Background: Condensin and CTCF regulate the state of rDNA repeats.
Results: Condensin interacts with CTCF and negatively regulates CTCF-mediated rRNA gene transcription.
Conclusion: Condensin and CTCF form a complex and play a negative role in regulation of rRNA gene transcription.
Significance: Regulation of rRNA gene transcription by condensin and CTCF indicates that chromatin binding protein and the status of the chromosome play important roles in the regulation of rRNA gene transcription.
Keywords: Chromosomes/Non-histone Chromosomal Proteins, Histone Modification, Ribosomal RNA (rRNA), RNA Interference (RNAi), RNA Polymerase I
Abstract
CCCTC-binding factor (CTCF) is a ubiquitously expressed “master weaver” and plays multiple functions in the genome, including transcriptional activation/repression, chromatin insulation, imprinting, X chromosome inactivation, and high-order chromatin organization. It has been shown that CTCF facilitates the recruitment of the upstream binding factor onto ribosomal DNA (rDNA) and regulates the local epigenetic state of rDNA repeats. However, the mechanism by which CTCF modulates rRNA gene transcription has not been well understood. Here we found that wild-type CTCF augments the pre-rRNA level, cell size, and cell growth in cervical cancer cells. In contrast, RNA interference-mediated knockdown of CTCF reduced pre-rRNA transcription. CTCF positively regulates rRNA gene transcription in a RNA polymerase I-dependent manner. We identified an RRGR motif as a putative nucleolar localization sequence in the C-terminal region of CTCF that is required for activating rRNA gene transcription. Using mass spectrometry, we identified SMC2 and SMC4, two subunits of condensin complexes that interact with CTCF. Condensin negatively regulates CTCF-mediated rRNA gene transcription. Knockdown of SMC2 expression significantly facilitates the loading of CTCF and the upstream binding factor onto the rDNA locus and increases histone acetylation across the rDNA locus. Taken together, our study suggests that condensin competes with CTCF in binding to a specific rDNA locus and negatively regulates CTCF-mediated rRNA gene transcription.
Introduction
Chromosomes carry genetic information and undergo essential structural changes during the cell cycle. Chromatin is dynamically organized to form distinct transcriptionally active or silent domains in eukaryotic cells to execute different functions (1). Chromatin insulators are proposed to play a role in the establishment of such domains, within which proper enhancer-promoter interactions occur and improper ones are excluded (2). In mammalian cells, insulators are bound by CCCTC-binding factor (CTCF),6 which is a highly conserved zinc finger and ubiquitously expressed protein involved in diverse regulatory functions, including transcriptional activation or repression, blocking of enhancer-promoter communication, and regulation of local histone modifications (3, 4). It is likely that CTCF interferes with enhancer-promoter interaction when acting as an insulator (2). CTCF insulator function is also mediated by the proteins that interact with CTCF (5, 6). Although CTCF binding sites are proposed to be insulators, their in vivo functions may not be limited to insulation. CTCF can regulate the balance between active and repressive chromatin modifications near its binding sites, with different outcomes in terms of transcription. Importantly, CTCF helps to bring distant sites together in the genome to create distinct topological domains (7). From genome-wide studies, it has been reported that cohesin, which mediates sister chromatid cohesion in the dividing cells, colocalizes at most sites of CTCF and plays a role in transcriptional insulation (8, 9), which suggests that cohesin functions together with CTCF to regulate relevant gene transcription.
The evolutionarily conserved condensin and cohesin complexes are important for chromosome condensation and epigenetic regulation of gene transcription. They both contain structural maintenance of chromosomes (SMC) subunits and non-SMC subunits (10). SMC proteins are involved in a wide range of processes, including chromosome structure and dynamics, gene regulation, and DNA repair (11). There are two analogous condensin complexes, named condensin I and condensin II. These two complexes have different spatial and temporal distributions during the cell cycle (12). Condensin II is mainly located in the nucleus during interphase and plays an important role in the early stages of chromosome assembly in prophase. In contrast, the majority of condensin I resides in the cytoplasm during interphase and gains access to chromosomes only after the nuclear envelope breaks down in prometaphase. Although condensin complexes are ubiquitously expressed multiprotein complexes and play important roles in gene regulation, the role of condensin in gene regulation in vertebrates is still poorly understood.
Production of rRNA is a critical process for ribosome biogenesis and mediates protein synthesis in eukaryotic cells (13). Cells usually have multiple copies of such genes, ranging from 150 to 300 per haploid genome (14). These gene clusters then colocalize in the nucleolus, where rRNA synthesis and processing occurs. An abundance of rRNA gene copies is important to maintain genome integrity (15). The rDNA repeating unit with a total length of 42.9 kb contains the 5.8 S, 18 S, and 28 S ribosomal subunits separated by intergenic sequences. However, even in metabolically active cells, significant numbers of repeats are not transcribed through epigenetic modifications of chromatin structure.
The rate of rRNA gene transcription requires RNA polymerase I, which is highly regulated by multiple proteins in either a positive (16, 17) or negative way (18). It has been reported that rRNA and protein synthesis are elevated in wide varieties of human cancers (19–21). Therefore, augmented expression of rRNA is potentially accompanied by increased protein synthesis and cell growth, which can accelerate tumor development. CTCF has been shown to regulate the local epigenetic state of rDNA repeats and may load upstream binding factor (UBF) onto rDNA, thereby forming part of a network that maintains rRNA genes poised for transcription (22, 23). A reduction in the CTCF level in cells results in nucleolar fragmentation and reduced rDNA silencing (24). In Drosophila, CTCF contributes to the regulation of rDNA and nucleolar stability (24). In addition, condensin has been reported to regulate rDNA silencing by modulating nucleolar Sir2p and Cdc14 in yeast (25, 26). So far, how CTCF and condensin together regulate rRNA gene transcription in mammalian cells is still unknown.
In this study, we found that CTCF positively regulates rRNA gene transcription in an RNA polymerase I-dependent manner. We identified that an RRGR motif in the C-terminal region of CTCF is required for the activation of rRNA gene transcription and found that the condensin subunits SMC2 and SMC4 exist in CTCF protein complexes. Further, we discovered that condensin negatively regulates rRNA gene transcription. Knockdown of SMC2 expression displays a significant increase in pre-rRNA synthesis and facilitates the loading of CTCF onto the rDNA locus. Our data show that CTCF and condensin bind to specific regions of human rDNA in a competitive manner. We conclude that condensin competes with CTCF in binding to the rDNA locus and modulates CTCF-mediated rRNA gene transcription.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa-S3 cells and 293T cells were cultured in DMEM (Hyclone) supplemented with 10% fetal bovine serum. Cells were maintained at 37 °C in a 5% CO2 incubator.
