32) and obtained through Addgene (Cambridge, MA). The human Cdk2 cDNA was digested with BamHI and then the fragment was ligated into the BamHI site of the pGEX-6P-1 vector to generate a glutathione S-transferase (GST)-tagged Cdk2 expression plasmid. The mutations of Cdk2 (Y15A, K33A, L55R, L55R, and N59A) were generated by two-step PCR. The primer sets using for generating mutations are listed in supplemental Table 2. The plasmid constructs were confirmed by restriction mapping and DNA sequencing.
Flexible Protein Docking between Compound 69407 and Cdk2
The crystal structure of Cdk2 (Protein Data Bank code 3PXQ (33)) was used as the receptor model structure, and the conformation from the binding mode between compound 69407 and Cdk2 was used as the ligand structure. Protein-ligand docking was run using a high performance hierarchical docking algorithm Glide in Maestro version 9.2 and Glide version 5.7. The final binding model structure of compound 69407-Cdk2 was generated from Schrödinger Induced Fit Docking (34), which merges the predictive power of Prime with the docking and scoring capabilities of Glide for accommodating the possible protein conformational change upon ligand binding. More computational details are provided in the supplemental Methods.
In Vivo Tumor Growth Assay
Athymic nude mice (BALB/c nude mouse, 6 weeks old) were from Orient Bio Inc. (Jungwon-gu, Gyeonggi-Do, Republic of Korea). Animals were maintained under “specific pathogen-free” conditions, and all animal studies were conducted according to guidelines approved by the Korea Research Institute of Bioscience and Biotechnology-Institutional Animal Care and Use Committee). Mice were housed in climate-controlled quarters with a 12-h light/12-h dark cycle, and all animals were acclimated for 2 weeks before the study and had free access to food and water. Animals were randomly assigned to the following groups: vehicle group (n = 9) and 50 mg/kg compound 69407 group (n = 9). Each mouse in the corresponding group was administered compound 69407 (50 mg/kg body weight in 100 μl of 100% soybean oil as vehicle) or only vehicle three times per week by intraperitoneal injection. After 1 week of treatment, A431 cells (5 × 105) were then injected subcutaneously into the right flank of mice in the respective groups. Following injection, each mouse was administered compound 69407 (50 mg/kg body weight in 100 μl of 100% soybean oil as vehicle) or vehicle only and continued to be administered compound 69407 or vehicle every other day by intraperitoneal injection. The dose of compound 69407 was based on preliminary pilot studies and also extrapolated from cell culture experiments. Mice were weighed and tumors measured by caliper every 3 days. Tumor volume was calculated from measurements of two diameters of the individual tumor according to the following formula: tumor volume (mm3) = (length × width × width/2). Mice were monitored until day 21, and at that time mice were euthanized and tumors extracted.
Statistical Analysis
All quantitative data are expressed as means ± S.E. or S.D. as indicated. The Student's t test or a one-way analysis of variance was used for statistical analysis. A probability of p < 0.05 was used as the criterion for statistical significance.
RESULTS
Compound 69407, a Derivative of the Natural Compound Chrysin, Suppresses EGF-induced Anchorage-independent Growth of JB6 P+ Cells
We first compared the cytotoxicity of chrysin (Fig. 1A) and compound 69407 (Fig. 1B) in JB6 P+ cells. Compound 69407 at concentrations up to 80 μm and exposure times up to 48 h had little effect on cell viability (Fig. 1C). However, chrysin treatment at 20 μm for 48 h began to show cytotoxicity (Fig. 1C), and at 72 h, a significant cytotoxicity could be observed (data not shown). Therefore, the highest dose of chrysin used in additional experiments was 10 μm. When comparing the effect of compound 69407 and chrysin on anchorage-independent cell growth, we found that compound 69407 at 10 μm inhibited EGF-induced neoplastic transformation of JB6 P+ cells by 60% (Fig. 1D). However, the same dose of chrysin inhibited EGF-induced neoplastic transformation of JB6 P+ cells by only 25% (Fig. 1D). These data indicated that compound 69407 is more potent than chrysin at suppressing cell transformation.
FIGURE 1.
Compound 69407 inhibits EGF-induced anchorage-independent growth of JB6 P+ cells. A, chemical structure of chrysin, and B, the chrysin derivative, compound 69407. C, cytotoxicity of compound 69407 or chrysin measured in JB6 P+ cells. JB6 P+ cells were seeded (1 × 104) into 96-well plates in 100 μl of 5% FBS/MEM and incubated in a 37 °C, 5% CO2 incubator overnight. The cells were treated with different doses of compound 69407 (0, 5, 10, 20, 40, 60, or 80 μm) or chrysin (0, 5, 10, 20, 40, 60, or 80 μm) for 24 or 48 h. Viability of JB6 P+ cells was measured as described under “Experimental Procedures.” D, effect of chrysin and its derivative compound 69407 on anchorage-independent growth of JB6 P+ cells stimulated by EGF. JB6 P+ cells (8 × 103/ml) were exposed to mixtures of EGF (10 ng/ml) containing the indicated concentrations of compound 69407 or chrysin in 1 ml of 0.33% basal medium Eagle's agar containing 10% FBS. The cultures were maintained at 37 °C in a 5% CO2 incubator for 7 days, and then cell colonies were counted. Data are represented as means ± S.D. The asterisk indicates a statistically significant (p < 0.01) decrease in number of colonies formed in treated cells compared with untreated control cells. The pound symbol indicates a significant difference between compound 69407 and chrysin-treated groups (p < 0.01).
Effects of Compound 69407 on Cell Cycle Progression and Rb Phosphorylation Levels in EGF-stimulated JB6 P+ Cells
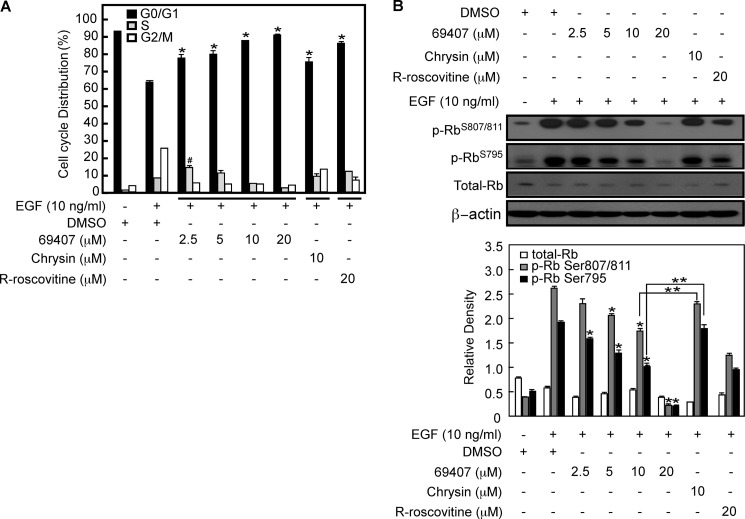
To reveal the mechanism of compound 69407's inhibition of EGF-induced anchorage-independent growth of JB6 P+ cells under the same conditions as for Fig. 1D, the effect of compound 69407 on cell cycle response in EGF-stimulated JB6 P+ cells was evaluated using flow cytometry (Fig. 2A). JB6 P+ cells were synchronized at G0 phase by serum starvation. Cells were treated for 1 h with various concentrations of compound 69407 and then stimulated with 10 ng/ml EGF. Cell cycle distribution analysis showed an accumulation of cells in the G1 phase of the cell cycle. At low concentrations of 2.5 or 5 μm treatment, some cells still could enter the S phase after exposure to EGF. However, cells treated with at least a 10 μm concentration of compound 69407 could not enter S phase after exposure to EGF. Results of cell cycle analysis indicated that compound 69407 causes G1 arrest and inhibits the G1 to S phase transition, which is consistent with Cdk4 and Cdk2 inhibition. Compared with the same dose of compound 69407, the inhibitory effect of 10 μm chrysin on G1 phase and the G1 to S transition was significantly lower. (R)-Roscovitine, a known Cdks inhibitor, showed less potency than that of compound 69407 (Fig. 2A).
