Background: CD147 induces invadopodia activity and invasiveness in breast epithelial cells.
Results: CD147 forms complexes with CD44 and EGFR in lipid raftlike membrane domains and induces hyaluronan-CD44-dependent EGFR-Ras-ERK signaling that promotes invadopodia activity and invasiveness.
Conclusion: CD147 induces signaling complexes critical to invasiveness in breast epithelial cells.
Significance: CD147 is a potential therapeutic target and disease marker in breast cancer.
Keywords: Breast Cancer, CD44, Epidermal Growth Factor Receptor (EGFR), Hyaluronate, Ras, CD147, Invadopodia
Abstract
The immunoglobulin superfamily glycoprotein CD147 (emmprin; basigin) is associated with an invasive phenotype in various types of cancers, including malignant breast cancer. We showed recently that up-regulation of CD147 in non-transformed, non-invasive breast epithelial cells is sufficient to induce an invasive phenotype characterized by membrane type-1 matrix metalloproteinase (MT1-MMP)-dependent invadopodia activity (Grass, G. D., Bratoeva, M., and Toole, B. P. (2012) Regulation of invadopodia formation and activity by CD147. J. Cell Sci. 125, 777–788). Here we found that CD147 induces breast epithelial cell invasiveness by promoting epidermal growth factor receptor (EGFR)-Ras-ERK signaling in a manner dependent on hyaluronan-CD44 interaction. Furthermore, CD147 promotes assembly of signaling complexes containing CD147, CD44, and EGFR in lipid raftlike domains. We also found that oncogenic Ras regulates CD147 expression, hyaluronan synthesis, and formation of CD147-CD44-EGFR complexes, thus forming a positive feedback loop that may amplify invasiveness. Last, we showed that malignant breast cancer cells are heterogeneous in their expression of surface-associated CD147 and that high levels of membrane CD147 correlate with cell surface EGFR and CD44 levels, activated EGFR and ERK1, and activated invadopodia. Future studies should evaluate CD147 as a potential therapeutic target and disease stratification marker in breast cancer.
Introduction
CD147 (emmprin; basigin) is a variably glycosylated transmembrane protein that is involved in both physiological and pathological cellular processes (1–4). CD147 expression is elevated in a wide range of cancer types, including breast cancer (5, 6). Earlier studies employing in situ hybridization and immunohistochemistry techniques found that CD147 is expressed at preinvasive and invasive areas as well as proliferative regions in breast lesions; although CD147 was also identified in normal breast tissue, it is expressed at lower levels (7, 8). CD147 expression gradually increases during progression from atypical ductal hyperplasia to invasive breast cancer and is correlated with hormone receptor-negative and ErbB2-overexpressing breast cancers (9). In accord with these correlative studies in human patients, the importance of CD147 in tumor growth and invasion has been demonstrated in several model systems, including a study in vivo in which CD147-transfected breast cancer cells injected into mammary fat pads of nude mice were found to form larger tumors than control-transfected cells, to be more locally invasive, and, in several animals, to metastasize to various sites (10).
Originally identified as a tumor cell-associated factor that induces stromal fibroblasts to synthesize and secrete matrix metalloproteinases (MMPs)2 (11–14), CD147 has since been shown to have pleiotropic functions. In addition to inducing MMP synthesis in stromal, tumor, and endothelial cells, CD147 contributes to therapy resistance, angiogenesis, inflammatory signaling, cytoskeletal remodeling, migration/invasion, and trafficking of monocarboxylate transporters to the cell surface (1–4). CD147 can also induce synthesis of the large extracellular polysaccharide, hyaluronan, the main ligand for the cell surface receptor CD44 (15–18). CD147-induced hyaluronan-CD44 interactions modulate various signaling pathways and potentiate tumorigenic properties in various cancer cell types (19). CD147 has also been shown to cooperate with cyclophilins to induce intracellular signaling pathways (3). However, in each case, the exact mechanisms by which CD147 activates signaling cascades are not fully understood.
Dysregulated expression of Ras genes has been identified in many cancer types and oncogenic Ras expression is associated with aggressive cancer phenotypes, such as proliferation, invasion/metastasis, and therapy resistance (20). Although the common point mutations identified in oncogenic forms of Ras are a rare occurrence in breast cancer (21), chronic Ras activity has been documented in breast cancer cell lines and patient tumor tissues (22, 23). In the absence of oncogenic Ras signaling, up-regulation of normal Ras activity can facilitate similar transformed phenotypes (24), which may be due to amplified expression and activation of receptor tyrosine kinases, such as epidermal growth factor receptor (EGFR) family members, mutations in modulators of the Ras activation state, or effectors downstream of Ras (25, 26).
In a recent study, we demonstrated that up-regulation of CD147 is sufficient to induce the formation of active invadopodia and invasiveness in the non-transformed human breast epithelial cell line, MCF-10A (27). In this study, we have identified novel signaling associations between CD147, hyaluronan-CD44 interactions, and the EGFR-Ras-ERK pathway that regulate the invasive properties of breast epithelial cells.
EXPERIMENTAL PROCEDURES
Cell Culture
The human breast adenocarcinoma cell lines MDA-MB-231 and MCF-7 were obtained from American Type Culture Collection (ATCC) and were cultured in RPMI 1640 (R-8755) with 2.38 g/liter HEPES, 2 g/liter sodium bicarbonate, and 10% FBS (pH 7.4). The spontaneously immortalized human breast epithelial cell line MCF-10A was obtained from ATCC. MCF-10A cells stably expressing a lentivirus construct containing K-RasV12 (10A-K-RasV12) or empty vector (10A-EV) were generously provided by Dr. Ben Ho Park (28). Low passage MCF-10A cells and MCF-10A derivatives were maintained in mammary epithelial cell growth medium with BulletKit supplements (Lonza) unless noted otherwise in the figure legends. All cells were cultured in a humidified 95% air, 5% CO2 incubator at 37 °C.
Antibodies and Reagents
The following primary antibodies were employed for these studies: CD147 (catalog no. 555961), CD147-FITC (catalog no. 55962), IgG-FITC, caveolin-1 (catalog no. 610059), and Flotillin-1 (catalog no. 610820) (BD Biosciences); Alexa-Fluor 647-conjugated CD147, Alexa-Fluor 488-conjugated CD147 (catalog no. 306208), and phycoerythrin-conjugated EGFR (catalog no. 352903) (Biolegend); cortactin (clone 4F11; catalog no. 05-180) and membrane type-1 MMP (MT1-MMP)/hinge region polyclonal (catalog no. AB6004) (Millipore); phospho-ERK1/2 (Thr-202/Tyr-204) (catalog no. 9106), pan-ERK1/2, phospho-EGFR (Tyr-1173)(catalog no. 4407), phospho-EGFR (Tyr-1068) (catalog no. 3777), and EGFR (catalog no. 4267) (Cell Signaling Technology, Inc); CD44/HCAM (mouse) and Alexa-Fluor 647-conjugated CD44 (catalog no. sc-7297) (Santa Cruz Biotechnology, Inc.); transferrin receptor and β-coatomer protein 1 (Abcam); phycoerythrin-conjugated CD44 (catalog no. CL8946PE) (Cedarlane Laboratories); CD44 (rabbit) (Epitomics); and β-actin and β-tubulin (Sigma). Goat anti-mouse HRP and Goat anti-rabbit HRP (Chemicon) were used as secondary antibodies for immunoblotting. Western blotting detection reagent enhanced chemiluminescence (ECL) was purchased from Pierce. The inhibitors tyrphostin (Calbiochem), U0126, and PD98059 (Cell Signaling Technology, Inc.) were used at concentrations indicated in the figure legends.
Recombinant Adenovirus Infections
Recombinant human CD147 adenovirus and control β-galactosidase (β-gal) adenovirus were constructed and used as described previously (27, 29). In each experiment, cells were incubated with these adenoviruses for 24 h in full medium prior to other treatments.
Surface Biotinylation
Cell surface biotinylation was performed using a Cell Surface Protein Isolation Kit according to the manufacturer's protocol (Pierce). Briefly, cells were treated with EZ-Link sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate for 45 min at 4 °C, followed by quenching and harvesting of cell lysates. Equivalent amounts of protein from each group were added to spin columns containing NeutrAvidin resin, incubated for several h with rocking at 4 °C, washed, eluted, and analyzed by immunoblotting.
Ras Activity Assay
Ras activation status was evaluated by employing the Active Ras Pull-Down and Detection Kit according to the manufacturer's protocol (Pierce). This assay is based on the observation that Ras-GTP (active), but not Ras-GDP (inactive), binds Raf-1 with high affinity. Briefly, fresh lysates from MCF-10A cells, non-treated or treated with adenovirus, were collected using standard procedures. GTPγS (0.1 mm) or GDP (1 mm) was added to control lysates for a positive and negative control, respectively. GST-Raf1-Ras-binding domain fusion protein was added to the spin column containing a glutathione resin, followed by the addition of 400–500 μg of protein from positive/negative controls and β-gal- or CD147-treated cells. Lysates were incubated for 1 h at 4 °C with rocking, followed by elution and subsequent immunoblotting with a pan-Ras antibody.
Hyaluronan Assay
Hyaluronan concentrations in media were determined after a 24-h incubation, using an ELISA-like assay described previously (30).
Preparation of Hyaluronan Oligosaccharides
Hyaluronan oligosaccharides were prepared as described previously (31). Preparations were mainly composed of 6–12 monosaccharide units. Average oligosaccharide size was assessed from the ratio of terminal N-acetyl-d-glucosamine (32) to total uronic acid (33).
Matrigel Invasion Assay
Matrigel invasion was performed as described previously (27), using inserts (8-μm pores) for 24-well tissue culture plates (BD Biosciences). MCF-10A-EV cells, MCF-10A K-RasV12 cells, or MCF10A-K-RasV12 cells (7.0 × 105) pretreated with siRNA (72 h) or MEK inhibitors (24 h) were detached and seeded in the upper chamber of the inserts for these assays. Cells that invaded through the membrane overnight were counted in four separate fields in three independent experiments.
Invadopodia Analyses
The gelatin degradation assay was set up as described previously (27). Briefly, gelatin was conjugated with Alexa-Fluor-568 dye (Molecular Probes) using the manufacturer's protocol. 18-mm round glass coverslips were acid-washed with 1 m HCl overnight, followed by extensive washing in water. Coverslips were sterilized with 70% ethanol and then coated with 50 μg/ml poly-l-lysine for 20 min at room temperature, washed with PBS, and fixed with ice-cold 0.5% glutaraldehyde for 15 min, followed by extensive washing. Fixation with glutaraldehyde allows cross-linking of polylysine to the glass slide as well as subsequent cross-linking of the gelatin layer. The coverslips were then inverted on an 80-μl drop of fluorescent gelatin matrix (0.2% gelatin and Alexa-Fluor-gelatin at an 8:1 ratio) and incubated for 15 min at room temperature. The coverslips were washed with PBS and quenched in 5 mg/ml sodium borohydride in PBS for 10 min, followed by further washing in PBS. The coated coverslips were then placed in 12-well plates, sterilized in 70% ethanol for 10 min, washed in PBS, equilibrated in serum-free mammary epithelial cell growth medium, and switched to fully supplemented mammary epithelial cell growth medium 30 min before the addition of cells.