Antibodies
The antibodies used in this study were rabbit anti-CTCF polyclonal antibody (Millipore, catalog no. 07-729), mouse anti-CTCF monoclonal antibody (Abcam, catalog no. ab37477), rabbit anti-SMC2 polyclonal antibody (Abcam, catalog no. ab10412), rabbit anti-SMC4 polyclonal antibody (Abcam, catalog no. ab17958), mouse anti-actin monoclonal antibody (Sigma, catalog no. A2228), mouse anti-FLAG monoclonal antibody (Sigma, catalog no. F1804), mouse anti-UBF monoclonal antibody (Santa Cruz Biotechnology, catalog no. sc13125), and rabbit anti-acetyl-histone H4 polyclonal antibody (Millipore, catalog no. 06-598).
Generation of Stable Cell Lines
Stably transduced cell lines with N-terminal FLAG/HA-tagged CTCF with different deletions or point mutations were generated. Positively expressing cells also coexpressed IL2R, and several rounds of selection using anti-IL2R magnetic sorting were performed to obtain stably expressing cells.
Western Blot Analysis
The cells were resuspended and sonicated in lysis buffer (1% Nonidet P-40, 150 mm NaCl, 0.25% Na-deoxycholate, 50 mm Tris-Cl (pH 7.4), 1% SDS, 1 mm PMSF, and 1× protease inhibitor mixture). Total soluble proteins were obtained by centrifugation at 12,000 rpm for 10 min. Samples were separated on an SDS-PAGE gel and transferred onto a PVDF membrane (Millipore). The PVDF membrane was blocked with 5% milk in TBS-T (TBS with 0.1% Tween 20). An immunoblot analysis was performed with the indicated antibodies.
Lentivirus-mediated shRNA Knockdown
The recombinant construct (pLKO.1 empty, pLKO.1-CTCF, and pLKO.1-SMC2) as well as two assistant vectors (CMV R8.91 and vesicular stomatitis virus G) were transiently transfected into 6 × 106 cells/10-cm dish of 293T cells by using the calcium phosphate method. 12 h after transfection, the fresh medium was added. After 48 h, viral supernatants were collected and filtered and were mixed with 8 μg/ml Polybrene to infect the target cells. Stable cell lines were selected with 4 μg/ml puromycin after 2 days of infection. RNAi knockdown efficiencies were determined by Western blot analysis and RT-qPCR.
RNA Extract and RT-qPCR
Total RNA was isolated with TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (Ambion, catalog no. AM1906). cDNA was synthesized by using a random primer and reverse transcriptase (Toyobo). Real-time PCR was performed using SYBR Premix Ex Tag (TAKARA) in a CFX96 real-time PCR system (Bio-Rad) according to the instructions of the manufacturer. Transcript levels were normalized to HPRT1 or GAPDH levels. The primers used for RT-qPCR are shown in supplemental Table 5.
GST Pull-down Assays
GST and GST-CTCF full-length were expressed in Escherichia coli strain BL21 cells. The expressed fusion proteins from bacteria were purified using glutathione-Sepharose 4 fast-flow beads (GE Healthcare) and incubated with HeLa-S3 nuclear extracts (NEs) in PBS-T buffer supplemented with protease inhibitors (protease inhibitor mixture and 1 mm PMSF) for 4 h or overnight at 4 °C. RNase A or DNase I was added to examine the effects on the interaction between CTCF and SMC2. Beads were washed four times with PBS-T buffer, boiled in SDS loading buffer, resolved on SDS-PAGE, and transferred to a PVDF membrane for Western blot analysis.
Immunoprecipitation
Cleared NEs were incubated with the indicated antibodies plus protein A magnetic beads (Active Motif) in immunoprecipitation buffer for 4 h. Then the antibody/protein/beads complexes were washed four times with immunoprecipitation wash buffer. Samples were separated on an SDS-PAGE gel and transferred onto a PVDF membrane. An immunoblot analysis was performed with the indicated antibodies.
Cell Size Analysis
The HeLa-S3 cells were suspended in PBS buffer and used for cell size analysis on an Acurri C6 flow cytometer (BD Biosciences) following the protocol of the manufacturer. Data were analyzed by using FloJo software.
ChIP
About 5 × 107 HeLa-S3 cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Then the reaction was stopped by adding glycine (final concentration, 0.125 m). The cells were sonicated in SDS lysis buffer to achieve a chromatin size of 100–500 bp. The sonicated chromatin was diluted by using ChIP dilution buffer (0.01% SDS, 1.1% Triton-100, 1.2 mm EDTA, 16.7 mm Tris-Cl (pH 8.0), and 167 mm NaCl). 10 μg of antibodies was coupled with Dynabead protein A and G (1:1 mixed). Then, the mixture was incubated with chromatin lysates overnight at 4 °C with rotation. Immune complexes were washed with the following buffers: low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-Cl (pH 8.0), and 150 mm NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-Cl (pH 8.0), and 500 mm NaCl), LiCl wash buffer (0.25 m LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid (sodium salt), 1 mm EDTA, and 10 mm Tris-Cl (pH 8.0)) and TE buffer (10 mm Tris-Cl (pH 8.0) and 1 mm EDTA). Antibody-bound chromatin was reverse-cross-linked, and the ChIP DNA samples were purified for PCR reaction. Primers used for ChIP-qPCR are shown in supplemental Table 6.