FIGURE 2.
Compound 69407 inhibits G1 to S phase transition and Rb phosphorylation levels in EGF-stimulated JB6 P+ cells. A, JB6 P+ cells were serum-starved in 0.1% FBS/MEM for 36 h to synchronize the cells at G0 phase and then treated for 1 h with DMSO or the indicated concentrations of compound 69407, chrysin, or (R)-roscovitine. Cells were then exposed to EGF (10 ng/ml) for 24 h. Trypsinized cells were stained with propidium iodide using the CycleTESTTM PLUS DNA reagent kit (BD Biosciences), and the cell cycle phases were analyzed in a FACSCalibur flow cytometer (BD Biosciences) using the CellQuest Pro (BD Biosciences) software package. The percentage of cells occupying G0/G1, S, or G2/M phase is shown as a bar graph. Data are represented as means ± S.D. of triplicate samples from two independent experiments. The known Cdk inhibitor (R)-roscovitine (CYC202) was used as a positive control. Statistical significance is as follows: #, p < 0.05, and *, p < 0.01 compared with the DMSO plus EGF control (Student's t test). B, JB6 P+ cells were treated using the same protocol as described in A. Rb protein expression and phosphorylation at Ser-795 and Ser-807/811 were analyzed by Western blotting using specific primary and HRP-conjugated secondary antibodies. β-Actin was used as a loading control. Band density was measured using the ImageJ program (National Institutes of Health, version 1.41), and the band intensities of total Rb, phospho-Rb (Ser-795), or phospho-Rb (Ser-807/811) were normalized against β-actin. Statistical significance was as follows: *, p < 0.05 compared with the DMSO plus EGF control. **, p < 0.05, compound 69407-treated group (10 μm) compared with chrysin-treated group (10 μm) (lower panel).
As one of the known natural substrates for Cdks, Rb would be expected to become hypophosphorylated following treatment with compound 69407. Phosphorylation of Rb is triggered by Cdk4/6 and probably completed by Cdk2 as cells enter S phase (35, 36). The phosphorylation of the endogenous Rb protein at Ser-795 is one of the preferred sites phosphorylated by Cdk4/6 (37–39). The phosphorylation of Rb protein at Ser-807/811 is mediated by Cdk2, and these phosphorylations are thought necessary for the G1 to S transition (40, 41). Therefore, we assessed the phosphorylation status of the endogenous Rb protein at both Ser-795 and Ser-807/811. Western blotting results indicated that treatment with compound 69407 resulted in a concentration-dependent loss of Rb phosphorylation at both Ser-795 and Ser-807/811 in EGF-stimulated JB6 P+ cells (Fig. 2B). When comparing the same dose (10 μm) of compound 69407 with chrysin, the result demonstrated that compound 69407 is more potent than chrysin in inhibiting Rb phosphorylation at both Ser-795 and Ser-807/811 in EGF-stimulated JB6 P+ cells (Fig. 2B, p < 0.05), which corresponds with the FACS results (Fig. 2A). Moreover, results of real time PCR (supplemental Table 1) and Western blotting analysis showed that the expression of Cdk2, Cdk4, Cdk6, or upstream Cdk-activating kinase Cdk7, in response to EGF stimulation, was unaffected by compound 69407 treatment (Fig. 3, A and B). These results indicate that treatment with compound 69407 causes inhibition of Cdk2 and Cdk4/6 downstream substrate phosphorylation in EGF-stimulated JB6 P+ cells.
FIGURE 3.

Effect of compound 69407 on cell cycle-regulated genes in EGF-stimulated JB6 P+ cells. JB6 P+ cells were serum-starved in 0.1% FBS/MEM for 36 h and treated for 1 h with DMSO or the indicated concentrations of compound 69407 and then exposed to 10 ng/ml EGF for 24 h. Expression of mRNAs and proteins was analyzed by real time RT-PCR (A) and Western blotting (B), respectively. Data are shown as means ± S.D. and significant differences were determined by t test. *, p < 0.05, compared with the DMSO plus EGF control.
Compound 69407 Inhibits Endogenous Cdk2 and Cdk4 Kinase Activities in EGF-stimulated JB6 P+ Cells
Treatment with compound 69407 inhibits the phosphorylation levels of both Cdk2 and Cdk4/6 downstream substrates in EGF-stimulated JB6 P+ cells, which suggested that compound 69407 might directly target Cdk2 and Cdk4/6. To test this hypothesis, immunoprecipitation kinase assays were performed to assess endogenous Cdk2 and Cdk4 activities in EGF-stimulated JB6 P+ cells. Cdk6 was excluded because compound 69407 had no effect on Cdk6 activity in vitro (Fig. 5C). Using the same protocol as described for Fig. 2A, cells were harvested, and lysates were immunoprecipitated with a Cdk2 or Cdk4 antibody. The immunocomplexes were then incubated with histone H1 (for Cdk2) or a recombinant Rb protein (773–928 amino acids; for Cdk4) in the presence of [γ-32P] ATP. Cdk4-associated kinase assay results indicated that Cdk4 activity was dose-dependently inhibited by compound 69407. However, chrysin at 10 μm had no effect on Cdk4 activity, and (R)-roscovitine at 20 μm had little effect (Fig. 4A). Cdk2-associated kinase assay results indicated that Cdk2 activity was dose-dependently inhibited by compound 69407. However, neither chrysin at 10 μm nor (R)-roscovitine at 20 μm had an effect on Cdk2 activity (Fig. 4B). Because Cdks form active complexes with their respective cyclins (e.g. Cdk2 with cyclin E or cyclin A, and Cdk4 with cyclin D), the effect of compound 69407 on cyclin-associated kinase activities was examined. Results (Fig. 4, C and D) indicated that compound 69407 dose-dependently inhibited cyclin D1-associated and cyclin A-associated kinase activities and, chrysin at 10 μm had no effect, which is consistent with the Cdk4- and Cdk2-associated kinase assay results. Additionally, co-immunoprecipitation data confirmed that cyclin D1 and cyclin A antibodies imunoprecipitated the active complexes efficiently (Fig. 4, E and F).