For each assay, 5.5 × 104 cells were incubated on the gelatin-coated coverslips at 37 °C for 15–18 h. Cells were fixed and permeabilized with 3.7% paraformaldehyde and 0.1% Triton X-100 for 10 min, washed with PBS, blocked in 3% BSA in PBS for 1 h, and incubated with primary and secondary antibodies or Alexa-Fluor-647-phalloidin for actin in 3% BSA in PBS.
Assessment of invadopodia characteristics was performed as before (27), using at least 10 randomly chosen fields per experiment. Fields were evaluated for degraded matrix foci, which appear as dark “holes” in the fluorescent matrix, and for actin puncta that colocalize with cortactin. Results were presented as the percentage of cells with at least one degradation foci and as numbers per cell of, or percentage of cells displaying, cortactin-actin aggregates with or without degradation foci.
In Situ Proximity Ligation Assay (PLA)
Protein interactions were detected in situ, as described previously (27), using the Duolink II secondary antibodies and detection kit (Olink Bioscience, Uppsala, Sweden) according to the manufacturer's instructions. Briefly, primary antibodies targeting CD147 (mouse; BD Pharmingen), EGFR (rabbit; Cell Signaling), and CD44 (mouse (Santa Cruz Biotechnology) and rabbit (Santa Cruz Biotechnology)) were applied using standard procedures. Oligonucleotide-conjugated Duolink secondary antibodies were then added, followed by Duolink ligation solution. The oligonucleotides ligate together in a closed circle only when the secondary antibodies are in close proximity (<40 nm) (34). Polymerase was added, which amplified any existing closed circles, and detection was achieved with complementary, fluorescently labeled oligonucleotides. Specificity of the reaction was determined for each experimental group by treating cells with a single primary antibody toward one of the proteins of interest followed by Duolink secondary antibodies, which results in minimal to no signal amplification because of the absence of a second oligonucleotide needed for ligation and subsequent closed circle amplification.
Microscopy and Image Analysis
Images were acquired with a Leica Total Confocal System SP5 acoustic optical beam splitter confocal (TCS SP5 AOBS) microscope using a ×63, 1.4 numerical aperture oil objective at the Josh Spruill Molecular Morphology and Imaging Center (Medical University of South Carolina). Images were captured at high confocality (pinhole = 1 Airy unit) to achieve the thinnest possible optical slices at the cell-matrix interface (for invadopodia) or at the cell center (for PLA). All images are single optical sections. Potential overlaps in emission spectra were eliminated by sequential scanning and tuning of the acoustic optical beam splitter. Differential interference contrast microscopy was performed using the Leica SP5 confocal system. Image processing and compilation were performed with Canvas software (Deneba Systems Inc.) and Adobe Photoshop.
Quantitative RT-PCR
For quantitative RT-PCR experiments, total RNA was isolated using the RNeasy minikit according to the manufacturer's instructions (Qiagen, Valencia, CA), and quality control and quantification were performed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA) and Agilent RNA 6000 Pico LabChip kits. Complementary DNA (cDNA) was synthesized from equivalent concentrations of total RNA using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Coding sequences for CD147 and β-actin (internal control) were amplified from synthesized cDNA using iQ SYBR Green Supermix (Bio-Rad). Custom primer sequences used for amplification were as follows: CD147 sense, 5′-CAGAGTGAAGGCTGTGAAGTCG-3′; CD147 antisense, 5′-TGCGAGGAACTCACGAAGAA-3′; β-actin sense, 5′-GGAAATCGTGCGTGACATT-3′; β-actin antisense, 5′-GACTCGTCATACTCCTGCTTG-3′. Amplification was performed using an iCycler IQ real-time PCR detection system, and cycle threshold (Ct) values were determined in duplicate for CD147 and β-actin transcripts for each experiment. “No template” (water) and “no-RT” controls were used to ensure minimal background signal from DNA contamination. Using mean Ct values tabulated for different experiments and Ct values for β-actin as loading controls, -fold changes for experimental groups relative to assigned controls were calculated using automated iQ5 2.0 software (Bio-Rad).
RNA Interference
Control non-silencing siRNA and target-specific pooled siRNAs for CD147 and CD44 were purchased from Santa Cruz Biotechnology and used according to the manufacturer's protocol. For all experiments, cells were transfected with siRNA 72 h before other treatments.
Detergent-resistant Membrane Fractionation
Detergent-resistant membrane domains (e.g. lipid rafts) were isolated as described previously (27). Briefly, cells cultured in D150 plates were washed with ice-cold PBS three times. Cells were lysed in 500 μl of lysis buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm dithiothreitol, 10% sucrose, 1% Triton X-100, 1 mm PMSF, 10 μg/ml aprotinin and leupeptin, 2 mm sodium orthovanadate, and 10 mm sodium fluoride) on ice for 30 min. The cells were scraped and mechanically disrupted by passaging 20 times through a 3-inch 22-gauge needle. The lysates were mixed directly with iodixanol stock solution (60% solution of Optiprep iodixanol) to yield a 40% (v/v) iodixanol-lysate solution, which was placed at the bottom of an ultracentrifuge tube. Equal volumes of 35, 30, 25, 20, and 0% Optiprep in lysis buffer without Triton X-100 were carefully overlaid above the iodixanol-lysate solution. The samples were centrifuged at 160,000 × g for 8 h at 4 °C in a SW41-Ti rotor (Beckman Coulter). Equal fractions were collected from the top of the tube, and equivalent aliquots of each fraction were subjected to immunoblotting using antibodies recognizing proteins of interest.
Labeling of Live Cells with Cholera Toxin-B (Ctx-B) to Identify Lipid Rafts
Lipid raft labeling was performed using the Vybrant Alexa-Fluor-488 Lipid Raft labeling kit using the manufacturer's protocol (Invitrogen). Briefly, live cells were washed with ice-cold PBS, labeled with Alexa-Fluor-488-conjugated Ctx-B, and cross-linked with anti-Ctx-B antibody, followed by standard immunofluorescence staining and mounting techniques.
Cell Surface Analysis and Fluorescence-activated Cell Sorting of CD147Lo and CD147Hi Subpopulations
For cell surface analysis, 2.0 × 105 cells were trypsinized to achieve single cell suspensions, counted, blocked with 3% BSA in PBS, and incubated with primary conjugated antibodies for 30 min on ice. Unbound antibody was removed in subsequent washes, and cells were analyzed no longer than 1 h poststaining. Cell sorting of CD147Lo and CD147Hi subpopulations was achieved in the following manner. Cells (10A-K-RasV12, MDA-MB-231, and MCF-7) were trypsinized into single cell suspensions, counted, blocked with 3% BSA in PBS, and treated with antibodies in 3% BSA in PBS. Cells were incubated with anti-CD147-FITC or IgG-FITC (negative control) for 30 min on ice. Unbound antibody was washed off, and cells were resuspended in mammary epithelial cell growth medium with supplements and DNase (Promega), filtered, and sorted immediately after processing. Cells expressing the lowest surface levels of CD147 (bottom 20%) and the highest surface levels of CD147 (top 20%) were selected as CD147Lo and CD147Hi subpopulations, respectively. For subsequent invadopodia or immunoblotting analyses, cells were directly seeded on gelatin or lysed postsorting, respectively. All FACS analysis and cell sorting was performed with a Beckman Coulter MoFlo Astrios instrument in the Department of Regenerative Medicine and Cell Biology Flow Cytometry Facility (Medical University of South Carolina).
Immunoblotting
Whole cell lysates were prepared for immunoblotting using a radioimmune precipitation assay buffer modified to contain 1 mm PMSF; 10 μg/ml aprotinin, pepstatin, and leupeptin; 2 mm sodium orthovanadate; and 10 mm sodium fluoride. Protein content was quantified using a BCA assay (Pierce), and aliquots were solubilized in reducing sample buffer, resolved on Pierce 4–20% polyacrylamide gels, transferred to nitrocellulose (Osmonics, Westborough, MA) with a Pierce apparatus, blocked in 3% nonfat dry milk or 3% BSA with Tris-buffered saline and 0.1% Tween 20, and stained with primary antibodies. HRP-linked secondary antibodies were detected with ECL, and sizes of proteins were estimated from molecular weight standards electrophoresed on the same gel as the samples. Band densitometric analysis was performed with ImageJ version 1.43s software, and values were recorded with Microsoft Excel software.
Co-immunoprecipitation
MDA-MB-231 cells were lysed as described previously (35), using buffer containing 1% (v/v) Nonidet P-40, 0.5 mm EGTA, 2 mm sodium orthovanadate, 10% (v/v) glycerol, 1 mm PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 10 μg/ml aprotinin, 2 mm benzamidine HCl, 2 mm sodium fluoride, and 50 mm HEPES, pH 7.5, at 4 °C. Aliquots of cell lysates containing 1 mg of protein were used for co-immunoprecipitation with antibodies against human CD147 (mouse IgG; BD Biosciences) or CD44 (mouse IgG; Santa Cruz Biotechnology), which were pretreated with the Pierce antibody clean-up kit to remove gelatin, according to the manufacturer's instructions. Co-immunoprecipitation was performed with the Pierce co-Immunoprecipitation kit according to the manufacturer's instructions. After the addition of lysate to the antibody-conjugated resins, the resins were incubated overnight and then washed five times to remove non-bound antigens. Control immunoprecipitations were carried out in a similar manner using mouse IgG. The control and test resins were then eluted with the lysis buffer employed under “Immunoblotting,” and the eluates were used for immunoblotting with antibodies against CD147, CD44, and EGFR. Resin washes were also subject to immunoblotting with the same antibodies to ensure that non-bound antigens were completely removed.
Statistical Analysis
Statistical tests and plots were performed with GraphPad Prism software. Error bars depict S.E. of three or more independent experiments unless noted otherwise in the figure legends. Statistical significance was determined by using one-way analysis of variance (α = 0.05) followed by comparison of all experimental groups with Bonferroni's post-test and two-sided Student's t test. Differences were considered significant if p was <0.05.
RESULTS
Up-regulation of CD147 Induces Activation of EGFR-Ras-ERK Signaling in Breast Epithelial Cells
As shown in our previous study (27), a multiplicity of infection (MOI) of 2 of our CD147 adenovirus construct gives rise to levels of CD147 in MCF-10A cells that are commonly found in aggressive human breast cancer cells. In addition, we have now shown that the level of CD147 at the surface of MCF-10A cells resulting from treatment with an equivalent MOI of virus is similar to that expressed on the surface of the malignant breast carcinoma cell line, MDA-MB-231, but as shown previously, the adenovirus induces mainly the less glycosylated form of CD147 found endogenously in cancer cells (Fig. 1A). As in our previous study, we used cells infected with β-gal adenovirus as a control; this adenovirus did not cause significant changes in CD147 expression. An MOI of 2 was used in all subsequent adenovirus experiments.