RESULTS
CTCF Enhances RNA Polymerase I-mediated Transcription of rRNA Genes
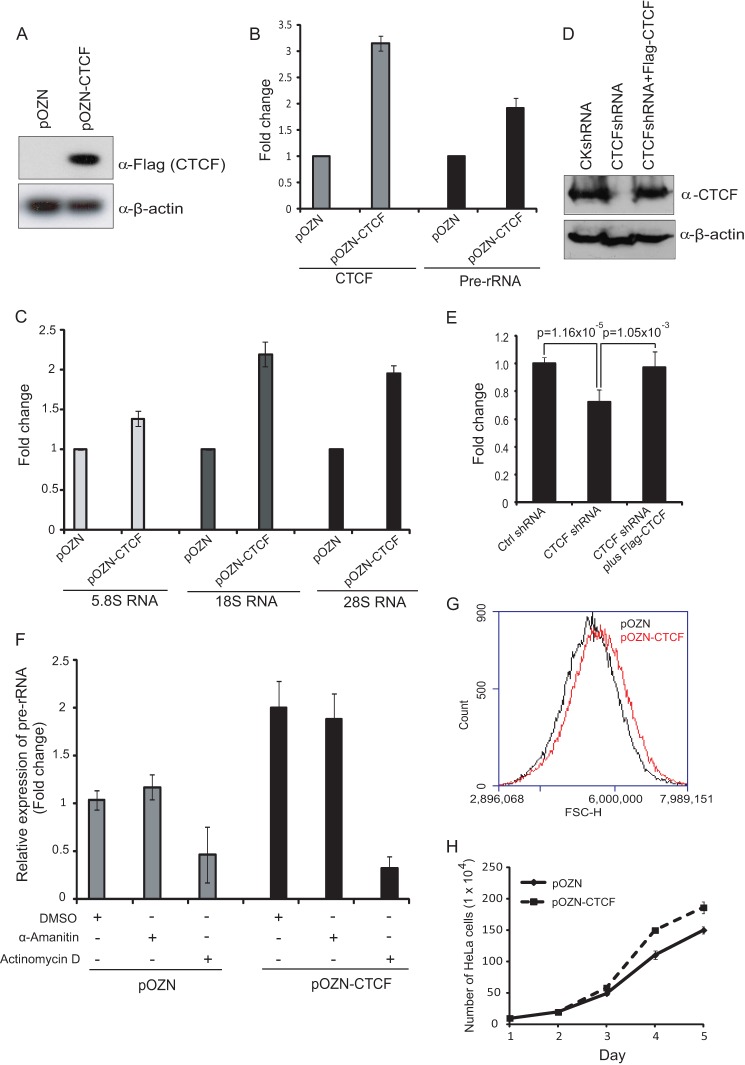
It has been shown that CTCF facilitates the recruitment of UBF onto rDNA and regulates the local epigenetic state of rDNA repeats (22). To determine whether induction of CTCF stimulates rRNA gene transcription, we generated HeLa-S3 cell lines with CTCF overexpression and tested this hypothesis. We conducted an RT-PCR analysis of 45 S pre-rRNA whose levels reflect the rate of RNA polymerase I transcription (16, 18). RT-PCR analysis of the expression of pre-rRNA revealed that ectopically expressed CTCF increases 45 S pre-rRNA levels around 2-fold (Fig. 1, A and B). A significant increase in transcription of 5.8 S, 18 S, and 28 S rRNA was also observed in HeLa-S3 cells with pOZN-CTCF overexpression compared with HeLa-S3 cells with pOZN alone (Fig. 1C). Subsequently, to investigate the role of CTCF in regulating rRNA gene transcription by CTCF knockdown, silencing of CTCF in HeLa-S3 cells using shRNA resulted in a significant decrease in the expression of pre-rRNA compared with control shRNA, which suggests that CTCF positively regulates rRNA gene transcription (Fig. 1, D and E). To rule out the possibility of off-target by shRNA, we further performed a rescue experiment and overexpressed mouse CTCF, which is resistant to the shRNA targeting to human CTCF back into CTCF depleted HeLa-S3 cells. As expected, we found that rescue of CTCF in CTCF depleted HeLa-S3 cells could rescue the expression of pre-rRNA (Fig. 1, D and E). These results suggest that CTCF stimulates rRNA gene transcription.
FIGURE 1.
CTCF activates RNA polymerase I-mediated rRNA gene transcription and increases cell size and proliferation. A, Western blot analysis of FLAG-CTCF expression in NEs with CTCF stably expressed HeLa-S3 cells. β-actin was used as a loading control. B, RT-qPCR analysis of pre-rRNA and CTCF expression after CTCF overexpression. Vertical bars represent mean ± S.D. C, RT-qPCR analysis of 5.8 S, 18 S, and 28 S RNA levels after ectopic CTCF expression. Vertical bars represent mean ± S.D. D, Western blot analysis of the CTCF level in whole cell extracts from control, CTCF-depleted HeLa-S3 cells, and CTCF-depleted HeLa-S3 cells with ectopic expression of mouse CTCF. β-actin was used as a loading control. E, RT-qPCR analysis of pre-rRNA expression from control (Ctrl), CTCF-depleted HeLa-S3 cells, and CTCF-depleted HeLa-S3 cells with ectopic expression of mouse CTCF. GAPDH was used as an internal control. Mean values of three independent experiments ± S.D. are included. F, RT-qPCR analysis of pre-rRNA expression in cells with pOZN or pOZN-CTCF overexpression after either actinomycin D or α-amanitin treatment. Vertical bars represent mean ± S.D. DMSO, dimethyl sulfoxide. G, cell size was determined by FACS (forward scatter) in HeLa-S3 cells retrovirally infected with empty vector (black) or constructs encoding wild-type FLAG-CTCF (red), as indicated. FSC-H, forward scatter height. H, cell growth analysis in pOZN- and pOZN-CTCF-overexpressed HeLa-S3 cells.
To further investigate whether activation of pre-rRNA transcription is RNA polymerase I-dependent, we treated HeLa-S3 cells with either the RNA polymerase I inhibitor actinomycin D or the RNA polymerase II inhibitor α-amanitin and examined the expression of pre-rRNA. Interestingly, we found that expression of pre-rRNA is significantly reduced in both pOZN-control cells and pOZN-CTCF cells by actinomycin D but not by α-amanitin (Fig. 1F). These results indicate that the induction of pre-rRNA transcription is RNA polymerase I-dependent but not RNA polymerase II-dependent or a secondary indirect effect. Because the synthesis of rRNAs is highly associated with cell growth (19), we further examined whether CTCF affects cell size and cell proliferation. A forward scatter analysis indicated that HeLa-S3 cells exogenously expressing CTCF are relatively larger than control HeLa-S3 cells (Fig. 1G and supplemental Fig. 1). CTCF-overexpressed cells proliferate at a faster rate than control cells (Fig. 1H).
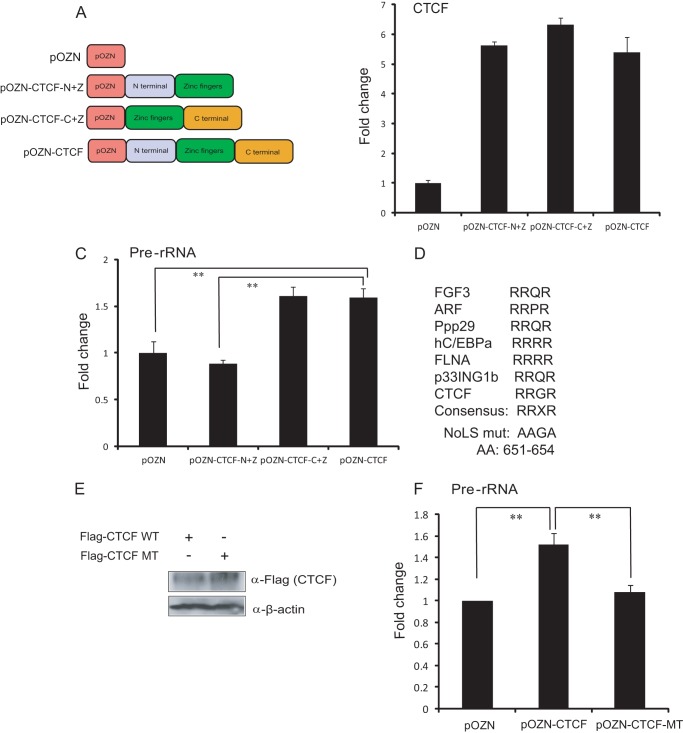
The Zinc Finger and the C Terminus of CTCF Are Sufficient for the Activation of rRNA Genes
To assess which part of CTCF is sufficient for activation of rRNA gene transcription, we made CTCF deletion constructs that contained only the N terminus plus the zinc finger or the C terminus plus the zinc finger and generated stable cell lines for both deletions (Fig. 2A). Then we examined rRNA gene expression levels after overexpressing both deletions and the full-length construct in HeLa-S3 cells. The results indicated that rRNA gene expression is increased in cells with overexpression of both the zinc finger plus the C terminus and the full-length construct (Fig. 2, B and C). Because the zinc finger plus the C-terminal fragment of CTCF are sufficient for rRNA gene activation, and nucleolar retention of proteins is often determined by a nucleolar localization sequence, we asked whether a nucleolar localization sequence existed in this fragment. Examination of the protein sequence of CTCF revealed a putative conserved amino acid motif (RRGR) located in the C-terminal region of CTCF (Fig. 2D). To further determine whether this RRGR motif in CTCF is required for activating rRNA gene transcription, we mutated the sequence from RRGR to AAGA on the basis of the HA-CTCF full-length construct and generated a mutant stable cell line. Then we examined whether this point mutation affects rRNA gene transcription. A Western blot analysis indicated that the expression of mutant CTCF is almost equivalent to that of wild-type CTCF (Fig. 2E). Interestingly, we did not observe the activation of rRNA gene expression after mutation of RRGR to AAGA in the C-terminal region of CTCF compared with the wild type (Fig. 2F), suggesting that mutation of the RRGR to AAGA abolishes the effects of CTCF on rRNA gene expression. Taken together, we speculate that the RRGR motif in the C-terminal fragment of CTCF allows CTCF to enter the nucleolus and then facilitates the zinc finger of CTCF to bind to rDNA and stimulate rRNA gene expression.