FIGURE 5.
Compound 69407 inhibits Cdk2 and Cdk4 kinase activities in vitro and binds with Cdk2 and Cdk4 in an ATP-noncompetitive manner. A, compound 69407 inhibits Cdk2 kinase activity. The inhibitory effect of compound 69407 on Cdk2 kinase activity was assessed by an in vitro kinase assay using a recombinant histone H1 protein as substrate and [γ-32P]ATP. The 32P-labeled histone H1 was visualized by autoradiography. B, compound 69407 inhibits Cdk4 kinase activity. The inhibitory effect of compound 69407 on Cdk4 kinase activity was assessed by an in vitro kinase assay using a recombinant Rb protein (773–928 amino acids) and [γ-32P]ATP. The 32P-labeled Rb protein was visualized by autoradiography. C, Cdk6 kinase activity is unaffected by compound 69407. The effect of compound 69407 on Cdk6 kinase activity was assessed by an in vitro kinase assay using a recombinant Rb protein (773–928 amino acids) as substrate and [γ-32P]ATP. The 32P-labeled Rb proteins were visualized by autoradiography. D, Cdk7 kinase activity is unaffected by compound 69407. The effect of compound 69407 on Cdk7 kinase activity was assessed by an in vitro kinase assay using a recombinant MBP as substrate and [γ-32P]ATP. The 32P-labeled MBPs were visualized by autoradiography. Data shown are representative of two independent experiments. Band density was measured using the ImageJ program (National Institutes of Health version 1.41), and the band intensities of active Cdk2 and histone H1 (A, lower panel), active Cdk4 and Rb protein (B, lower panel), active Cdk6 and Rb protein (C, lower panel), or active Cdk7 and MBP (D, lower panel) were compared. E, compound 69407 binds with Cdk2 in vitro. The active Cdk2-cyclin E complex (100 ng) was subjected to a pulldown assay with compound 69407-conjugated CNBr-Sepharose 4B beads. Compound 69407 binding with Cdk2 was visualized by Western blotting (WB) with anti-Cdk2. F, compound 69407 binds with Cdk4 in vitro. The active Cdk4-cyclin D1 complex (100 ng) was subjected to a pulldown assay with compound 69407-conjugated CNBr-Sepharose 4B beads. Compound 69407 binding of Cdk4 was visualized by Western blotting with anti-Cdk4. G, compound 69407 binds with Cdk2 ex vivo. The cellular protein fraction (500 μg) of JB6 P+ cells was used for the pulldown assay with CNBr-DMSO or CNBr-compound 69407-conjugated beads. The Cdk2 proteins pulled down were visualized by Western blotting with anti-Cdk2. H, compound 69407 binds with Cdk4 ex vivo. The cellular protein fraction (500 μg) of JB6 P+ cells was used for the pulldown assay with CNBr-DMSO or CNBr-compound 69407-conjugated beads. The Cdk4 proteins pulled down were visualized by Western blotting with anti-Cdk4. I and J, chrysin does not bind to Cdk2 and Cdk4 ex vivo. Protein fractions (500 μg) of JB6 P+ cells were used for a pulldown assay with CNBr-DMSO or CNBr-chrysin-conjugated beads. The pulled down Cdk2 (I) and Cdk4 (J) proteins were visualized by Western blotting with anti-Cdk2 and anti-Cdk4, respectively. K and L, compound 69407 binds to Cdk2 in an ATP noncompetitive manner. Active Cdk2/cyclin E (K) or GST-Cdk2 (L) was incubated with ATP at different concentrations (0, 0.01, 0.1, or 1 mm) and compound 69407-conjugated Sepharose 4B beads or with Sepharose 4B beads alone (as a negative control) in reaction buffer. The pulled down proteins were analyzed by Western blotting. M, compound 69407 binds to Cdk4 in an ATP-noncompetitive manner. The active Cdk4-cyclin D1 complex was incubated with ATP at different concentrations (0, 0.01, 0.1, or 1 mm) and compound 69407-conjugated Sepharose 4B beads or with Sepharose 4B beads alone (as a negative control) in reaction buffer. The pulled down proteins were analyzed by Western blotting. Results shown are representative of at least two independent experiments.
FIGURE 4.
Compound 69407 inhibits endogenous Cdk2 and Cdk4 kinase activities in EGF-stimulated JB6 P+ cells. JB6 P+ cells were serum-starved in 0.1% FBS/MEM for 36 h and treated for 1 h with DMSO or the indicated concentrations of compound 69407, chrysin, or (R)-roscovitine and then exposed to EGF (10 ng/ml) for 24 h. Cell lysates were subjected to IP using the appropriate antibodies. A, cell lysates were immunoprecipitated with anti-Cdk4 and subjected to an in vitro kinase assay using a recombinant Rb protein (773–928 amino acids) as substrate. The phosphorylated Rb protein from the reaction was resolved by SDS-PAGE, and the blots were developed by autoradiography. B, cell lysates were immunoprecipitated with anti-Cdk2 and subjected to a histone H1 kinase assay. Phosphorylated histone H1 was electrophoretically separated by SDS-PAGE and subjected to autoradiography. C, cell lysates were immunoprecipitated with anti-cyclin D1 and subjected to an in vitro kinase assay using a recombinant Rb protein (773–928 amino acids) as substrate. Phosphorylated Rb proteins from the reactions were resolved by SDS-PAGE, and the blots were developed by autoradiography. D, cell lysates were immunoprecipitated with anti-cyclin A and subjected to a histone H1 kinase assay. Phosphorylated histone H1 was electrophoretically separated by SDS-PAGE and subjected to autoradiography. E, co-immunoprecipitation of endogenous cyclin D1 and Cdk4. Cell lysates were immunoprecipitated with anti-cyclin D1 or a normal IgG antibody. The immune complexes and the input were analyzed by immunoblotting with a Cdk4 antibody. F, co-immunoprecipitation of endogenous cyclin A and Cdk2. Cell lysates were immunoprecipitated with anti-cyclin A or a normal IgG antibody. The immune complexes and input were analyzed by immunoblotting with a Cdk2 antibody. WB, Western blot; CB, Coomassie blue staining.