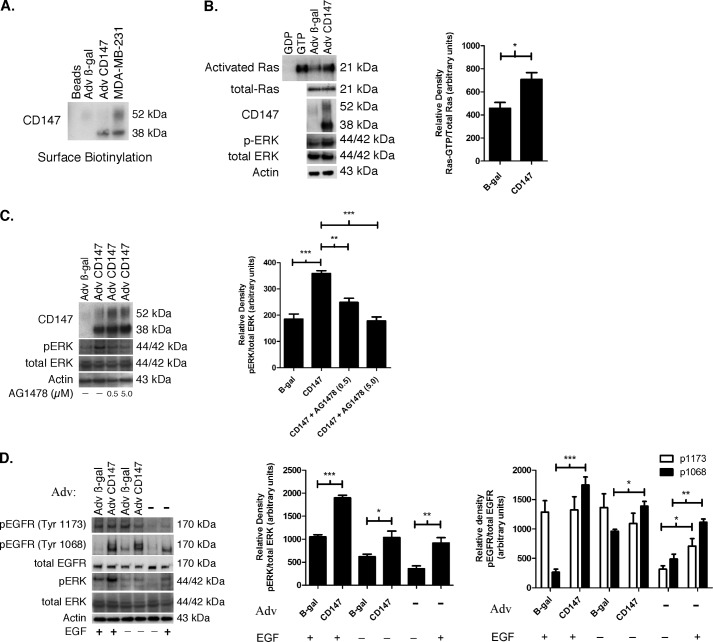
FIGURE 1.
Up-regulation of CD147 induces activation of EGFR-Ras-ERK signaling. A, Western blot depicting surface biotinylation of MCF-10A cells, treated with an MOI of 2 of β-gal or CD147 adenovirus (Adv), compared with untreated MDA-MB-231 cells. An MOI of 2 was used in all subsequent adenovirus experiments. For a negative control, a lane was loaded with eluate from NeutrAvidin resin only (labeled Beads), which was prepared similarly to experimental groups but without cell lysate; n = 3. B, evaluation of Ras activity and ERK phosphorylation in lysates of MCF-10A cells that had been pretreated in culture with β-gal or CD147 adenovirus. Activated Ras was identified by probing with a pan-Ras antibody. Parental MCF-10A whole cell lysates treated directly with GDP and GTP were used as negative and positive controls, respectively. β-Actin was used as a loading control. Left, a representative gel; right, densitometric quantitation of Ras-GTP versus total Ras. Columns are means ± S.E. (error bars); n = 4; *, p < 0.05. C, effect of inhibition of EGFR activity on ERK phosphorylation in MCF-10A cells treated with β-gal or CD147 adenovirus. After treatment with adenovirus for 24 h in full medium, cells were incubated in supplement-free medium for 4 h and subsequently treated with vehicle (DMSO) or 0.5 and 5.0 μm tyrphostin (AG1478) for 12 h. β-Actin was used as a loading control. Left, a representative gel; right, densitometric quantitation of p-ERK versus total ERK. Columns are means ± S.E.; n = 3; **, p < 0.01; ***, p < 0.001. D, EGFR and ERK phosphorylation in MCF-10A cells treated with β-gal or CD147 adenovirus for 24 h in fully supplemented medium, followed by incubation in medium containing supplements with and without EGF (20 ng/ml) for 6 h; EGFR activation was evaluated by probing for phosphorylation of the tyrosine 1173 or 1068 residues. For controls, parental MCF-10A cells were incubated in medium containing supplements with and without EGF (20 ng/ml) for 6 h. β-Actin was used as a loading control. Left, a representative gel; middle, densitometric quantitation of p-ERK versus total ERK; right, densitometric quantitation of p-EGFR versus total EGFR. Columns are means ± S.E.; n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Prior evidence suggests that CD147 activates ERK signaling (15, 36–38), although the upstream regulators of this pathway have not been fully characterized. Here, we first evaluated the effect of increased CD147 on the activation status of Ras in MCF-10A cells because Ras is frequently upstream of ERK activation (39). We found that up-regulation of CD147 led to a ∼1.5–2-fold increase in Ras activation, measured as an increased proportion of Ras-bound GTP, as well as a concomitant increase in phosphorylated ERK (p-ERK) (Fig. 1B). Next, we investigated EGFR signaling because it is often dysregulated in breast cancer and is commonly upstream of Ras and ERK (40). We found that treatment of MCF-10A cells with increasing concentrations of the specific EGFR kinase inhibitor AG1478, subsequent to up-regulation of CD147, resulted in attenuated p-ERK (Fig. 1C), suggesting that CD147 may be involved in EGFR activation. Because phosphorylation of tyrosine 1173 or 1068 on the cytoplasmic tail of EGFR has been associated with regulation of Ras-MAPK signaling (41, 42), we determined whether these tyrosine residues were phosphorylated in response to increased CD147 expression. Interestingly, we found that increased expression of CD147 resulted in a large increase in phosphorylation of the Tyr-1068 residue but not Tyr-1173 (Fig. 1D); increased Tyr-1068 phosphorylation occurred whether or not EGF was present in the medium used for analysis of EGFR activation subsequent to adenoviral transfection (Fig. 1D). Also, in the MCF-10A cells studied here, EGF alone without preincubation with adenoviral vectors caused a greater increase in phosphorylation of Tyr-1068 than Tyr-1173 (Fig. 1D).
Hyaluronan-CD44 Interaction Mediates CD147-induced Activation of EGFR Signaling
Previously we demonstrated that CD147 stimulates the synthesis of hyaluronan in breast carcinoma cells (15). Here we found that up-regulation of CD147 in non-transformed MCF-10A breast epithelial cells also led to a ∼2-fold increase in hyaluronan production compared with control cells (Fig. 2A). Several studies (43) have demonstrated that endogenous or exogenous hyaluronan can elicit activation of multiple signaling pathways, including those involving receptor tyrosine kinases (e.g. EGFR) and downstream effectors (e.g. Ras and ERK), which are frequently dysregulated in breast cancer. Consequently, we determined whether the effects of CD147 on activation of EGFR and ERK were dependent on hyaluronan-receptor interactions. We first employed treatment with oligosaccharides of hyaluronan that are 6–12 residues in length, which compete with multivalent hyaluronan-receptor interactions by replacing them with monovalent hyaluronan oligomer-receptor interactions (44, 45), thus attenuating endogenous hyaluronan-receptor signaling (35, 46). We found that treatment of MCF-10A cells with hyaluronan oligomers, subsequent to up-regulation of CD147, resulted in attenuated phosphorylation of EGFR Tyr-1068 and ERK (Fig. 2B). Next, to determine whether this perturbation of hyaluronan-receptor signaling is dependent on CD44, the main receptor for hyaluronan in breast cancer cells, we tested the effects of pooled siRNA-mediated knockdown of CD44 in MCF-10A cells subsequent to up-regulation of CD147. The partial depletion of CD44 resulted in corresponding decreases in activation of EGFR Tyr-1068 and p-ERK levels, whereas significant differences were not seen in expression of CD147 or MT1-MMP (MMP-14) (Fig. 2C), an enzyme that has previously been shown to be induced by CD147 (27, 47).
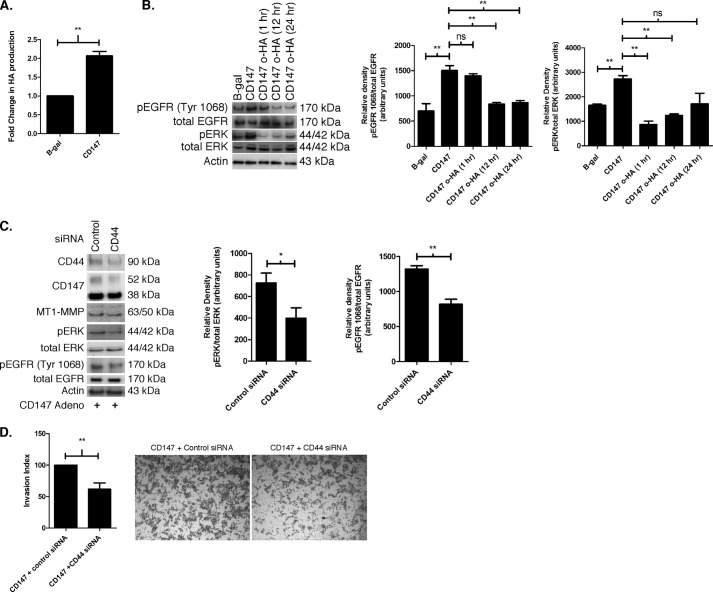
FIGURE 2.
Hyaluronan-CD44 interaction mediates CD147-induced activation of EGFR signaling. A, hyaluronan secretion in MCF-10A cells treated with β-gal or CD147 adenovirus. After seeding the adenovirus-treated cells into 24-well plates and incubating for 24 h, the media were replaced with fresh media. After another 24-h incubation, media were collected and analyzed for hyaluronan concentration using an ELISA-like assay. Hyaluronan (HA) in the media was normalized to cell number and depicted as mean -fold change. Columns are means ± S.E. (error bars); n = 3; **, p < 0.01. B, effects of hyaluronan oligomers (o-HA) on activation of EGFR and ERK. MCF-10A cells treated with β-gal or CD147 adenovirus were subsequently treated with 100 μg/ml o-HA for 1, 12, and 24 h and probed for p-EGFR (Tyr-1068) and p-ERK. β-Actin was used as a loading control. Left, a representative gel; middle, densitometric quantitation of p-EGFR (Tyr-1068) versus total EGFR; right, densitometric quantitation of p-ERK versus total ERK. Columns are means ± S.E.; n = 3; *, p < 0.05; **, p < 0.01; ns, not significant. C, effects of CD44 knockdown on activation of EGFR and ERK. MCF-10A cells treated with CD147 adenovirus were subsequently treated with nonspecific control siRNA or pooled CD44-specific siRNA as described under “Experimental Procedures” and probed for the indicated proteins. β-Actin was used as a loading control. Left, a representative gel; middle, densitometric quantitation of p-ERK versus total ERK; right, densitometric quantitation of p-EGFR (Tyr-1068) versus total EGFR. Columns are means ± S.E.; n = 3; *, p < 0.05; **, p < 0.01. D, effect of CD44 knockdown on invasion. Shown are quantitation and representative images of cell invasion through Matrigel by CD147-up-regulated MCF-10A cells after treatment with nonspecific control siRNA or pooled CD44-specific siRNA. The cell invasion index was calculated as the number of invasive cells that had been treated with CD147 adenovirus and then pooled CD44-specific siRNA, normalized to the number of invasive cells that had been treated with CD147 adenovirus then nonspecific control siRNA. Columns are means ± S.E.; n = 3; **, p < 0.01.