FIGURE 2.
The zinc finger and the C-terminal region of CTCF are sufficient for rRNA gene activation. A, schematic of pOZN-CTCF full-length and deletions used in examining rRNA gene transcription. B, RT-qPCR analysis of CTCF expression after overexpression of CTCF full-length and deletions. Primers were designed in the zinc finger region. C, RT-qPCR analysis of pre-rRNA expression after overexpression of CTCF full-length and deletions. **, p < 0.01. D, alignment of the RRXR motif within the domains mediating nucleolar localization of CTCF and other proteins with known nucleolar localization. NoLS, nuclear localization sequence; mut, mutant; AA, amino acids. E, Western blot analysis of the expressions of mutant CTCF and wild-type CTCF. β-actin was used as a loading control. F, comparative analysis of pre-rRNA expression in HeLa-S3 cells with both wild-type CTCF overexpression and mutation of RRGR to AAGA in the C-terminal region of CTCF. **, p < 0.01.
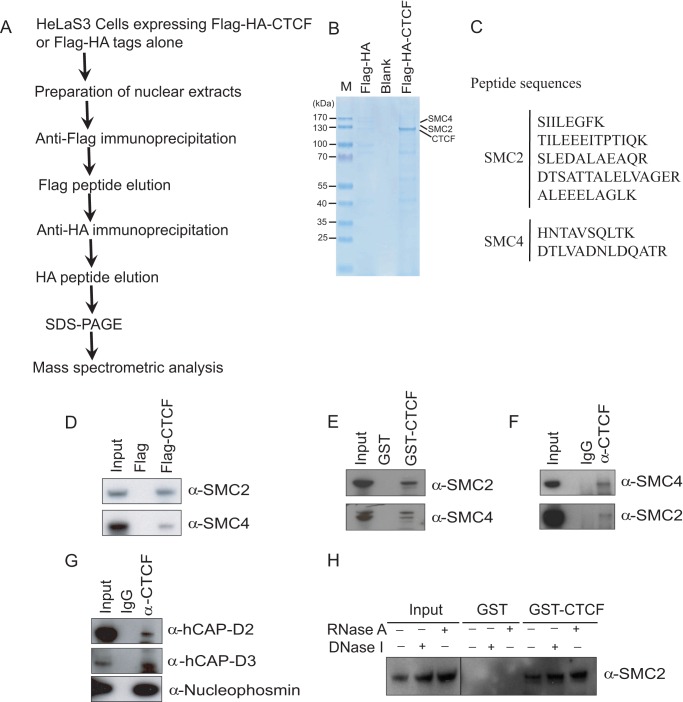
Condensin Complexes Interact with CTCF Both in Vivo and in Vitro
To further understand the function of CTCF in gene regulation, especially in rRNA gene transcription, we employed FLAG and HA tandem affinity purification using HeLa-S3 cell lines with stably expressed human FLAG-HA-CTCF. We further identified proteins that associate with CTCF and regulate CTCF function. NEs from these cells were subjected to double-step immuno-affinity purification, and the immunopurified CTCF complexes were separated by SDS-PAGE (Fig. 3, A and B). CTCF-copurifying factors were further identified by MS. We found that ectopic CTCF was copurified with previously reported interaction partners, including CTCF (supplemental Table 1), cohesin complex (SMC1A, SMC3, and Rad21) (supplemental Tables 2, 3, and 4), DDX5, poly(ADP-ribose) polymerase 1, and so on. Interestingly, we found that multiple peptides of SMC2 and SMC4 exist in CTCF protein complexes that have not been reported before (Fig. 3C). SMC2 and SMC4 form a heterodimer that is the catalytic subunit of both condensin I and II complexes, which play roles in chromosome condensation during mitosis and meiosis, removal of cohesin during mitosis and meiosis, and interphase rDNA compaction (27–29).
FIGURE 3.
CTCF interacts with the condensin complex both in vivo and in vitro. A, two-step purification schematic for identifying CTCF-associated proteins. B, immunopurified human FLAG-CTCF complexes were fractionated on SDS-PAGE. C, mass spectrometry analysis identifying peptide sequences of SMC2 and SMC4 in the CTCF complex. D, HeLa-S3 nuclear extract with human FLAG-CTCF or FLAG alone were tested for coimmunoprecipitation. The precipitates were analyzed by immunoblot analysis, with antibodies shown on the right. E, GST pull-down assays confirmed that CTCF interacts with SMC2 and SMC4. GST-CTCF and GST fusion proteins were expressed and purified in BL21. F, MCF-7 NEs were precipitated with the antibodies shown at the top, and precipitates were immunoblotted with the antibodies shown on the right. G, CTCF interacts with both condensin I and condensin II. MCF-7 NEs were precipitated with the antibodies shown at the top, and precipitates were immunoblotted with the antibodies shown on the right. CAP-D2 and CAP-D3 are the specific subunits of condensin I and condensin II, respectively. H, DNase and RNase treatment have no effect on the interaction between CTCF and condensin.