Compound 69407 Inhibits Cdk2 and Cdk4 Kinase Activities in Vitro and Binds with Cdk2 and Cdk4 in Vitro and Ex Vivo
Treatment with compound 69407 causes G1 arrest and inhibits the G1 to S phase transition as well as attenuates the phosphorylation levels of both Cdk2 and Cdk4/6 downstream substrates in EGF-stimulated JB6 P+ cells. This suggested that Cdk2 and Cdk4/6 might be molecular targets of compound 69407 for the inhibition of cell transformation. Results of in vitro kinase assays indicated that compound 69407 inhibited Cdk2 (Fig. 5A) and Cdk4 (Fig. 5B) kinase activities with IC50 values of 8.3 and 8.4 μm, respectively, compared with those for (R)-roscovitine at 0.7 μm for Cdk2 (42) and 15.3 μm for Cdk4 (43). However, Cdk6 kinase activity was not obviously changed by treatment with compound 69407 (Fig. 5C). To determine whether compound 69407 specifically inhibits Cdk2 and Cdk4 kinase activities, we investigated the effect of compound 69407 on the kinase activity of the upstream Cdk-activating kinase, Cdk7. Compound 69407 had no effect on Cdk7 kinase activity in vitro (Fig. 5D). Because compound 69407 attenuates Cdk2 and Cdk4 kinase activities, we conducted pulldown assays to determine whether this compound interacts directly with Cdk2 or Cdk4 kinase. Results indicated that Cdk2 directly binds to compound 69407-conjugated Sepharose 4B beads, but it does not bind to the Sepharose 4B beads alone (Fig. 5E). A direct interaction between compound 69407 with Cdk4 was also observed (Fig. 5F). Additionally, we confirmed in cell lysates that this compound bound with endogenous Cdk2 (Fig. 5G) or Cdk4 (Fig. 5H). In contrast, chrysin did not bind with either Cdk2 or Cdk4 (Fig. 5, I and J), suggesting that such modification may increase its binding affinity with its target protein. To further investigate the binding of compound 69407 with Cdk2 and Cdk4, we performed a pulldown assay with increasing concentrations of ATP. The binding ability of compound 69407 with Cdk2 was not altered by increasing the ATP concentration (Fig. 5, K and L), suggesting that compound 69407 did not compete with ATP for binding to Cdk2. Furthermore, compound 69407 also did not compete with ATP for binding to Cdk4 (Fig. 5M). These results showed that compound 69407 is an ATP-noncompetitive inhibitor that reduces Cdk2 and Cdk4 kinase activities.
Molecular Modeling of the Binding of Compound 69407 Inside the Cdk2 Allosteric Binding Site
To assess the possible binding mode between compound 69407 and Cdks, the hierarchical docking algorithm Glide (Glide version 5.7) (33) in the Schrödinger package was used for docking experiments. Currently, 268 crystal structures of Cdk2 are available, which is many more than for other Cdks, and therefore, it is the most likely Cdk2 to use for building an accurate binding model with compound 69407. To capture the ligand-induced conformational changes in the receptor allosteric binding site, we performed flexible-ligand flexible-protein docking by using the induced fit docking module (Schrödinger Suite 2011 Induced Fit Docking protocol; Glide version 5.7; Prime version 3.0). The binding pose of compound 69407-Cdk2 obtained from the induced fit docking results suggested that compound 69407 bound inside the Cdk2 allosteric binding site (Fig. 6A), which was similar to that for two 1-anilino-8-naphthalene sulfonate molecules that bound at a region away from the ATP site and located in a large pocket that extends from the DFG region above the C-helix (26). Compound 69407 formed five hydrogen bonds with Cdk2. Four bonds were involved in the backbone atoms of three residues, Phe-146, Val-64, and Leu-58, and the other hydrogen bond was formed with the side chain atoms (i.e. hydroxyl group) of Thr-14 (Fig. 6B). In addition, 11 residues around the allosteric site, including Val-69, Leu-78, Leu-66, Phe-80, Val-64, Leu-58, Phe-146, Leu-55, Leu-148, Ile-52, and Phe-152, formed hydrophobic interactions with the carbons of compound 69407 (Fig. 6C). Specifically, Leu-55 showed a very strong hydrophobic interaction with compound 69407. The side chains of Leu-55 were closely packed with the three phenyl rings of compound 69407 (Fig. 6D). Therefore, Leu-55 should play an important role in the binding of compound 69407 with Cdk2. Experimental results confirmed this idea because mutation of Leu-55 to Arg-55 significantly decreased the binding ability (Fig. 6E). However, the side chains of Lys-33 and Tyr-15 were a little bulky for accommodating compound 69407 nearby (Fig. 6D). Thus, a mutation of either residue to alanine could increase the binding affinity slightly (Fig. 6E). The computational results were in good agreement with the experimental results. Moreover, immunoblotting results indicated that compound 69407 had no effect on the activities of numerous other kinases (Fig. 7). This supports the hypothesis that compound 69407 functions as a Cdk inhibitor at a site other than the ATP pocket, which is well conserved in all kinases. These results indicated that compound 69407 binds to a Cdk2 allosteric site, suggesting that compound 69407 might be a type III inhibitor with ATP noncompetitive inhibitory effects on Cdks.
FIGURE 6.
Computational modeling of the binding of compound 69407 inside the Cdk2 allosteric binding site. A, binding pose of compound 69407 inside the allosteric binding site of Cdk2. B, hydrogen bonds formed between compound 69407 and four residues in the allosteric site. C, hydrophobic interactions between compound 69407 and 11 residues of Cdk2 (for clarity, the oxygens, nitrogens, and hydrogens of the protein residues are not shown). D, function of protein residues, Lys-33, Tyr-15, and Leu-55, in the docking of compound 69407 to Cdk2. Note: the α-helices are drawn as cylinders and the β-strands as arrows. Compound 69407 is shown in stick model, and protein residues are shown in line model. In addition, Leu-55 in D is shown in CPK model. The figures were generated using Maestro version 9.2. E, Leu-55 plays an important role in the binding of compound 69407 inside the Cdk2 allosteric binding site. To confirm the predicted model of the Cdk2-compound 69407 complex, Cdk2 proteins harboring mutations at Tyr-15, Lys-33, Leu-55, or Leu-55/Asn-59 were expressed in the BL21 bacterial strain, and the purified GST, GST-Cdk2, and Cdk2 mutants were incubated with compound 69407-conjugated Sepharose 4B beads or with Sepharose 4B beads alone (as a negative control) in reaction buffer. The pulled down proteins were analyzed by Western blotting (WB) with anti-GST (top), and Coomassie Blue (C.B.) staining was used to verify equal loading (middle). Similar results were obtained from two independent experiments. Band density was measured using the ImageJ program (Version 1.41) (bottom). Data are shown as means ± S.D., and significant differences were determined by t test. #, p < 0.01, compared with the wild type Cdk2.
FIGURE 7.

Inhibitory effect of compound 69407 on other signaling pathways in EGF-stimulated JB6 P+ cells. JB6 P+ cells were serum-starved in 0.1% FBS/MEM for 36 h and treated for 1 h with DMSO or indicated concentrations of compound 69407, and then exposed to 10 ng/ml EGF for 30 min. Individual levels of phosphorylated and total proteins were visualized by Western blotting with specific antibodies. β-Actin was used as an internal control to verify equal protein loading.