Because CD147 induces invasiveness, we next evaluated whether depletion of CD44 in MCF-10A cells, subsequent to up-regulation of CD147 expression, altered invasiveness. We found that CD44 knockdown resulted in a ∼2-fold decrease in invasion through Matrigel compared with control siRNA (Fig. 2D). These results suggest that CD147-induced activation of the EGFR-ERK pathway is at least partially mediated by hyaluronan-CD44 interactions and that induction of MT1-MMP expression by CD147 may employ a separate pathway independent of CD44. Although CD147 does not appear to employ CD44 signaling for induction of MT1-MMP expression, CD44 does participate in CD147-mediated invasion through EGFR-ERK signaling.
CD147 Promotes the Formation of Signaling Complexes
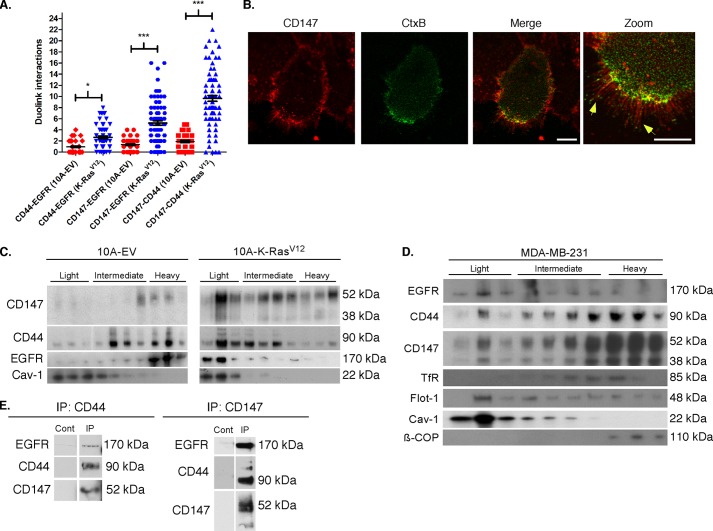
CD44 has been shown to interact with EGFR family members in a hyaluronan-dependent manner (35, 48, 49), and our above data show that up-regulation of CD147 results in increased hyaluronan-CD44-mediated EGFR activation. To evaluate whether up-regulation of CD147 increases EGFR-CD44, CD147-CD44, or CD147-EGFR interactions, we employed a PLA technique that detects associations of proteins occurring within 40 nm of each other (34). We found that there were significant differences between CD147-up-regulated and control MCF-10A cells in regard to the percentage of the cell population demonstrating close associations of CD44 with EGFR, CD147 with EGFR, and CD147 with CD44 as well as the numbers of such associations per optical section (Fig. 3, A and B).
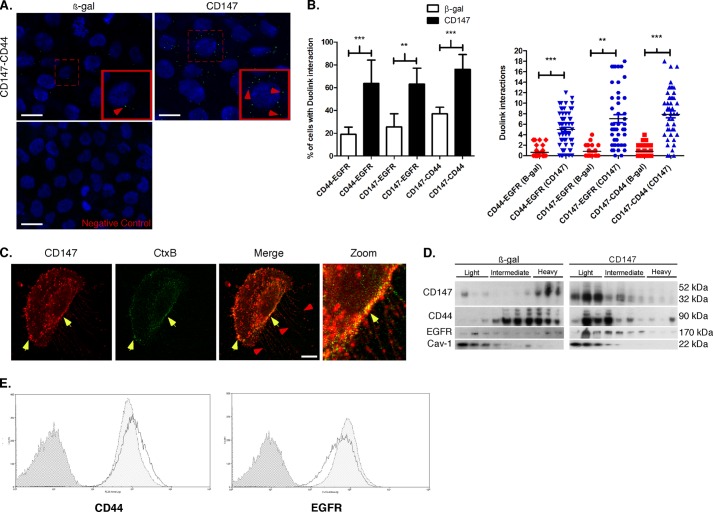
FIGURE 3.
CD147 induces assembly of signaling complexes. A, representative images demonstrating protein interactions (<40 nm) using PLA (see “Experimental Procedures” for details). CD147-treated MCF-10A cells were probed with primary antibodies in the following combinations to detect protein interactions in the cell population: CD44 (mouse) and EGFR (rabbit), CD147 (mouse) and EGFR (rabbit), and CD147 (mouse) and CD44 (rabbit). Protein interactions appear as small punctate green signals (i.e. dots), and cell nuclei were identified by co-staining with DAPI. Examples of these protein interactions in MCF-10A cells treated with β-gal or CD147 adenovirus, in this case interactions between CD147 and CD44, are indicated by the red arrowheads. The bottom panel shows a representative negative control where only one primary antibody was employed. B, quantitation of the indicated protein interactions in the cell population over five random fields with at least 30 cells/field. Left, percentage of cells showing interaction; column values are means ± S.E. (error bars) over three independent experiments. Right, interactions per optical slice, estimated to be at the center of the cell based on DAPI signal; scatter plot showing each interaction with mean ± S.E.; n = 3; **, p < 0.01; ***, p < 0.001. C, representative image of MCF-10A cells treated with CD147 adenovirus and evaluated for CD147 (red), cholera toxin-B (green), and colocalization (yellow; indicated by yellow arrowheads). Examples of filopodia are identified by red arrowheads. Scale bar, 10 μm. The CD147 channel was pseudocolored from blue to red for easier visualization of colocalization. D, MCF-10A cells treated with β-gal or CD147 adenovirus were subjected to detergent-resistant membrane isolation (see “Experimental Procedures”) and probed for the indicated proteins. Light fractions are from the gradient interface (0–20%), where detergent-resistant membrane domains, such as lipid rafts, localize; n = 3. Cav-1, caveolin (lipid raft marker). E, CD44 and EGFR surface expression was analyzed by FACS in CD147-treated MCF-10A cells. Distributions with dark gray shading, light shading, and no shading represent IgG-isotype control, β-gal, and CD147 treated cells, respectively; n = 3.
We and others have demonstrated that CD147, CD44, and EGFR partially localize in membrane fractions with properties of lipid rafts in malignant cancer cells (27, 35, 43, 50, 51), and that lipid rafts are critical to invadopodia function (52–54). Thus, we evaluated the relative localization of CD147 and Ctx-B, a lipid raft marker (55), and found that they colocalize at the leading edge of the cell and appear to be arranged along Ctx-B-enriched filopodial projections (Fig. 3C). We also found that CD147, CD44, and EGFR are enriched in caveolin-rich lipid raft domains after up-regulation of CD147, as compared with the control cells (Fig. 3D). To evaluate whether the enrichment of CD44 and EGFR in lipid rafts is due to increased trafficking to the cell surface, we utilized FACS analysis to evaluate CD44 and EGFR surface expression levels in β-gal or CD147 adenovirus-infected cells. We found that up-regulation of CD147 had no significant effects on levels of CD44 or EGFR at the cell surface compared with control cells (Fig. 3E), suggesting that the major effect is on organization of these proteins into raftlike complexes rather than increased trafficking to the surface.
Oncogenic Ras and ERK Regulate Hyaluronan, CD147, and MT1-MMP Expression and Invasiveness
Our data here indicate that up-regulation of CD147 increases Ras-ERK signaling via hyaluronan-CD44 interactions. However, other evidence indicates that the MEK-ERK pathway may also regulate CD147 expression (56–58), suggesting the possibility of a positive feedback loop. To evaluate whether the Ras-MEK-ERK signaling pathway influences CD147 expression in the system studied here, we employed an MCF-10A cell line stably expressing the oncogenic K-RasV12 mutant (10A-K-RasV12) because this cell line has been shown to activate ERK signaling to a greater extent than PI3K (28).
First, we found that the 10A-K-RasV12 cells produced ∼3.5-fold more hyaluronan and expressed significantly higher levels of CD147 and MT1-MMP protein than empty vector MCF-10A (10A-EV) cells (Fig. 4, A and B). Interestingly, quantitative PCR analysis demonstrated only a slight increase in CD147 message compared with 10A-EV cells, suggesting that K-RasV12 may regulate CD147 expression post-transcriptionally (Fig. 4C).
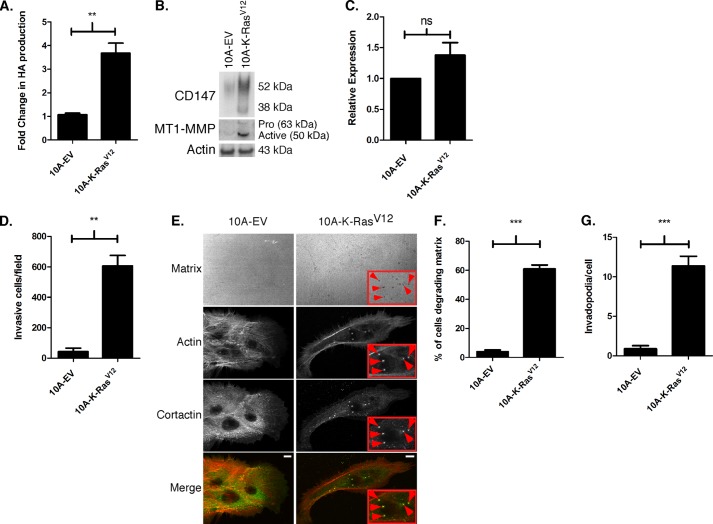
FIGURE 4.
Activated Ras regulates hyaluronan production, CD147 and MT1-MMP expression, and invasiveness. A, hyaluronan production over a 24-h period was quantitated in 10A-EV and 10A-K-RasV12 cells using an ELISA-like assay. Hyaluronan (HA) in the media was normalized to cell number and depicted as mean -fold change ± S.E.; each column represents three independent experiments; **, p < 0.01. B, representative Western blot depicting CD147 and MT1-MMP expression in 10A-EV and 10A-K-RasV12 cells. β-Actin was used a loading control; n = 4. C, comparison of CD147 mRNA levels in 10A-EV and 10A-K-RasV12 cells, as measured by quantitative PCR and determined by cycle threshold values (see “Experimental Procedures” for details); normalized to β-actin; n = 3; ns, not significant. D, quantitation of invasion through Matrigel by 10A-EV and 10A-K-RasV12 cells. Columns represent the mean number of invasive cells ± S.E. per field; n = 3; **, p < 0.01. E, representative micrographs demonstrating invadopodia in 10A-EV and 10A-K-RasV12 cells cultured on fluorescent gelatin matrix. After fixation, cortactin was detected by immunolabeling (primary 4F11 antibody followed by secondary Alexa-Fluor-488 antibody) and actin by staining with Alexa-Fluor-647 phalloidin. Actin and gelatin matrix were pseudocolored red and blue, respectively, to allow easier visualization of colocalization (yellow) of actin (red) and cortactin (green). Invadopodia-mediated matrix degradation appears as dark black foci in the bright fluorescent matrix field (see panels labeled Matrix). The boxed regions with red arrowheads depict actively degrading invadopodia. Scale bar, 10 μm. F, percentage of cells degrading the matrix; defined as a cell with at least one degradation spot underneath the cell or near the cell edge. G, quantification of invadopodia/cell; defined as actin-cortactin aggregate over degraded matrix. Each invadopodia parameter was calculated by evaluating random fields containing at least 15 cells/field over three independent experiments. Column values are means ± S.E. ***, p < 0.001.