To confirm the MS data, we performed FLAG coimmunoprecipitation experiments and found that SMC2 and SMC4 were retained by FLAG-CTCF but not by FLAG alone (Fig. 3D). To examine whether CTCF interacts with SMC2/SMC4 in vitro, we expressed and purified GST-CTCF and GST fusion proteins in BL21 cells and then carried out GST pull-down assays. The results indicated that both SMC2 and SMC4 bind GST-CTCF but not GST alone (Fig. 3E). To verify the endogenous interaction between CTCF and SMC2/SMC4, we performed endogenous coimmunoprecipitation experiments and found that SMC2 and SMC4 were coimmunoprecipitated from MCF-7 NEs by a CTCF mouse monoclonal antibody but not by the normal mouse immunoglobulin G (Fig. 3F). To further examine which condensin complex interacts with CTCF, we performed endogenous coimmunoprecipitation experiments and found that both the specific subunit, CAP-D3, of the condensin II complex and the specific subunit, CAP-D2, of condensin I interact with CTCF, although the relative amounts of condensin II in CTCF complex are much higher compared with the amounts of condensin I (Fig. 3G). In addition, the CTCF-condensin interaction seems to be both DNA- and RNA-independent because the bindings of SMC2 to GST-CTCF were not destroyed when we incubated them with either RNase A or DNase I in the GST pull-down assays (Fig. 3H).
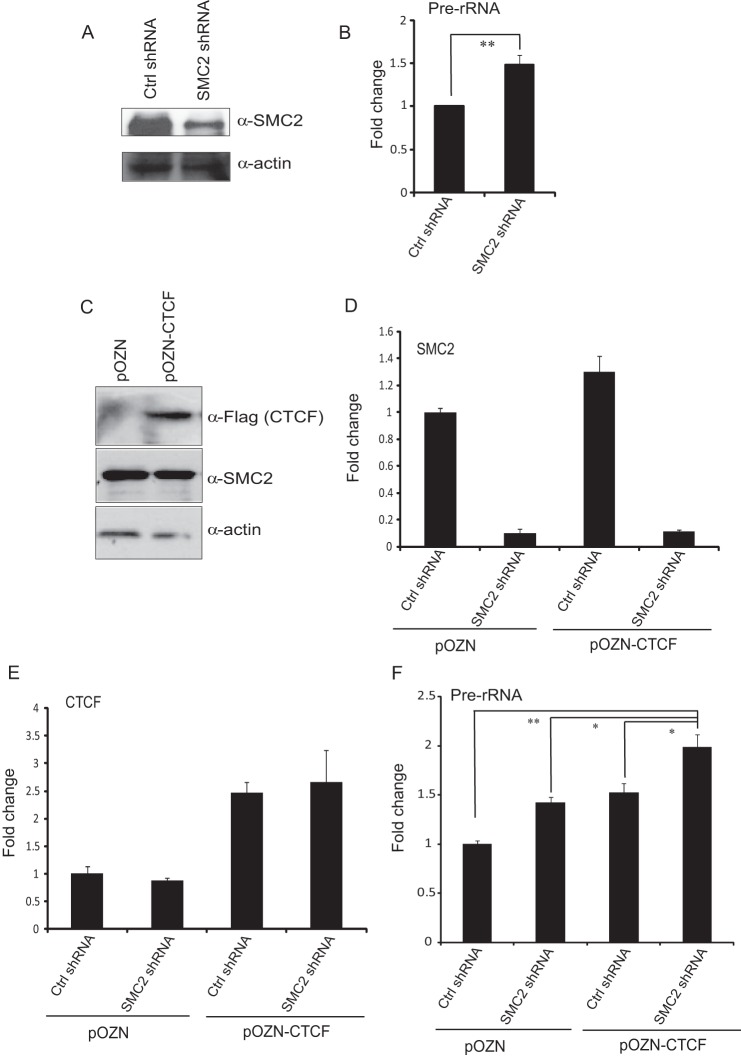
Condensin Negatively Regulates CTCF-mediated rRNA Gene Transcription
It has been reported that condensin regulates rDNA silencing in yeast (25). To investigate whether condensin regulates rRNA gene transcription in mammalian cells, lentiviruses expressing SMC2 shRNA were generated to knock down endogenous SMC2. A Western blot analysis showed that SMC2 was depleted more than 80% in HeLa-S3 cells expressing SMC2 shRNA compared with HeLa-S3 cells expressing control shRNA (Fig. 4A). SMC2 knockdown has no effect on CTCF expression (supplemental Fig. 2). Real-time RT-PCR experiments revealed that knockdown of SMC2 significantly enhances the expression of the pre-rRNA gene (Fig. 4B). To examine how condensin affects CTCF-mediated rRNA gene transcription, we depleted endogenous SMC2 expression in HeLa-S3 cells with the expression of either control vector pOZN or pOZN-CTCF (Fig. 4, C and D). Interestingly, we found that SMC2 knockdown facilitates the enhancement of pre-rRNA expression by CTCF (Fig. 4, E and F), suggesting that condensin negatively modulates CTCF-mediated rRNA gene transcription.
FIGURE 4.
The effects of SMC2 on CTCF-mediated regulation of rRNA gene transcription. A, Western blot analysis of SMC2 levels in NEs from SMC2 shRNA or empty vector stably expressed Hela-S3 cells. Ctrl, control. B, RT-qPCR analysis of pre-rRNA level after SMC2 knockdown. Mean values of three independent experiments ± S.D. are included. **, p < 0.01. C, Western blot analysis of FLAG and SMC2 levels in NEs from HeLa-S3 cells with either pOZN-CTCF or pOZN overexpression. β-actin was used as a loading control. D, RT-qPCR analysis of SMC2 expressions in pOZN-CTCF- or pOZN-overexpressed HeLa-S3 cells after SMC2 knockdown. E, RT-qPCR analysis of CTCF expressions in pOZN-CTCF- or pOZN-overexpressed HeLa-S3 cells after SMC2 knockdown. F, RT-qPCR analysis of pre-rRNA precursor expressions in pOZN-CTCF- or pOZN-overexpressed HeLa-S3 cells after SMC2 knockdown. *, p < 0.05; **, p < 0.01.