Compound 69407 Inhibits Human Epidermoid Carcinoma Cell Growth and Is Dependent on Cdk2 and Cdk4
Both JB6 P+ cells and A431 human epidermoid carcinoma cells are EGF receptor-overexpressing cells that are very responsive to EGF stimulation (44, 45). Therefore, we further determined whether compound 69407 could suppress the growth of the A431 human epidermoid carcinoma cell line. The experimental data indicated that compound 69407 inhibited A431 proliferation (Fig. 8A) and anchorage-independent cell growth (Fig. 8B) more potently than chrysin. In addition, we found that transient knockdown of endogenous Cdk2 and Cdk4 by si-Cdk2 and si-Cdk4 (Fig. 8C) inhibited A431 cell growth (Fig. 8D), indicating that both Cdk2 and Cdk4 are required for A431 cell proliferation. Furthermore, A431 cell viability was decreased more effectively by compound 69407 in si-Control cells compared with si-Cdk2 or si-Cdk4 cells (Fig. 8E). These findings showed that the anticancer activity induced by compound 69407 is dependent on Cdk2 and Cdk4.
FIGURE 8.
Compound 69407 suppresses the growth of A431 human epidermoid carcinoma cells in a Cdk2- and Cdk4-dependent manner. A, compound 69407 suppresses anchorage-dependent growth of A431 cells. Cell proliferation was estimated by WST-1 assay. Absorbance was read at 24-h intervals up to 96 h. Data are represented as means ± S.D. of three separate determinations, and significant differences were determined by t test. *, p < 0.05, and #, p < 0.01, compared with the DMSO group. **, p < 0.05, comparing compound 69407-treated group (10 μm) with chrysin-treated group (10 μm). B, compound 69407 inhibits anchorage-independent growth of A431 cells. Data are shown as means ± S.D. of two separate determinations, and significant differences were determined by t test. *, p < 0.05, and #, p < 0.01, compared with the DMSO group. **, p < 0.01, comparing compound 69407-treated group (10 μm) with chrysin-treated group. C, growing A431 cells were transiently transfected with the indicated siRNAs and cultured for 72 h as described under “Experimental Procedures.” Knockdown of Cdk2 or Cdk4 was analyzed by Western blotting. The results shown are representative of three independent experiments. β-Actin was used as a loading control. D, A431 cells were transfected with scrambled, Cdk2, or Cdk4 siRNA in 6-well plates as described under “Experimental Procedures” for 24 h, trypsinized, and transferred to a 96-well plate (3 × 103 cells per well) in 100 μl of complete DMEM. After culturing for another 24, 48, 72, or 96 h, 10 μl of WST-1 was added to each well, and cells were incubated for 2 h at 37 °C. Absorbance was read at 24-h intervals. Data are represented as means ± S.D. of three separate determinations, and significant differences were determined by t test. #, p < 0.01, compared with the si-Ctrl group. E, inhibition of A431 cell growth induced by compound 69407 is less apparent in knockdown Cdk2 or Cdk4 cells. A431 cells were transfected transiently for 24 h with si-Ctrl, si-Cdk2, or si-Cdk4, trypsinized, and transferred to a 96-well plate (3 × 103 cells per well) in 100 μl of complete DMEM without or with compound 69407 (20 μm). Cells were incubated for 72 h, and growth was determined by WST-1 assay. Data are represented means ± S.D. of three separate determinations, and significant differences were determined by t test. *, p < 0.05, and #, p < 0.01, compared with the DMSO group.
Compound 69407 Inhibits the Growth of Human Epidermoid Carcinoma Cells in a Xenograft Mouse Model
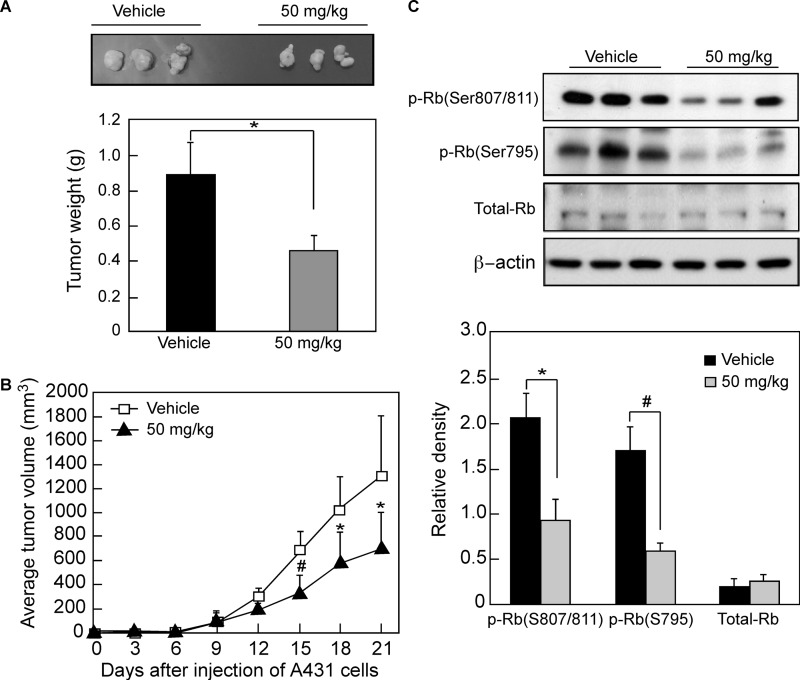
We next determined whether compound 69407 could suppress tumor growth in vivo. The A431 cell line was chosen as a model for evaluation of the in vivo compound efficacy based on the data shown above. Compound 69407 was injected into the right flank of individual athymic nude mice. The results showed that the mean tumor weight was decreased in the compound 69407-treated group (Fig. 9A), and the mean tumor volume in the vehicle-treated group increased faster than that in the compound 69407-treated group (Fig. 9B). The body weights of compound 69407- or vehicle-treated groups were similar throughout the study (data not shown). Tumor extracts from vehicle-treated and compound 69407-treated mice (i.e. euthanized on the same day of the experiment) were prepared, and phosphorylation of Rb (Ser-795 and Ser-807/811) was analyzed. Western blotting analysis revealed that the compound 69407-treated tumor extracts exhibited substantially decreased Rb phosphorylation at both Ser-795 and Ser-807/811 compared with vehicle-treated tumors (Fig. 9C). The protein levels of Rb between the two groups remained unchanged (Fig. 9C). These data suggested that A431 human epidermoid carcinoma development was suppressed by compound 69407 through the inhibition of Cdk2 and Cdk4 kinase activities.
FIGURE 9.