We also found that the 10A-K-RasV12 cells invaded Matrigel (Fig. 4D) and formed active invadopodia (Fig. 4E) to a greater extent than 10A-EV cells. In the invadopodia assay, ∼60% of the 10A-K-RasV12 cells degraded the underlying fluorescent matrix (Fig. 4F). The 10A-K-RasV12 cells had a mean of 10–12 invadopodia/cell and a range of 1–28 invadopodia/cell, whereas 10A-EV cells demonstrated no invadopodia (Fig. 4G). Thus, these data indicate that the K-RasV12 oncogene imparts an invasive phenotype in MCF-10A cells, similar to that demonstrated recently in pancreatic cells (59, 60).
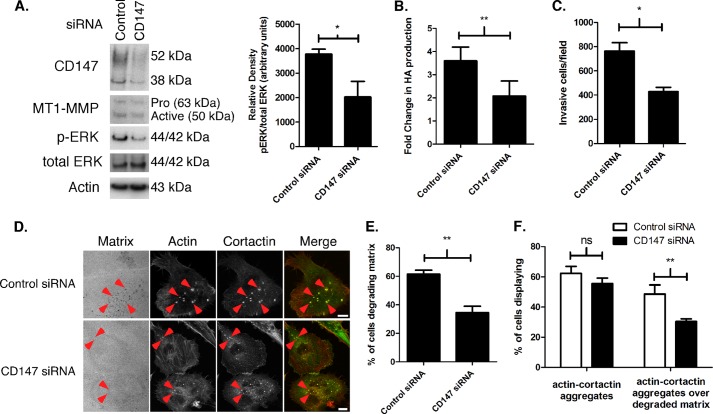
Because we and others have shown previously that CD147 regulates MT1-MMP expression (2, 27, 47) and hyaluronan synthesis (15–18) (Fig. 2A), we examined whether increased expression of MT1-MMP or hyaluronan in 10A-K-RasV12 cells was due to the observed increase in CD147. Partial knockdown of CD147, obtained using pooled siRNA, demonstrated that MT1-MMP expression (Fig. 5A) and hyaluronan production (Fig. 5B) were at least partially dependent on CD147. We also found that CD147 depletion attenuated activation of ERK in 10A-K-RasV12 cells (Fig. 5A). Because CD147 plays a prominent role in breast cancer cell invasion and invadopodia formation (10, 27), we evaluated effects on Matrigel invasion and invadopodia formation and activity in the 10A-K-RasV12 cells. We found that the partial knockdown of CD147 resulted in a partial, but significant, decrease in Matrigel invasion in these cells (Fig. 5C). Likewise, in the invadopodia assay, CD147 knockdown decreased the percentage of cells degrading matrix (Fig. 5, D and E), but most of this inhibition was due to decreased activity of invadopodia rather than decreased formation (Fig. 5F).
FIGURE 5.
Knockdown of CD147 in MCF-10 K-RasV12 cells results in decreased hyaluronan synthesis, MT1-MMP expression, ERK activation, and invasiveness. A, Western blot depicting CD147 and MT1-MMP protein levels and ERK phosphorylation in 10A-K-RasV12 cells depleted of CD147 by treatment with pooled siRNAs. Nonspecific siRNA was used as a control for CD147 knockdown, and β-actin was used as a loading control. Left, a representative gel. Right, densitometric quantitation of p-ERK versus total ERK. Columns, means ± S.E. (error bars); n = 3; *, p < 0.05. B, comparison of hyaluronan (HA) production in 10A-K-RasV12 cells treated with nonspecific control or pooled CD147-specific siRNA. Hyaluronan in the media was normalized to cell number and depicted as mean -fold change ± S.E.; each column represents three independent experiments; **, p < 0.01. C, quantitation of cell invasion through Matrigel by 10A-K-RasV12 cells treated with nonspecific control or pooled CD147-specific siRNA. Columns represent the mean number of invasive cells/field ± S.E.; n = 3; *, p < 0.05. D, representative micrographs demonstrating invadopodia in 10-K-RasV12 cells treated with nonspecific control or pooled CD147-specific siRNA cultured on fluorescent gelatin matrix. Red arrowheads, actin-cortactin aggregates. Scale bar, 10 μm. E, percentage of cells degrading underlying matrix or; F, percentage of cells with actin-cortactin aggregates with or without underlying degraded matrix in 10A-K-RasV12 cells treated with nonspecific control or pooled CD147-specific siRNA. Each invadopodia parameter was calculated by evaluating random fields containing at least 15 cells/field over three independent experiments. Column values are means ± S.E.; **, p < 0.01; ns, not significant.
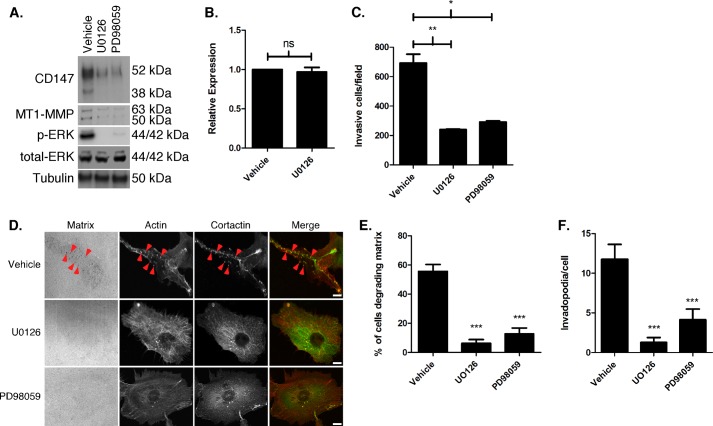
Next we determined whether increased CD147 expression in 10A-K-RasV12 cells was dependent on ERK signaling. Treatment of 10A-K-RasV12 cells with two different MEK inhibitors resulted in a decrease in CD147 and MT1-MMP protein levels as well as p-ERK (Fig. 6A), although this effect did not appear to be at the transcriptional level for CD147 (Fig. 6B). We next evaluated whether ERK signaling mediates the invasive characteristics that we described above in 10A-K-RasV12 cells. We found that MEK-ERK inhibition greatly decreased Matrigel invasion (Fig. 6C) as well as invadopodia formation and activity (Fig. 6, D–F). We monitored cell viability during the course of treatment with the MEK-ERK inhibitors and did not observe significant cell death (data not shown). In contrast to CD147 depletion in 10A-K-RasV12 cells, MEK-ERK inhibition greatly decreased invadopodia formation as well as activity.
FIGURE 6.
Increased CD147 and MT1-MMP expression and invasiveness in MCF-10A-K-RasV12 cells is mediated by MEK-ERK signaling. A, Western blot depicting 10A-K-RasV12 cells treated with vehicle, U0126 (10 μm), or PD98059 (25 μm) for 12 h. β-Tubulin was used as a loading control; n = 3. B, CD147 mRNA levels of 10A-K-RasV12 cells treated with vehicle or UO126 (10 μm) as measured by quantitative PCR and determined by cycle threshold values (see “Experimental Procedures” for details); normalized to β-actin; n = 3; ns, not significant. C, quantitation of cell invasion through Matrigel by 10A-K-RasV12 cells pretreated with vehicle, U0126 (10 μm), or PD98059 (25 μm) for 24 h. Columns, mean number of invasive cells/field ± S.E. (error bars); n = 3; *, p < 0.05; **, p < 0.01. D, representative micrographs showing invadopodia in 10A-K-RasV12 cells pretreated for 30 min with vehicle, U0126 (10 μm), or PD98059 (25 μm) and then seeded on fluorescent gelatin matrix in the presence of vehicle or inhibitors; see “Experimental Procedures” for further details. Red arrowheads, examples of invadopodia. Scale bar, 10 μm. E, percentage of cells degrading underlying matrix; or F, quantitation of invadopodia/cell in 10A-K-RasV12 cells treated with vehicle, U0126 (10 μm), or PD980539 (25 μm). Each invadopodia parameter was calculated by evaluating random fields containing at least 15 cells/field over three independent experiments. Column values are means ± S.E.; ***, p < 0.001.
Similar to our approach with CD147-up-regulated MCF-10A cells (Fig. 3, A and B), we evaluated associations of CD44 with EGFR, CD147 with EGFR, and CD147 with CD44 in 10A-K-RasV12 cells using PLA. We found that these cells also exhibited significant increases in these associations as compared with control 10A-EV cells (Fig. 7A), which had an average of <1 association per optical section, similar to β-gal control cells (Fig. 3B). In the 10A-K-RasV12 cells, the incidence of close associations varied in the following manner: CD147-CD44 > CD147-EGFR > CD44-EGFR (Fig. 7A). These data suggest a relationship similar to that identified in MCF-10A cells after adenoviral up-regulation of CD147 (Fig. 3, A and B). Also, we found that endogenous CD147 colocalizes with Ctx-B (Fig. 7B) and that CD147, CD44, and EGFR are enriched in caveolin-rich raft domains in 10A-K-RasV12 cells compared with the 10A-EV cells (Fig. 7C), as is also observed in MCF-10A cells with adenovirally up-regulated CD147.
FIGURE 7.
Identification and localization of endogenous protein interactions in MCF-10A-EV, MCF-10A-K-RasV12, and MDA-MB-231 cells. A, endogenous protein interactions (<40 nm) were analyzed by PLA (see “Experimental Procedures” for details) and quantitated for the number of indicated interactions/optical slice in a similar manner as in Fig. 3. Shown is a scatter plot of the indicated protein interactions in MCF-10A-EV and MCF-10A-K-RasV12 cells with mean ± S.E.; n = 3; *, p < 0.05; ***, p < 0.001. B, representative image demonstrating colocalization (yellow) of CD147 (red) and cholera toxin-B (green) in 10A-K-RasV12 cells. Scale bar, 10 μm; the CD147 channel was pseudocolored from blue to red for easier visualization. Filopodia are highlighted with yellow arrowheads. C, Western blot comparing protein distribution between 10A-EV and 10A-K-RasV12 cells subjected to detergent-resistant membrane isolation and probed for indicated proteins. Light fractions are from the gradient interface (0–20%), where detergent-resistant membrane domains, such as lipid rafts, localize; n = 2. Cav-1, caveolin (lipid raft marker). D, Western blot showing protein distribution for MDA-MB-231 cells subjected to lipid raft analysis as in C; n = 2. Tfr, transferrin receptor (non-raft plasma membrane marker); Flot, flotillin-1 (lipid raft marker); β-COP, coatomer protein (Golgi marker). E, co-immunoprecipitation of CD147, CD44, and EGFR from lysates of MDA-MB-231 cells. Whole cell lysates from MDA-MB-231 cells were co-immunoprecipitated (IP) with mouse Ig antibodies against human CD147 or CD44, as described under “Experimental Procedures”; controls (Cont) were performed in a similar manner with mouse Ig. Immunoblotting of aliquots of test and control eluates from the antibody-conjugated resins was performed with antibodies against CD147, CD44, and EGFR; similar results were obtained in two independent experiments.