CTCF and Condensin Bind to a Specific rDNA Locus in a Competitive Manner
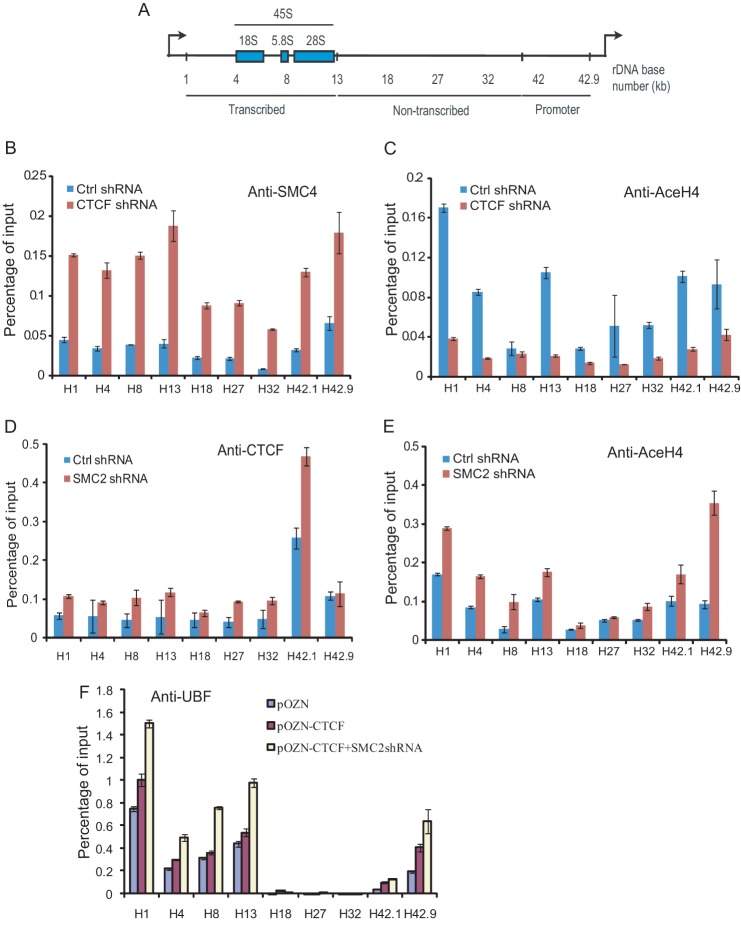
The interaction of CTCF with the condensin complex indicates that recruitment of CTCF to rDNA may influence the occupancy of the condensin complex. To test this hypothesis, we performed ChIP assays followed by quantitative real-time PCR using primer pair sets that span the entire human rDNA repeat to examine the distribution of the condensin complex in HeLa-S3 cells expressing either control shRNA or CTCF shRNA (Fig. 5A). The results show that repression of CTCF by shRNA causes a general increase of SMC4 binding within the entire rDNA locus but preferentially in the transcription initiation region and throughout the transcribed region (Fig. 5B). ChIP assays with anti-acetylated histone H4 antibody indicate that there is a significant reduction of the acetylated histone H4 level in the rDNA locus after CTCF knockdown (Fig. 5C). These results support the transcriptional reduction of pre-rRNA after CTCF knockdown (Fig. 1, D and E). In addition, we performed a ChIP assay using anti-CTCF antibody after SMC2 knockdown to investigate whether condensin regulates the binding of CTCF in the rDNA locus. Interestingly, we found that SMC2 knockdown causes a slight increase in binding of CTCF within the promoter region at H1 but a significant enrichment of CTCF binding in the region H42.1, which was identified as a CTCF binding site (Fig. 5D) (22, 31). These results suggest that CTCF and condensin bind to a specific rDNA locus in a competitive manner. In addition, SMC2 knockdown significantly enhances the enrichment of acetylated histone H4 around the promoter region (H1 and H42.9) (Fig. 5E).
FIGURE 5.
Functional relationships of CTCF, condensin, and histone acetylation across the rDNA locus. A, schematic of the human 45 S rRNA precursor. B and C, ChIP-qPCR analyses were performed in HeLa-S3 cells transduced with either control (Ctrl) shRNA or CTCF shRNA. The antibodies are shown at the top, and the indicated primers are shown at the bottom. Mean values of three independent experiments ± S.D. are included. D and E, ChIP-qPCR analyses were performed in HeLa-S3 cells transduced with either control shRNA or SMC2 shRNA. The antibodies are shown at the top, and the indicated primers are shown at the bottom. Mean values of three independent experiments ± S.D. are included. F, ChIP-qPCR analyses of UBF distributions across the rDNA locus in pOZN- or pOZN-CTCF-overexpressed HeLa-S3 cells after SMC2 knockdown. Mean values of three independent experiments ± S.D. are included.
SMC2 Depletion Facilitates the Loading of UBF onto the rDNA Locus by CTCF
UBF is an important factor for efficient transcription from the ribosomal gene promoter (32). To investigate how condensin and CTCF regulate the recruitment of UBF onto the rDNA locus, we measured the occupancy of UBF within the entire rDNA locus by ChIP analysis after CTCF overexpression or CTCF overexpression followed by SMC2 depletion (Fig. 5F). We found that overexpression of CTCF enhances the loading of UBF onto the transcribed and promoter regions of the rDNA locus. Interestingly, a further increase in the recruitment of UBF was observed in SMC2-depleted cells with CTCF overexpression (Fig. 5F). Our data indicate that enrichment of UBF onto the rDNA locus is consistent with the expression of the rRNA gene (Fig. 4). Taken together, our study suggests that SMC2 knockdown facilitates CTCF-enhanced rRNA gene transcription.
DISCUSSION
Stimulation of RNA polymerase I-dependent rRNA gene transcription is an important event in ribosome biogenesis, which is prerequisite for enhanced protein synthesis (19). RNA polymerase I-mediated rRNA gene transcription occurs in the nucleolus (13). Regulation of rRNA gene transcription is critical to ribosome production, cell growth, and proliferation, and this process is highly regulated by multiprotein complexes.
CTCF has been shown to localize to the nucleolus and repress nucleolar transcription in UR61 cells (33). However, in this study, we found that CTCF positively regulates rRNA gene transcription in HeLa-S3 cells. Ectopic expression of CTCF enhances pre-rRNA levels in cells, whereas CTCF knockdown has the opposite effect. The importance of CTCF in the regulation of cell proliferation has been documented. In previous studies, CTCF overexpression was found to be strongly associated with growth suppression in several cellular systems (34–36). However, on the other hand, it has been reported that CTCF positively regulates cell growth in the rapidly dividing thymocytes (37). In different cell types, the chromatin environment, transcriptional machinery, and metabolic state are quite different. Therefore, CTCF may regulate rRNA gene transcription and cell growth in both the positive and negative directions in a context-dependent manner.
In this report, we found that ectopically expressed CTCF promotes HeLa-S3 cell proliferation (Fig. 1). It seems that CTCF is an oncogene and not a tumor suppressor in cervical cancer cells. Therefore, CTCF-promoted rRNA gene transcription may play important roles in cervical cancer development. It has been reported that CTCF has many roles in normal and abnormal physiological functions, such as cancer angiogenesis (38–40). However, whether CTCF-mediated rRNA gene transcription regulates different types of cancers or angiogenesis needs to be explored further. Because CTCF plays multiple different roles and is a master weaver in the genome. Therefore, the mechanisms in which CTCF mediates rRNA transcription in different contexts might be different.
It has been reported that translocation of CTCF from nucleoplasm to the nucleolus was observed after differentiation of K562 myeloid cells or induction of apoptosis in MCF7 breast cancer cells (33). Nucleolar protein nucleophosmin/B23 has been shown to interact with CTCF and is present in vivo at insulator sites in the chicken β-globin locus (31). Here, we have shown that the zinc finger and the C-terminal region of CTCF are sufficient for the induction of rRNA gene transcription. Nucleolar targeting of proteins is usually determined by a nucleolar localization sequence, which is characterized by the consensus sequence RRXR and mediates nucleolar retention of well characterized nucleolar proteins, such as FGF3, ARF, Ppp29, hC/EBPα, p33ING1b, and filamin A (41). We examined the primary sequence of CTCF and identified an RRGR motif in the C-terminal region. This motif is consistent with the consensus nucleolar localization sequence (42) and is required for activating rRNA gene transcription (Fig. 2).