Compound 69407 suppresses tumor growth by inhibiting CDK activity. A, total average tumor weight in the compound 69407-treated group is significantly less than that of the vehicle-treated group. Tumors were extracted and weighed after mice were sacrificed. Data are shown as means ± S.D., and significant differences were determined by one-way analysis of variance. *, p < 0.05, compared with the vehicle group. B, total average tumor volume in the compound 69407-treated group increased significantly less compared with the vehicle-treated group. Tumor volume was measured and recorded every 3 days for the duration of the study. Data are shown as means ± S.D., and significant differences were determined by one-way analysis of variance. *, p < 0.05, and #, p < 0.01, compared with the vehicle group. C, expression of Rb and phosphorylated Rb (Ser-795 and Ser-807/811) was assessed by Western blotting in vehicle- and compound 69407-treated tumor tissues. β-Actin was as a loading control. Data are represented as means ± S.D. of three separate experiments, and significant differences were determined by t test. *, p < 0.05, and #, p < 0.01, compared with the vehicle-treated group.
DISCUSSION
In this study, a chrysin derivative, compound 69407, was synthesized and the activity compared with that of its prototype, chrysin. Our results showed that compound 69407 induces G0/G1 cell cycle arrest and inhibits the G1/S transition in quiescent JB6 P+ cells stimulated with EGF (Fig. 2A). Because the progression of cells through G1 phase is regulated by Cdk4/6 complexes with cyclin D and the transition from G1 into S phase is regulated by Cdk2 in complex with cyclin E, and the transition through S phase is regulated by Cdk2 in complex with cyclin A (46, 47), we further analyzed kinase activities under the same conditions. Our results showed that the G0/G1 arrest and the inhibition of the G1/S transition by compound 69407 in quiescent JB6 P+ cells stimulated with EGF were associated with the attenuation of endogenous Cdk4 and Cdk2 activities (Fig. 4). The results supported the idea that compound 69407 is a Cdk inhibitor. Furthermore, results showed that compound 69407 exhibited a more potent inhibitory effect than that of chrysin and appeared to exert its effect by targeting Cdk2 and Cdk4. Importantly, compound 69407 was less toxic than the parent compound. The results suggested that inhibition of Cdk4, which controls the entrance into the cell cycle, and Cdk2, which controls the cell through S phase, might be important molecular targets explaining the antitumor promoting effects of compound 69407.
Key checkpoint targets in G1/S arrest are thought to be the G1/S Cdks, including Cdk2, Cdk4, and Cdk6, which activate the G1/S transition by phosphorylating the Rb protein. This results in the release of the E2F transcription factors, which promotes the transcription of genes required for the transition. Activation of E2F-dependent transcription is believed to be one of the primary cell cycle-related functions of the G1/S Cdks (48). When evaluating whether compound 69407 affects cell cycle-related Cdk and cyclin abundance, the results of real time PCR and Western blotting analysis showed that compound 69407 significantly attenuates the expression of Cdk1 in response to EGF stimulation at both mRNA and protein levels, whereas Cdk2, Cdk4, Cdk6, or Cdk7 remain unaffected. The EGF-induced cyclin A and cyclin B expression was also reduced at both the mRNA and protein levels. Cyclin E was also affected, but to a lesser extent (Fig. 3, A and B). Because E2F-binding sites are found in the promoters of many genes, including the Cdk1, cyclin E, cyclin A, and cyclin B promoters (49–51), we believe this effect to be an indirect rather than a direct effect. This observation might reflect the fact that initiation of DNA replication is regulated through multiple coherent pathways. Re-replication prior to mitosis might require over-riding more than one control mechanism (52, 53).
The allosteric pocket in Cdk2 is relatively unique compared with allosteric sites in other protein kinases. Docking experimental results showed that compound 69407 binds away from the ATP site and locates itself in a large pocket that extends from the DFG region above the C-helix (Fig. 6A). In contrast, allosteric sites in other protein kinases, such as the Abl-Imatinib complex or the MEK1-PD318088-ATP complex, extend along the DFG region underneath the C-helix. In addition, the residues of Cdk2 around the allosteric site include Val-69, Leu-78, Leu-66, Phe-80, Val-64, Leu-58, Phe-146, Leu-55, Leu-148, Ile-52, and Phe-152, which form hydrophobic interactions with the carbons of compound 69407 (Fig. 6, B and C). When performing multisequence alignments for Cdk2 and Cdk1, and -3–7, which are the Cdks involved in cell cycle control (54), results indicated that residues comprising the allosteric pocket are largely conserved among Cdk1 through Cdk7 (data not shown). These characteristics of the allosteric pocket in Cdk2 may provide an opportunity for developing a general pharmacophore model by which to design a new generation of inhibitors selectively targeting Cdk2 and possibly other Cdks as well, but not other kinases.
We found that compound 69407 showed a higher binding affinity with its target proteins compared with its progenitor, chrysin (Figs. 5 and 6). Similarly, an analog of resveratrol, RSVL2, was shown to strongly bind MEK1, whereas resveratrol bound only very weakly (55). This was also true of a derivative of indole 3-carbinol, 3CAI, which bound strongly to recombinant Akt1 and Akt2, whereas indole 3-carbinol showed no binding (23). In addition, compared with resveratrol and quercetin, the addition of the one or two hydroxyl groups in the resveratrol analog RSVL2 and in myrcetin, respectively, seemed to increase their binding affinity with MEK (56). These results support the idea that compounds with only a small difference in molecular structure can lead to substantial differences in the target protein and a subtle difference in phytochemical structure, such as the addition of hydroxyl groups, will affect the binding affinity of a compound with a target protein (56). This raises the possibility of increasing the affinity between a compound and its target and lowering unwanted side effects by modifying the natural phytochemical structure.
In conclusion, we found that compound 69407, a derivative of chrysin, is an effective agent for inhibiting neoplastic transformation and tumor growth and acts by targeting Cdk2 and Cdk4 in an ATP-noncompetitive manner. The possible mechanism by which ATP did not compete with compound 69407 for binding to Cdk2 is that compound 69407 binds inside the Cdk2 allosteric pocket. This study provides information for deriving a general pharmacophore model through which the design and development of the new ATP-noncompetitive agents with chemopreventive or chemotherapeutic potency can be performed on the basis of prototype phytochemicals.
Acknowledgment
We thank Dr. Robert Huber (Chemistry Nobel Prize winner in 1988) for the enlightening discussion on the molecular modeling of the Cdk2 compound 69407 complex.
This work was supported, in whole or in part, by National Institutes of Health Grants CA120388, CA1666011, R37 CA081064, CA172457, and ES016548. This work was also supported by The Hormel Foundation, National Research Foundation of Korea Grant 2010-0029233 funded by the Korea Government (MSIFP), Leap Research Program Grant 2010-0029233), and World Class Institute Program (WCI 2009-002) funded by the Ministry of Science, ICT and Future Planning, Korea.

This article contains supplemental Tables S1 and S2, Methods, and additional references.