To ensure that similar interactions occur endogenously in invasive breast cancer cells without experimental manipulation, we analyzed MDA-MB-231 breast carcinoma cells, which possess an endogenous K-RasG13D mutation (61), for lipid raft components. We found that these cells also contained CD147, CD44, and EGFR in discrete fractions from gradient interfaces that correspond to lipid raftlike complexes (light fraction) (Fig. 7D). Note that the non-raft plasma membrane protein, transferrin receptor, co-distributes with the bulk of CD44 and CD147 in the intermediate and heavy fractions but does not co-distribute in the lipid raft-containing light fraction with CD147, CD44, and EGFR (Fig. 7D).
To further confirm that CD147, CD44, and EGFR form complexes in MDA-MB-231 cells, we determined whether CD147, CD44, and EGFR co-immunoprecipitate after treatment of cell lysates with antibody against either CD147 or CD44. We found that subfractions of CD44 and EGFR co-immunoprecipitate with CD147 and that subfractions of CD147 and EGFR co-immunoprecipitate with CD44 (Fig. 7E). These results imply that all three proteins participate in forming a common complex.
10A-K-RasV12 Cells with High Constitutive Surface Levels of CD147 Are More Invasive than Those with Low Constitutive Surface Levels of CD147
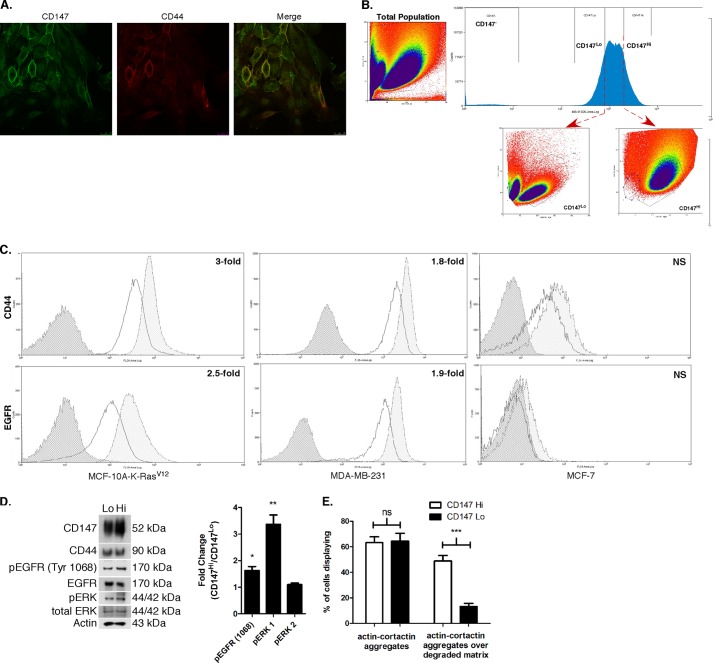
It has been clearly demonstrated that breast cancer cells exhibit heterogeneity in cell surface marker expression and contain subpopulations with different functional capacities (62). In recent studies from our laboratory (63, 64), we found that various types of malignant cancer cells exhibit heterogeneity in levels of cell surface CD147 and CD44 expression with corresponding functional differences. Therefore, we extended our current findings by evaluating whether subpopulations of 10A-K-RasV12 cells with different levels of expression of constitutive CD147 on the cell surface could be isolated from the parent cells. First, we found that immunostaining of 10A-K-RasV12 cells demonstrates a subpopulation of cells with greater CD147 and CD44 surface intensity (Fig. 8A). Next, by employing FACS, we isolated cells with relatively low surface expression of CD147 (CD147Lo) versus relatively high surface expression of CD147 (CD147Hi). We found that these two subpopulations exhibited distinct properties, as indicated by scatter analysis (Fig. 8B). The CD147Hi population also demonstrated 3-fold more CD44 and 2.5-fold more EGFR on the surface compared with the CD147Lo population (Fig. 8C, left panels), although total levels of these proteins were similar in the two fractions (Fig. 8D). Similar analysis of the highly invasive MDA-MB-231 breast cancer cells demonstrated a 1.8-fold increase in surface CD44 and a 1.9-fold increase in surface EGFR in CD147Hi versus CD147Lo cells (Fig. 8C, middle panels), whereas much less invasive MCF-7 cells demonstrated very little CD44 or EGFR surface staining, and subpopulation analysis for differential CD147 expression revealed no significant differences (Fig. 8C, right panels).
FIGURE 8.
MCF-10A-K-RasV12 cells with high constitutive surface levels of CD147 have elevated activation of EGFR and ERK and are more invasive. A, representative micrographs demonstrating heterogeneity in CD147 and CD44 staining intensity across the 10A-K-RasV12 cell population. Scale bar, 25 μm. B, FACS scatter distribution of total 10A-K-RasV12 cell population (left) and delineation of CD147Lo and CD147Hi cells (bottom and top 20%, respectively) in the distribution of CD147-expressing cells (top right) with corresponding scatter characteristics (bottom right). Note that this cell sorting scheme identifies two distinct cell populations. C, FACS analysis of cell surface expression of CD44 and EGFR in CD147Lo and CD147Hi cells in 10A-K-RasV12 (left; n = 3), MDA-MB-231 (middle; n = 3), and MCF-7 (right; n = 2) cells. Distributions with dark gray shading, no shading, and light gray shading represent IgG-isotype control, CD147Lo, and CD147Hi cells, respectively. D, left, immunoblot comparing protein expression of indicated proteins in CD147Lo and CD147Hi 10A-K-RasV12 cells. β-Actin was used as a loading control; n = 2. Right, histogram demonstrating -fold change between CD147Hi and CD147Lo cells for pEGFR (Tyr-1068), pERK1, and pERK2 protein levels; densitometric intensities were normalized to total EGFR and ERK1/2 protein; n = 2. E, quantitation of the percentage of cells with actin-cortactin aggregates with or without underlying degraded gelatin matrix in CD147Lo and CD147Hi 10A-K-RasV12 cells was calculated by evaluating random fields containing at least 15 cells/field over three independent experiments. Column values are means ± S.E. (error bars); ***, p < 0.001; ns, not significant.
We also measured levels of activated ERK and EGFR in the two 10A-K-RasV12 subpopulations by immunoblotting and found that CD147Hi cells expressed increased p-ERK1 and p-EGFR (Y1068) levels compared with CD147Lo cells, whereas the total levels of CD44 and EGFR were similar (Fig. 8D). Finally, we evaluated the invasive characteristics of the CD147Hi versus CD147Lo cells. Similar to our above results in CD147-depleted 10A-K-RasV12 cells (Fig. 5F), we found that CD147Lo cells have a much lower percentage of cells with actively degrading invadopodia than CD147Hi cells, whereas no significant difference was observed between CD147Lo and CD147Hi cells in regard to total numbers of inactive plus active invadopodia (actin-cortactin aggregates) (Fig. 8E).
DISCUSSION
Increased ERK activation has been observed at the advancing margin of tumors derived from xenografts of CD147-overexpressing breast cancer cells (38). CD147-mediated ERK activation has also been observed in numerous cellular studies, but several different mechanisms have been invoked to explain these effects, including cis homodimerization of CD147 (37) and cooperation with cyclophilins (3), syndecan-1 (65), or hyaluronan-CD44 interactions (15). Our studies provide further evidence for cooperation between CD147 and hyaluronan-CD44 interactions, in this case through regulating the EGFR-Ras-ERK signaling pathway leading to invasiveness.
We and others have demonstrated that CD147 influences migration and invasion via protease induction and cytoskeletal remodeling (10, 27, 66, 67), although the exact signaling mechanisms regulating these processes are not understood. Here we have shown that up-regulation of CD147 in non-transformed breast epithelial cells leads to increased EGFR, Ras, and ERK activation. Moreover, we show that the activation of EGFR and ERK by CD147 is mediated in large part by increased hyaluronan-CD44 signaling and is accompanied by enhanced interactions between CD147, CD44, and EGFR. These protein interactions most likely take place within lipid raftlike regions of the plasma membrane, which are known to participate in signaling complex formation (68) and invadopodia formation/activity (52–54). Our data suggest that standard CD44 (∼90 kDa) is the major isoform involved in these interactions. The relative functions of standard and variant isoforms of CD44 are controversial. For example, some recent studies indicate that standard CD44 is essential for certain malignant characteristics (69, 70), whereas others indicate an important role for variant isoforms (71, 72). Clearly, this long argued issue requires further work before it is settled. Although we have shown that CD147 regulates hyaluronan production in the systems studied here and elsewhere (15, 16), we have not determined here whether this is due to increased expression of hyaluronan synthases. However, in a previous study, we found that CD147 regulates expression of hyaluronan synthases in breast carcinoma cells (15), suggesting that this is the mechanism underlying increased hyaluronan production induced by CD147.
In this study, we observed that a relatively low glycosylated form of CD147 was induced by treatment with recombinant CD147 adenovirus and that this form was targeted to the cell surface (Fig. 1A) and to lipid raft domains (Fig. 3D). However, in K-RasV12-transfected MCF-10A cells (Fig. 7C), both high and low glycosylated forms enter the raft fractions, accompanied by similar CD147-dependent effects on hyaluronan production, invadopodia activity, and invasiveness (Figs. 4 and 5). This implies that factors other than glycosylation regulate these phenomena. It should be noted that only a small fraction of high and low glycosylated CD147, as well as CD44 and EGFR, are present in lipid raft fractions in MDA-MB-231 cells (Fig. 7D), but interactions between these proteins clearly occur in these cells without experimental manipulations (Fig. 7E). We have also shown in MDA-MB-231 cells (27) that CD147 regulates expression of and interacts with MT1-MMP in invadopodia, most likely within lipid rafts therein, suggesting strongly that a small but critical subfraction of CD147 is preferentially distributed along with its binding partners in these structures.
The cytoplasmic tail of EGFR contains six tyrosine residues, which can be trans-autophosphorylated via the intrinsic tyrosine kinase activity of EGFR (40). Of these, the tyrosine residues 1173 (42) and 1068 (41) are important for EGFR-mediated Ras-MAPK signaling by acting as docking sites for the scaffold proteins Shc and Grb2, respectively. Here we found that CD147 induces phosphorylation of Tyr-1068 but not Tyr-1173. The reason for this differential phosphorylation is not known, but it may depend on spatio-temporal interactions between the phosphorylated tyrosine residues and associated scaffold proteins or on other cell-specific contexts (73–76). Supporting this possibility, we found that treatment of MCF-10A cells with EGF also resulted in greater activation of Tyr-1068 than Tyr-1173.
Ras mutations are infrequent in breast cancer (21), but chronic Ras activity is evident in breast cancer cell lines and patient tumor tissues without any apparent Ras mutations (22, 23). The present study suggests that CD147 stimulates wild-type Ras activity, as MCF-10A cells do not have mutant Ras forms (77). CD147 expression progressively increases as breast cancer transitions from a benign proliferation to an invasive carcinoma (9), but it is not known whether CD147 is a driver or passenger oncogene. To our knowledge, no mutations or intrinsic signaling motifs have been identified in CD147; thus, it is probable that CD147 mediates breast cancer progression by facilitating the assembly of protumorigenic signaling complexes on the cell surface.