In a previous report, affinity purification of SMC2 protein complex by streptavidin-binding peptide-tagged SMC2 could not detect CTCF (43). In our study, because of technical improvements, we performed FLAG and HA double-affinity protein purification for FLAG-HA-CTCF expressed in HeLa-S3 cells, cut the whole lane of PAGE gel, and performed MS. In this case, we could identify many CTCF-interacting cofactors. From the MS data, we found that condensin core subunits SMC2 and SMC4 were copurified with insulator binding protein CTCF. We confirmed the interaction between CTCF and condensin not only by immunoprecipitation of CTCF and Western blot analysis with anti-SMC2 and anti-SMC4 both in vivo and in vitro but also by GST pull-down of CTCF and Western blot analysis with anti-SMC2 and anti-SMC4 (Fig. 3).
Condensin is important for the timely compaction and resolution of chromosomes to remove and prevent catenations that would otherwise inhibit segregation during mitosis (10). Recently, condensin has been reported to play critical roles in interphase genome organization. In yeast, it has been reported that condensin binds all yeast tDNA genes, and disruption of any condensin subunit causes the dispersal of tDNA clusters (45). Moreover, condensin regulates rDNA silencing by modulating yeast nucleolar sir2p (25). However, how condensin regulates rRNA gene transcription in mammalian systems is still unclear. Our data indicate that down-regulation of SMC2 significantly elevates CTCF-regulated rRNA gene transcription in HeLa-S3 cells (Fig. 4), indicating that condensin represses CTCF-enhanced rRNA gene transcription.
Van de Nobelen et al. (22) identified the region of H42.1 as one of the CTCF binding sites in the human rDNA repeat, which is 0.9 kb upstream of the ribosomal gene promoter. In this study, our ChIP experiments showed that the binding of CTCF at the H42.1 locus is more significant than any other region of rDNA locus (Fig. 5D). Although our data indicate that CTCF forms a complex with condensin, interestingly, in the knockdown experiments we have shown that CTCF and condensin bind to a specific rDNA locus and affect rRNA gene transcription in a competitive way (Fig. 5). These may suggest that the complex formation between CTCF and condensin occurs only when not bound to DNA. CTCF and condensin may dissociate from each other when binding to DNA. Histone acetylation is highly correlated with transcriptional activity (46). Our data indicate that both CTCF- and condensin- regulated histone acetylation at the rDNA locus are consistent with the transcriptional status of the rRNA gene.
The upstream binding factor UBF is an activator of RNA polymerase I transcription. It has been reported that UBF interacts with CTCF and that CTCF binding enhances the recruitment of UBF to the rDNA locus (47). Nucleolar UBF, in turn, ensures that rDNA repeats remain accessible to RNA polymerase I (47). Our experiments showed that the binding of UBF onto the rDNA repeat increases significantly after CTCF overexpression and that down-regulation of SMC2 further enhances this effect, which suggests that CTCF, as a mediator, regulates RNA polymerase I-mediated rRNA gene transcription.
Consistent with the report from Grandori et al. (16), our UBF ChIP results also show that UBF preferentially binds to the transcription initiation region and throughout the transcribed region but was not detectable in the intergenic region (Fig. 5F). Although CTCF interacts with UBF, UBF is present abundantly in the nucleolus, where it binds rDNA with relatively low specificity (48) and is highly dynamic (30, 44), and the interaction of UBF and CTCF might be transient. Therefore, we do not expect that the bindings of CTCF and UBF always exist in the same sites at the rDNA locus, although CTCF binding enhances UBF binding. Taken together, recruitment of UBF onto the rDNA locus depends on not only CTCF but also on other unknown possibilities that may need to be further characterized.
Acknowledgments
We thank Dr. Gary Felsenfeld at the National Institutes of Health and Dr. Christopher Zhang at Guangzhou Institutes of Biomedicine and Health for critical reading and comments on this paper.
This work was supported in part by National Natural Science Foundation of China (Grants 31271391 and 81201580), by Guangdong Science and Technology Planning Project of Province, China (Grant 2011A060901019), and by Chinese Academy of Sciences (Grant KSZD-EW-Z-003-1-5).

This article contains supplemental Figs. 1 and 2 and Tables 1–7.
- CTCF
- CCCTC-binding factor
- SMC
- structural maintenance of chromosomes
- UBF
- upstream binding factor
- RT-qPCR
- quantitative RT-PCR
- NE
- nuclear extract.
REFERENCES
- 1. Fraser P., Bickmore W. (2007) Nuclear organization of the genome and the potential for gene regulation. Nature 447, 413–417 [DOI] [PubMed] [Google Scholar]
- 2. Wallace J. A., Felsenfeld G. (2007) We gather together. Insulators and genome organization. Curr. Opin. Genet. Dev. 17, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phillips J. E., Corces V. G. (2009) CTCF. Master weaver of the genome. Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bell A. C., West A. G., Felsenfeld G. (2001) Insulators and boundaries. Versatile regulatory elements in the eukaryotic genome. Science 291, 447–450 [DOI] [PubMed] [Google Scholar]
- 5. Yao H., Brick K., Evrard Y., Xiao T., Camerini-Otero R. D., Felsenfeld G. (2010) Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J. M. (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- 7. Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Göndör A., Ohlsson R. (2008) Chromatin insulators and cohesins. EMBO Rep. 9, 327–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H. C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., Cobb B. S., Yokomori K., Dillon N., Aragon L., Fisher A. G., Merkenschlager M. (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 [DOI] [PubMed] [Google Scholar]
- 10. Wood A. J., Severson A. F., Meyer B. J. (2010) Condensin and cohesin complexity. The expanding repertoire of functions. Nat. Rev. Genet. 11, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griese J. J., Witte G., Hopfner K. P. (2010) Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 38, 3454–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ono T., Fang Y., Spector D. L., Hirano T. (2004) Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15, 3296–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grummt I. (2003) Life on a planet of its own. Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 14. Worton R. G., Sutherland J., Sylvester J. E., Willard H. F., Bodrug S., Dubé I., Duff C., Kean V., Ray P. N., Schmickel R. D. (1988) Human ribosomal RNA genes. Orientation of the tandem array and conservation of the 5′ end. Science 239, 64–68 [DOI] [PubMed] [Google Scholar]
- 15. Ide S., Miyazaki T., Maki H., Kobayashi T. (2010) Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327, 693–696 [DOI] [PubMed] [Google Scholar]