- CDK
- cyclin-dependent kinase
- MBP
- myelin basic protein
- IP
- immunoprecipitation
- MEM
- minimum essential medium
- Rb
- retinoblastoma.
REFERENCES
- 1. Lee K. W., Kang N. J., Heo Y. S., Rogozin E. A., Pugliese A., Hwang M. K., Bowden G. T., Bode A. M., Lee H. J., Dong Z. (2008) Raf and MEK protein kinases are direct molecular targets for the chemopreventive effect of quercetin, a major flavonol in red wine. Cancer Res. 68, 946–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kang N. J., Lee K. W., Lee D. E., Rogozin E. A., Bode A. M., Lee H. J., Dong Z. (2008) Cocoa procyanidins suppress transformation by inhibiting mitogen-activated protein kinase kinase. J. Biol. Chem. 283, 20664–20673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bode A. M., Ma W. Y., Surh Y. J., Dong Z. (2001) Inhibition of epidermal growth factor-induced cell transformation and activator protein 1 activation by [6]-gingerol. Cancer Res. 61, 850–853 [PubMed] [Google Scholar]
- 4. He Z., Tang F., Ermakova S., Li M., Zhao Q., Cho Y. Y., Ma W. Y., Choi H. S., Bode A. M., Yang C. S., Dong Z. (2008) Fyn is a novel target of (−)-epigallocatechin gallate in the inhibition of JB6 Cl41 cell transformation. Mol. Carcinog. 47, 172–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bode A. M., Dong Z. (2006) Molecular and cellular targets. Mol. Carcinog. 45, 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malumbres M., Barbacid M. (2001) To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer 1, 222–231 [DOI] [PubMed] [Google Scholar]
- 7. Zou X., Ray D., Aziyu A., Christov K., Boiko A. D., Gudkov A. V., Kiyokawa H. (2002) Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev. 16, 2923–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haas K., Staller P., Geisen C., Bartek J., Eilers M., Möröy T. (1997) Mutual requirement of CDK4 and Myc in malignant transformation: evidence for cyclin D1/CDK4 and p16INK4A as upstream regulators of Myc. Oncogene 15, 179–192 [DOI] [PubMed] [Google Scholar]
- 9. Lazarov M., Kubo Y., Cai T., Dajee M., Tarutani M., Lin Q., Fang M., Tao S., Green C. L., Khavari P. A. (2002) CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat. Med. 8, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 10. Berthet C., Aleem E., Coppola V., Tessarollo L., Kaldis P. (2003) Cdk2 knockout mice are viable. Curr. Biol. 13, 1775–1785 [DOI] [PubMed] [Google Scholar]
- 11. Ortega S., Prieto I., Odajima J., Martín A., Dubus P., Sotillo R., Barbero J. L., Malumbres M., Barbacid M. (2003) Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35, 25–31 [DOI] [PubMed] [Google Scholar]
- 12. Sotillo E., Garriga J., Kurimchak A., Graña X. (2008) Cyclin E and SV40 small T antigen cooperate to bypass quiescence and contribute to transformation by activating CDK2 in human fibroblasts. J. Biol. Chem. 283, 11280–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Woo K. J., Jeong Y. J., Inoue H., Park J. W., Kwon T. K. (2005) Chrysin suppresses lipopolysaccharide-induced cyclooxygenase-2 expression through the inhibition of nuclear factor for IL-6 (NF-IL6) DNA-binding activity. FEBS Lett. 579, 705–711 [DOI] [PubMed] [Google Scholar]
- 14. Zhang T., Chen X., Qu L., Wu J., Cui R., Zhao Y. (2004) Chrysin and its phosphate ester inhibit cell proliferation and induce apoptosis in HeLa cells. Bioorg. Med. Chem. 12, 6097–6105 [DOI] [PubMed] [Google Scholar]
- 15. Galijatovic A., Walle U. K., Walle T. (2000) Induction of UDP-glucuronosyltransferase by the flavonoids chrysin and quercetin in Caco-2 cells. Pharm. Res. 17, 21–26 [DOI] [PubMed] [Google Scholar]
- 16. Otake Y., Hsieh F., Walle T. (2002) Glucuronidation versus oxidation of the flavonoid galangin by human liver microsomes and hepatocytes. Drug Metab. Dispos. 30, 576–581 [DOI] [PubMed] [Google Scholar]
- 17. Li H. Q., Shi L., Li Q. S., Liu P. G., Luo Y., Zhao J., Zhu H. L. (2009) Synthesis of C(7) modified chrysin derivatives designing to inhibit β-ketoacyl-acyl carrier protein synthase III (FabH) as antibiotics. Bioorg. Med. Chem. 17, 6264–6269 [DOI] [PubMed] [Google Scholar]
- 18. Lv P. C., Wang K. R., Li Q. S., Chen J., Sun J., Zhu H. L. (2010) Design, synthesis, and biological evaluation of chrysin long-chain derivatives as potential anticancer agents. Bioorg. Med. Chem. 18, 1117–1123 [DOI] [PubMed] [Google Scholar]
- 19. Zheng X., Meng W. D., Xu Y. Y., Cao J. G., Qing F. L. (2003) Synthesis and anticancer effect of chrysin derivatives. Bioorg. Med. Chem. Lett. 13, 881–884 [DOI] [PubMed] [Google Scholar]
- 20. Cheng H., Force T. (2010) Molecular mechanisms of cardiovascular toxicity of targeted cancer therapeutics. Circ. Res. 106, 21–34 [DOI] [PubMed] [Google Scholar]
- 21. Ohren J. F., Chen H., Pavlovsky A., Whitehead C., Zhang E., Kuffa P., Yan C., McConnell P., Spessard C., Banotai C., Mueller W. T., Delaney A., Omer C., Sebolt-Leopold J., Dudley D. T., Leung I. K., Flamme C., Warmus J., Kaufman M., Barrett S., Tecle H., Hasemann C. A. (2004) Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 11, 1192–1197 [DOI] [PubMed] [Google Scholar]
- 22. Lamba V., Ghosh I. (2012) New directions in targeting protein kinases: focusing upon true allosteric and bivalent inhibitors. Curr. Pharm. Des. 18, 2936–2945 [DOI] [PubMed] [Google Scholar]
- 23. Kim D. J., Reddy K., Kim M. O., Li Y., Nadas J., Cho Y. Y., Kim J. E., Shim J. H., Song N. R., Carper A., Lubet R. A., Bode A. M., Dong Z. (2011) (3-Chloroacetyl)-indole, a novel allosteric AKT inhibitor, suppresses colon cancer growth in vitro and in vivo. Cancer Prev. Res. 4, 1842–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okram B., Nagle A., Adrián F. J., Lee C., Ren P., Wang X., Sim T., Xie Y., Wang X., Xia G., Spraggon G., Warmuth M., Liu Y., Gray N. S. (2006) A general strategy for creating “inactive-conformation” abl inhibitors. Chem. Biol. 13, 779–786 [DOI] [PubMed] [Google Scholar]