Breast cancer is a heterogeneous disease and, as with other cancers, metastases to distant organs remain the primary cause of mortality (78). Currently, evaluation of hormone receptor expression, ErbB2 amplification, and specific genetic profiling represent the mainstay utilized clinically for treatment stratification, although much effort has also been placed on characterizing distinct subpopulations isolated from the tumor bulk with differing cell surface expression profiles. One of the main surface markers that has emerged in identifying invasive and therapy-resistant cancer cells is CD44 (79). Hyaluronan, the primary CD44 ligand, facilitates construction of pericellular matrix scaffolds that stabilize signaling and transporter complexes (31, 35, 80), and multiple studies have shown that hyaluronan-CD44 interactions are intimately involved in the integration of signaling pathways leading to receptor tyrosine kinase activation, therapy resistance, and invasion (43, 81). We and others have demonstrated that CD147 and hyaluronan receptors interact in various cancer cells (16, 80, 82); thus, we evaluated whether differential expression patterns of CD147 and CD44 occur in breast cancer cells. Our initial evaluation demonstrated heterogeneous staining intensity for CD147 and CD44 in the 10A-K-RasV12 cell population, and subsequent sorting of CD147Hi and CD147Lo cells demonstrated distinct cell subpopulations. We found that CD147Hi cells had significantly more CD44 and EGFR on the cell surface than their CD147Lo counterparts, albeit total expression levels were similar. Notably, increased CD147 expression induced by treatment with our CD147 adenovirus did not cause an increase in cell surface CD44 or EGFR but did cause redistribution into lipid raft complexes (Fig. 3). Additionally, CD147Hi 10A-K-RasV12 cells had increased pEGFR Tyr-1068 and pERK1 levels as well as more active invadopodia compared with CD147Lo cells. In a previous study (63), we also found that CD147Hi cells derived from MDA-MB-231 or primary mouse MMTV-PyMT mammary carcinoma cells were more invasive than the corresponding CD147Lo cells. Interestingly, in the current study (data not shown) and our previous study (63), densitometric analysis of CD147 in the CD147Hi and CD147Lo immunoblots demonstrated small changes in total CD147 protein, suggesting that the presence of increased CD147 on the cell surface may be due, at least in some cases, to enhanced retention and/or rapid recycling rather than increased protein synthesis.
This study shows that up-regulation of CD147 in non-transformed, non-invasive breast epithelial cells leads to chronic activation of the EGFR-Ras-ERK signaling cascade. Our data indicate that wild-type Ras activity is increased in MCF-10A cells after up-regulation of CD147 and is probably involved in the mechanism whereby up-regulation of CD147 elicits an invasive phenotype, as identified here and in our previous study (27). Also, although we saw differences in protein levels of CD147 with MEK inhibition and in our comparison of 10A-EV with 10A-K-RasV12 cells, negligible changes in the mRNA level of CD147 were observed in either case, suggesting that the influence of Ras-MAPK signaling on CD147 expression occurs mainly at a post-transcriptional level. Similar post-transcriptional regulation of CD147 has been observed in earlier studies (83–87). Thus, although CD147 expression is clearly increased in most aggressive cancers, identification of CD147 as a differentially expressed message in microarray analyses may not always be apparent. CD147 may act as an independent prognostic factor in some breast cancer types (9, 88), and further studies should evaluate the utility of CD147 surface expression in stratifying subpopulations in breast cancer. Moreover, attenuation of CD147 expression or activity may be a promising therapeutic intervention in breast cancer patients.
Acknowledgments
We thank Dr. Ben Ho Park for providing the MCF-10A cells stably expressing K-RasV12 and empty vector. We thank Dr. Lu Dai and Dr. Zhiqiang Qin for assistance with quantitative RT-PCR and Chris Fuchs for FACS analysis and cell sorting expertise.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA073839 and R01 CA082867. This work was also supported by Department of Defense Predoctoral Fellowship W81XWH-10-1-0083 (to G. D. G.) and IDEA Grant OC050368 (to B. P. T.) and by the South Carolina Clinical and Translational Research Institute (National Institutes of Health, NCRR, Grant UL1RR029882).
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type-1 matrix metalloproteinase
- Ctx-B
- cholera toxin B subunit
- EGFR
- epidermal growth factor receptor
- MOI
- multiplicity of infection
- PLA
- proximity ligation assay
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- p-ERK and p-EGFR
- phosphorylated ERK and EGFR, respectively.
REFERENCES
- 1. Nabeshima K., Iwasaki H., Koga K., Hojo H., Suzumiya J., Kikuchi M. (2006) Emmprin (basigin/CD147). Matrix metalloproteinase modulator and multifunctional cell recognition molecule that plays a critical role in cancer progression. Pathol. Int. 56, 359–367 [DOI] [PubMed] [Google Scholar]
- 2. Yan L., Zucker S., Toole B. P. (2005) Roles of the multifunctional glycoprotein, emmprin (basigin; CD147), in tumour progression. Thromb. Haemost. 93, 199–204 [DOI] [PubMed] [Google Scholar]
- 3. Yurchenko V., Constant S., Eisenmesser E., Bukrinsky M. (2010) Cyclophilin-CD147 interactions. A new target for anti-inflammatory therapeutics. Clin. Exp. Immunol. 160, 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weidle U. H., Scheuer W., Eggle D., Klostermann S., Stockinger H. (2010) Cancer-related issues of CD147. Cancer Genomics Proteomics 7, 157–169 [PubMed] [Google Scholar]
- 5. Riethdorf S., Reimers N., Assmann V., Kornfeld J. W., Terracciano L., Sauter G., Pantel K. (2006) High incidence of EMMPRIN expression in human tumors. Int. J. Cancer 119, 1800–1810 [DOI] [PubMed] [Google Scholar]
- 6. Li Y., Xu J., Chen L., Chen L., Zhong W. D., Zhang Z., Mi L., Zhang Y., Liao C. G., Bian H. J., Jiang J. L., Yang X. M., Li X. Y., Fan C. M., Zhu P., Fu L., Chen Z. N. (2009) HAb18G (CD147), a cancer-associated biomarker and its role in cancer detection. Histopathology 54, 677–687 [DOI] [PubMed] [Google Scholar]
- 7. Caudroy S., Polette M., Tournier J. M., Burlet H., Toole B., Zucker S., Birembaut P. (1999) Expression of the extracellular matrix metalloproteinase inducer (EMMPRIN) and the matrix metalloproteinase-2 in bronchopulmonary and breast lesions. J. Histochem. Cytochem. 47, 1575–1580 [DOI] [PubMed] [Google Scholar]
- 8. Polette M., Gilles C., Marchand V., Lorenzato M., Toole B., Tournier J. M., Zucker S., Birembaut P. (1997) Tumor collagenase stimulatory factor (TCSF) expression and localization in human lung and breast cancers. J. Histochem. Cytochem. 45, 703–709 [DOI] [PubMed] [Google Scholar]
- 9. Liu F., Cui L., Zhang Y., Chen L., Wang Y., Fan Y., Lei T., Gu F., Lang R., Pringle G. A., Zhang X., Chen Z., Fu L. (2010) Expression of HAb18G is associated with tumor progression and prognosis of breast carcinoma. Breast Cancer Res. Treat. 124, 677–688 [DOI] [PubMed] [Google Scholar]
- 10. Zucker S., Hymowitz M., Rollo E. E., Mann R., Conner C. E., Cao J., Foda H. D., Tompkins D. C., Toole B. P. (2001) Tumorigenic potential of extracellular matrix metalloproteinase inducer (EMMPRIN). Am. J. Pathol. 158, 1921–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biswas C. (1984) Collagenase stimulation in cocultures of human fibroblasts and human tumor cells. Cancer Lett. 24, 201–207 [DOI] [PubMed] [Google Scholar]
- 12. Ellis S. M., Nabeshima K., Biswas C. (1989) Monoclonal antibody preparation and purification of a tumor cell collagenase-stimulatory factor. Cancer Res. 49, 3385–3391 [PubMed] [Google Scholar]
- 13. Biswas C., Zhang Y., DeCastro R., Guo H., Nakamura T., Kataoka H., Nabeshima K. (1995) The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 55, 434–439 [PubMed] [Google Scholar]
- 14. Guo H., Zucker S., Gordon M. K., Toole B. P., Biswas C. (1997) Stimulation of matrix metalloproteinase production by recombinant extracellular matrix metalloproteinase inducer from transfected Chinese hamster ovary cells. J. Biol. Chem. 272, 24–27 [PubMed] [Google Scholar]
- 15. Marieb E. A., Zoltan-Jones A., Li R., Misra S., Ghatak S., Cao J., Zucker S., Toole B. P. (2004) Emmprin promotes anchorage-independent growth in human mammary carcinoma cells by stimulating hyaluronan production. Cancer Res. 64, 1229–1232 [DOI] [PubMed] [Google Scholar]
- 16. Qin Z., Dai L., Bratoeva M., Slomiany M. G., Toole B. P., Parsons C. (2011) Cooperative roles for emmprin and LYVE-1 in the regulation of chemoresistance for primary effusion lymphoma. Leukemia 25, 1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kato N., Kosugi T., Sato W., Ishimoto T., Kojima H., Sato Y., Sakamoto K., Maruyama S., Yuzawa Y., Matsuo S., Kadomatsu K. (2011) Basigin/CD147 promotes renal fibrosis after unilateral ureteral obstruction. Am. J. Pathol. 178, 572–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edward M., Quinn J. A., Sands W. (2011) Keratinocytes stimulate fibroblast hyaluronan synthesis through the release of stratifin. A possible role in the suppression of scar tissue formation. Wound Repair Regen. 19, 379–386 [DOI] [PubMed] [Google Scholar]
- 19. Toole B. P., Slomiany M. G. (2008) Hyaluronan, CD44 and Emmprin. Partners in cancer cell chemoresistance. Drug Resist. Updat. 11, 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. (2011) RAS oncogenes. Weaving a tumorigenic web. Nat. Rev. Cancer 11, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rochlitz C. F., Scott G. K., Dodson J. M., Liu E., Dollbaum C., Smith H. S., Benz C. C. (1989) Incidence of activating ras oncogene mutations associated with primary and metastatic human breast cancer. Cancer Res. 49, 357–360 [PubMed] [Google Scholar]
- 22. Eckert L. B., Repasky G. A., Ulkü A. S., McFall A., Zhou H., Sartor C. I., Der C. J. (2004) Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res. 64, 4585–4592 [DOI] [PubMed] [Google Scholar]