- 16. Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. (2005) c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 17. Holmström T. H., Mialon A., Kallio M., Nymalm Y., Mannermaa L., Holm T., Johansson H., Black E., Gillespie D., Salminen T. A., Langel U., Valdez B. C., Westermarck J. (2008) c-Jun supports ribosomal RNA processing and nucleolar localization of RNA helicase DDX21. J. Biol. Chem. 283, 7046–7053 [DOI] [PubMed] [Google Scholar]
- 18. Frescas D., Guardavaccaro D., Bassermann F., Koyama-Nasu R., Pagano M. (2007) JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450, 309–313 [DOI] [PubMed] [Google Scholar]
- 19. Ruggero D., Pandolfi P. P. (2003) Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 20. White R. J. (2005) RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6, 69–78 [DOI] [PubMed] [Google Scholar]
- 21. Li L. Y., Chen H., Hsieh Y. H., Wang Y. N., Chu H. J., Chen Y. H., Chen H. Y., Chien P. J., Ma H. T., Tsai H. C., Lai C. C., Sher Y. P., Lien H. C., Tsai C. H., Hung M. C. (2011) Nuclear ErbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res. 71, 4269–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van de Nobelen S., Rosa-Garrido M., Leers J., Heath H., Soochit W., Joosen L., Jonkers I., Demmers J., van der Reijden M., Torrano V., Grosveld F., Delgado M. D., Renkawitz R., Galjart N., Sleutels F. (2010) CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zentner G. E., Saiakhova A., Manaenkov P., Adams M. D., Scacheri P. C. (2011) Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 39, 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guerrero P. A., Maggert K. A. (2011) The CCCTC-binding factor (CTCF) of Drosophila contributes to the regulation of the ribosomal DNA and nucleolar stability. PloS ONE 6, e16401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Machín F., Paschos K., Jarmuz A., Torres-Rosell J., Pade C., Aragón L. (2004) Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr. Biol. 14, 125–130 [PubMed] [Google Scholar]
- 26. Clemente-Blanco A., Mayán-Santos M., Schneider D. A., Machín F., Jarmuz A., Tschochner H., Aragón L. (2009) Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 458, 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Losada A., Hirano T. (2005) Dynamic molecular linkers of the genome. The first decade of SMC proteins. Genes Dev. 19, 1269–1287 [DOI] [PubMed] [Google Scholar]
- 28. Ivanovska I., Orr-Weaver T. L. (2006) Histone modifications and the chromatin scaffold for meiotic chromosome architecture. Cell Cycle 5, 2064–2071 [DOI] [PubMed] [Google Scholar]
- 29. Tsang C. K., Wei Y., Zheng X. F. (2007) Compacting DNA during the interphase. Condensin maintains rDNA integrity. Cell Cycle 6, 2213–2218 [DOI] [PubMed] [Google Scholar]
- 30. O'Sullivan A. C., Sullivan G. J., McStay B. (2002) UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yusufzai T. M., Tagami H., Nakatani Y., Felsenfeld G. (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13, 291–298 [DOI] [PubMed] [Google Scholar]
- 32. Bell S. P., Learned R. M., Jantzen H. M., Tjian R. (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241, 1192–1197 [DOI] [PubMed] [Google Scholar]
- 33. Torrano V., Navascués J., Docquier F., Zhang R., Burke L. J., Chernukhin I., Farrar D., León J., Berciano M. T., Renkawitz R., Klenova E., Lafarga M., Delgado M. D. (2006) Targeting of CTCF to the nucleolus inhibits nucleolar transcription through a poly(ADP-ribosyl)ation-dependent mechanism. J. Cell Sci. 119, 1746–1759 [DOI] [PubMed] [Google Scholar]
- 34. Torrano V., Chernukhin I., Docquier F., D'Arcy V., León J., Klenova E., Delgado M. D. (2005) CTCF regulates growth and erythroid differentiation of human myeloid leukemia cells. J. Biol. Chem. 280, 28152–28161 [DOI] [PubMed] [Google Scholar]
- 35. Qi C. F., Martensson A., Mattioli M., Dalla-Favera R., Lobanenkov V. V., Morse H. C. (2003) CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc. Natl. Acad. Sci. U.S.A. 100, 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rasko J. E., Klenova E. M., Leon J., Filippova G. N., Loukinov D. I., Vatolin S., Robinson A. F., Hu Y. J., Ulmer J., Ward M. D., Pugacheva E. M., Neiman P. E., Morse H. C., 3rd, Collins S. J., Lobanenkov V. V. (2001) Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res. 61, 6002–6007 [PubMed] [Google Scholar]
- 37. Heath H., Ribeiro de Almeida C., Sleutels F., Dingjan G., van de Nobelen S., Jonkers I., Ling K. W., Gribnau J., Renkawitz R., Grosveld F., Hendriks R. W., Galjart N. (2008) CTCF regulates cell cycle progression of αβ T cells in the thymus. EMBO J. 27, 2839–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang M., Chen B., Lin T., Li Z., Pardo C., Pampo C., Chen J., Lien C. L., Wu L., Ai L., Wang H., Yao K., Oh S. P., Seto E., Smith L. E., Siemann D. W., Kladde M. P., Cepko C. L., Lu J. (2011) Restraint of angiogenesis by zinc finger transcription factor CTCF-dependent chromatin insulation. Proc. Natl. Acad. Sci. U.S.A. 108, 15231–15236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu J., Tang M. (2012) CTCF-dependent chromatin insulator as a built-in attenuator of angiogenesis. Transcription 3, 73–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Recillas-Targa F., de la Rosa-Velázquez I. A., Soto-Reyes E. (2011) Insulation of tumor suppressor genes by the nuclear factor CTCF. Biochem. Cell Biol. 89, 479–488 [DOI] [PubMed] [Google Scholar]
- 41. Deng W., Lopez-Camacho C., Tang J. Y., Mendoza-Villanueva D., Maya-Mendoza A., Jackson D. A., Shore P. (2012) Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription. Proc. Natl. Acad. Sci. U.S.A. 109, 1524–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Müller C., Bremer A., Schreiber S., Eichwald S., Calkhoven C. F. (2010) Nucleolar retention of a translational C/EBPα isoform stimulates rDNA transcription and cell size. EMBO J. 29, 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim J. H., Chang T. M., Graham A. N., Choo K. H., Kalitsis P., Hudson D. F. (2010) Streptavidin-binding peptide (SBP)-tagged SMC2 allows single-step affinity fluorescence, blotting or purification of the condensin complex. BMC Biochem. 11, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen D., Huang S. (2001) Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. D'Ambrosio C., Schmidt C. K., Katou Y., Kelly G., Itoh T., Shirahige K., Uhlmann F. (2008) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 22, 2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deckert J., Struhl K. (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21, 2726–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T. P., Quin J., Robb E., Wong L. H., Thomas W. G., Stefanovsky V., Moss T., Rothblum L., Hannan K. M., McArthur G. A., Pearson R. B., Hannan R. D. (2008) UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 183, 1259–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sanij E., Hannan R. D. (2009) The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 4, 374–382 [DOI] [PubMed] [Google Scholar]