- 25. Corsino P., Horenstein N., Ostrov D., Rowe T., Law M., Barrett A., Aslanidi G., Cress W. D., Law B. (2009) A novel class of cyclin-dependent kinase inhibitors identified by molecular docking act through a unique mechanism. J. Biol. Chem. 284, 29945–29955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Betzi S., Alam R., Martin M., Lubbers D. J., Han H., Jakkaraj S. R., Georg G. I., Schönbrunn E. (2011) Discovery of a potential allosteric ligand binding site in CDK2. ACS Chem. Biol. 6, 492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee D. E., Lee K. W., Song N. R., Seo S. K., Heo Y. S., Kang N. J., Bode A. M., Lee H. J., Dong Z. (2010) 7,3′,4′-Trihydroxyisoflavone inhibits epidermal growth factor-induced proliferation and transformation of JB6 P+ mouse epidermal cells by suppressing cyclin-dependent kinases and phosphatidylinositol 3-kinase. J. Biol. Chem. 285, 21458–21466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lun X. Q., Zhou H., Alain T., Sun B., Wang L., Barrett J. W., Stanford M. M., McFadden G., Bell J., Senger D. L., Forsyth P. A. (2007) Targeting human medulloblastoma: oncolytic virotherapy with myxoma virus is enhanced by rapamycin. Cancer Res. 67, 8818–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Colburn N. H., Wendel E. J., Abruzzo G. (1981) Dissociation of mitogenesis and late-stage promotion of tumor cell phenotype by phorbol esters: mitogen-resistant variants are sensitive to promotion. Proc. Natl. Acad. Sci. U.S.A. 78, 6912–6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cho Y. Y., Yao K., Pugliese A., Malakhova M. L., Bode A. M., Dong Z. (2009) A regulatory mechanism for RSK2 NH(2)-terminal kinase activity. Cancer Res. 69, 4398–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng D., Cho Y. Y., Lau A. T., Zhang J., Ma W. Y., Bode A. M., Dong Z. (2008) Cyclin-dependent kinase 3-mediated activating transcription factor 1 phosphorylation enhances cell transformation. Cancer Res. 68, 7650–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van den Heuvel S., Harlow E. (1993) Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262, 2050–2054 [DOI] [PubMed] [Google Scholar]
- 33. Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 34. Irwin J. J., Shoichet B. K. (2005) ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng M., Olivier P., Diehl J. A., Fero M., Roussel M. F., Roberts J. M., Sherr C. J. (1999) The p21(Cip1) and p27(Kip1) CDK “inhibitors” are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18, 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Su T. T., Stumpff J. (2004) Promiscuity rules? The dispensability of cyclin E and Cdk2. Sci. STKE 2004, pe11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Connell-Crowley L., Harper J. W., Goodrich D. W. (1997) Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell 8, 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo J., Sheng G., Warner B. W. (2005) Epidermal growth factor-induced rapid retinoblastoma phosphorylation at Ser-780 and Ser-795 is mediated by ERK1/2 in small intestine epithelial cells. J. Biol. Chem. 280, 35992–35998 [DOI] [PubMed] [Google Scholar]
- 39. Kitagawa M., Higashi H., Jung H. K., Suzuki-Takahashi I., Ikeda M., Tamai K., Kato J., Segawa K., Yoshida E., Nishimura S., Taya Y. (1996) The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15, 7060–7069 [PMC free article] [PubMed] [Google Scholar]
- 40. Brugarolas J., Moberg K., Boyd S. D., Taya Y., Jacks T., Lees J. A. (1999) Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G1 arrest after γ-irradiation. Proc. Natl. Acad. Sci. U.S.A. 96, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tetsu O., McCormick F. (2003) Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell 3, 233–245 [DOI] [PubMed] [Google Scholar]
- 42. Meijer L., Borgne A., Mulner O., Chong J. P., Blow J. J., Inagaki N., Inagaki M., Delcros J. G., Moulinoux J. P. (1997) Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2, and cdk5. Eur. J. Biochem. 243, 527–536 [DOI] [PubMed] [Google Scholar]
- 43. Ali S., Heathcote D. A., Kroll S. H., Jogalekar A. S., Scheiper B., Patel H., Brackow J., Siwicka A., Fuchter M. J., Periyasamy M., Tolhurst R. S., Kanneganti S. K., Snyder J. P., Liotta D. C., Aboagye E. O., Barrett A. G., Coombes R. C. (2009) The development of a selective cyclin-dependent kinase inhibitor that shows antitumor activity. Cancer Res. 69, 6208–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mizuno H., Cho Y. Y., Ma W. Y., Bode A. M., Dong Z. (2006) Effects of MAP kinase inhibitors on epidermal growth factor-induced neoplastic transformation of human keratinocytes. Mol. Carcinog. 45, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fan Z., Baselga J., Masui H., Mendelsohn J. (1993) Antitumor effect of anti-epidermal growth factor receptor monoclonal antibodies plus cis-diamminedichloroplatinum on well established A431 cell xenografts. Cancer Res. 53, 4637–4642 [PubMed] [Google Scholar]
- 46. Morgan D. O. (1995) Principles of CDK regulation. Nature 374, 131–134 [DOI] [PubMed] [Google Scholar]
- 47. Sherr C. J. (1993) Mammalian G1 cyclins. Cell 73, 1059–1065 [DOI] [PubMed] [Google Scholar]
- 48. Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262 [DOI] [PubMed] [Google Scholar]
- 49. Huet X., Rech J., Plet A., Vié A., Blanchard J. M. (1996) Cyclin A expression is under negative transcriptional control during the cell cycle. Mol. Cell. Biol. 16, 3789–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Royzman I., Whittaker A. J., Orr-Weaver T. L. (1997) Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 11, 1999–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu W., Giangrande P. H., Nevins J. R. (2004) E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23, 4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nguyen V. Q., Co C., Li J. J. (2001) Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 53. Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D. S., Dutta A. (2003) A p53-dependent checkpoint pathway prevents re-replication. Mol. Cell 11, 997–1008 [DOI] [PubMed] [Google Scholar]
- 54. Malumbres M., Pevarello P., Barbacid M., Bischoff J. R. (2008) CDK inhibitors in cancer therapy: what is next? Trends Pharmacol. Sci. 29, 16–21 [DOI] [PubMed] [Google Scholar]
- 55. Lee K. W., Kang N. J., Rogozin E. A., Oh S. M., Heo Y. S., Pugliese A., Bode A. M., Lee H. J., Dong Z. (2008) The resveratrol analogue 3,5,3′,4′,5′-pentahydroxy-trans-stilbene inhibits cell transformation via MEK. Int. J. Cancer 123, 2487–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee K. W., Bode A. M., Dong Z. (2011) Molecular targets of phytochemicals for cancer prevention. Nat. Rev. Cancer 11, 211–218 [DOI] [PubMed] [Google Scholar]