- 23. von Lintig F. C., Dreilinger A. D., Varki N. M., Wallace A. M., Casteel D. E., Boss G. R. (2000) Ras activation in human breast cancer. Breast Cancer Res. Treat. 62, 51–62 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J., Liu X., Datta A., Govindarajan K., Tam W. L., Han J., George J., Wong C., Ramnarayanan K., Phua T. Y., Leong W. Y., Chan Y. S., Palanisamy N., Liu E. T., Karuturi K. M., Lim B., Miller L. D. (2009) RCP is a human breast cancer-promoting gene with Ras-activating function. J. Clin. Invest. 119, 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Campbell P. M., Der C. J. (2004) Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin. Cancer Biol. 14, 105–114 [DOI] [PubMed] [Google Scholar]
- 26. Stamatakos M., Stefanaki C., Kontzoglou K., Masouridi S., Sakorafas G., Safioleas M. (2010) Recapitulation of ras oncogene mutations in breast cancer. Onkologie 33, 540–544 [DOI] [PubMed] [Google Scholar]
- 27. Grass G. D., Bratoeva M., Toole B. P. (2012) Regulation of invadopodia formation and activity by CD147. J. Cell Sci. 125, 777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Konishi H., Karakas B., Abukhdeir A. M., Lauring J., Gustin J. P., Garay J. P., Konishi Y., Gallmeier E., Bachman K. E., Park B. H. (2007) Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation. Cancer Res. 67, 8460–8467 [DOI] [PubMed] [Google Scholar]
- 29. Li R., Huang L., Guo H., Toole B. P. (2001) Basigin (murine EMMPRIN) stimulates matrix metalloproteinase production by fibroblasts. J. Cell. Physiol. 186, 371–379 [DOI] [PubMed] [Google Scholar]
- 30. Gordon L. B., Harten I. A., Calabro A., Sugumaran G., Csoka A. B., Brown W. T., Hascall V., Toole B. P. (2003) Hyaluronan is not elevated in urine or serum in Hutchinson-Gilford progeria syndrome. Hum. Genet. 113, 178–187 [DOI] [PubMed] [Google Scholar]
- 31. Slomiany M. G., Dai L., Bomar P. A., Knackstedt T. J., Kranc D. A., Tolliver L., Maria B. L., Toole B. P. (2009) Abrogating drug resistance in malignant peripheral nerve sheath tumors by disrupting hyaluronan-CD44 interactions with small hyaluronan oligosaccharides. Cancer Res. 69, 4992–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reissig J. L., Strominger J. L., Leloir L. F. (1955) A modified colometric method for the estimation of N-acetylamino sugars. J. Biol. Chem. 217, 959–966 [PubMed] [Google Scholar]
- 33. Bitter T., Muir H. M. (1962) A modified uronic acid carbazole reaction. Anal. Biochem. 4, 330–334 [DOI] [PubMed] [Google Scholar]
- 34. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 35. Ghatak S., Misra S., Toole B. P. (2005) Hyaluronan regulates constitutive ErbB2 phosphorylation and signal complex formation in carcinoma cells. J. Biol. Chem. 280, 8875–8883 [DOI] [PubMed] [Google Scholar]
- 36. Misra S., Ghatak S., Zoltan-Jones A., Toole B. P. (2003) Regulation of multi-drug resistance in cancer cells by hyaluronan. J. Biol. Chem. 278, 25285–25288 [DOI] [PubMed] [Google Scholar]
- 37. Cui H. Y., Guo T., Wang S. J., Zhao P., Dong Z. S., Zhang Y., Jiang J. L., Chen Z. N., Yu X. L. (2012) Dimerization is essential for HAb18G/CD147 promoting tumor invasion via MAPK pathway. Biochem. Biophys. Res. Commun. 419, 517–522 [DOI] [PubMed] [Google Scholar]
- 38. Tang Y., Nakada M. T., Rafferty P., Laraio J., McCabe F. L., Millar H., Cunningham M., Snyder L. A., Bugelski P., Yan L. (2006) Regulation of vascular endothelial growth factor expression by EMMPRIN via the PI3K-Akt signaling pathway. Mol. Cancer Res. 4, 371–377 [DOI] [PubMed] [Google Scholar]
- 39. Bodart J. F. (2010) Extracellular-regulated kinase-mitogen-activated protein kinase cascade. Unsolved issues. J. Cell. Biochem. 109, 850–857 [DOI] [PubMed] [Google Scholar]
- 40. Eccles S. A. (2011) The epidermal growth factor receptor/Erb-B/HER family in normal and malignant breast biology. Int. J. Dev. Biol. 55, 685–696 [DOI] [PubMed] [Google Scholar]
- 41. Rojas M., Yao S., Lin Y. Z. (1996) Controlling epidermal growth factor (EGF)-stimulated Ras activation in intact cells by a cell-permeable peptide mimicking phosphorylated EGF receptor. J. Biol. Chem. 271, 27456–27461 [DOI] [PubMed] [Google Scholar]
- 42. Sakaguchi K., Okabayashi Y., Kido Y., Kimura S., Matsumura Y., Inushima K., Kasuga M. (1998) Shc phosphotyrosine-binding domain dominantly interacts with epidermal growth factor receptors and mediates Ras activation in intact cells. Mol. Endocrinol. 12, 536–543 [DOI] [PubMed] [Google Scholar]
- 43. Toole B. P. (2009) Hyaluronan-CD44 interactions in cancer. Paradoxes and possibilities. Clin. Cancer Res. 15, 7462–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Underhill C. B., Toole B. P. (1979) Binding of hyaluronate to the surface of cultured cells. J. Cell Biol. 82, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lesley J., Hascall V. C., Tammi M., Hyman R. (2000) Hyaluronan binding by cell surface CD44. J. Biol. Chem. 275, 26967–26975 [DOI] [PubMed] [Google Scholar]
- 46. Ghatak S., Misra S., Toole B. P. (2002) Hyaluronan oligosaccharides inhibit anchorage-independent growth of tumor cells by suppressing the phosphoinositide 3-kinase/Akt cell survival pathway. J. Biol. Chem. 277, 38013–38020 [DOI] [PubMed] [Google Scholar]
- 47. Sameshima T., Nabeshima K., Toole B. P., Yokogami K., Okada Y., Goya T., Koono M., Wakisaka S. (2000) Glioma cell extracellular matrix metalloproteinase inducer (EMMPRIN) (CD147) stimulates production of membrane-type matrix metalloproteinases and activated gelatinase A in co-cultures with brain-derived fibroblasts. Cancer Lett. 157, 177–184 [DOI] [PubMed] [Google Scholar]
- 48. Bourguignon L. Y., Gilad E., Brightman A., Diedrich F., Singleton P. (2006) Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase Cϵ-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J. Biol. Chem. 281, 14026–14040 [DOI] [PubMed] [Google Scholar]
- 49. Wang S. J., Bourguignon L. Y. (2006) Hyaluronan and the interaction between CD44 and epidermal growth factor receptor in oncogenic signaling and chemotherapy resistance in head and neck cancer. Arch. Otolaryngol. Head Neck Surg. 132, 771–778 [DOI] [PubMed] [Google Scholar]
- 50. Tang W., Hemler M. E. (2004) Caveolin-1 regulates matrix metalloproteinases-1 induction and CD147/EMMPRIN cell surface clustering. J. Biol. Chem. 279, 11112–11118 [DOI] [PubMed] [Google Scholar]
- 51. Irwin M. E., Mueller K. L., Bohin N., Ge Y., Boerner J. L. (2011) Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J. Cell. Physiol. 226, 2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Caldieri G., Giacchetti G., Beznoussenko G., Attanasio F., Ayala I., Buccione R. (2009) Invadopodia biogenesis is regulated by caveolin-mediated modulation of membrane cholesterol levels. J. Cell Mol. Med. 13, 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamaguchi H., Takeo Y., Yoshida S., Kouchi Z., Nakamura Y., Fukami K. (2009) Lipid rafts and caveolin-1 are required for invadopodia formation and extracellular matrix degradation by human breast cancer cells. Cancer Res. 69, 8594–8602 [DOI] [PubMed] [Google Scholar]
- 54. Caldieri G., Capestrano M., Bicanova K., Beznoussenko G., Baldassarre M., Buccione R. (2012) Polarised apical-like intracellular sorting and trafficking regulates invadopodia formation and degradation of the extracellular matrix in cancer cells. Eur. J. Cell Biol. 91, 961–968 [DOI] [PubMed] [Google Scholar]
- 55. Blank N., Schiller M., Krienke S., Wabnitz G., Ho A. D., Lorenz H. M. (2007) Cholera toxin binds to lipid rafts but has a limited specificity for ganglioside GM1. Immunol. Cell Biol. 85, 378–382 [DOI] [PubMed] [Google Scholar]
- 56. Pons M., Cousins S. W., Alcazar O., Striker G. E., Marin-Castaño M. E. (2011) Angiotensin II-induced MMP-2 activity and MMP-14 and basigin protein expression are mediated via the angiotensin II receptor type 1-mitogen-activated protein kinase 1 pathway in retinal pigment epithelium. Implications for age-related macular degeneration. Am. J. Pathol. 178, 2665–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Venkatesan B., Valente A. J., Reddy V. S., Siwik D. A., Chandrasekar B. (2009) Resveratrol blocks interleukin-18-EMMPRIN cross-regulation and smooth muscle cell migration. Am. J. Physiol. Heart Circ. Physiol. 297, H874–H886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramos D. M., Dang D. (2011) EMMPRIN expression in oral SCC is regulated by FYN kinase. Anticancer Res. 31, 1205–1209 [PubMed] [Google Scholar]
- 59. Botta G. P., Reginato M. J., Reichert M., Rustgi A. K., Lelkes P. I. (2012) Constitutive K-RasG12D activation of ERK2 specifically regulates 3D invasion of human pancreatic cancer cells via MMP-1. Mol. Cancer Res. 10, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Neel N. F., Rossman K. L., Martin T. D., Hayes T. K., Yeh J. J., Der C. J. (2012) The RalB small GTPase mediates formation of invadopodia through a GTPase-activating protein-independent function of the RalBP1/RLIP76 effector. Mol. Cell. Biol. 32, 1374–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kozma S. C., Bogaard M. E., Buser K., Saurer S. M., Bos J. L., Groner B., Hynes N. E. (1987) The human c-Kirsten ras gene is activated by a novel mutation in codon 13 in the breast carcinoma cell line MDA-MB231. Nucleic Acids Res. 15, 5963–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ricardo S., Vieira A. F., Gerhard R., Leitao D., Pinto R., Cameselle-Teijeiro J. F., Milanezi F., Schmitt F., Paredes J. (2011) Breast cancer stem cell markers CD44, CD24 and ALDH1. Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 64, 937–946 [DOI] [PubMed] [Google Scholar]
- 63. Dai L., Guinea M. C., Slomiany M. G., Bratoeva M., Grass G. D., Tolliver L. B., Maria B. L., Toole B. P. (2013) CD147-dependent heterogeneity in malignant and chemoresistant properties of cancer cells. Am. J. Pathol. 182, 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Slomiany M. G., Dai L., Tolliver L. B., Grass G. D., Zeng Y., Toole B. P. (2009) Inhibition of functional hyaluronan-CD44 interactions in CD133-positive primary human ovarian carcinoma cells by small hyaluronan oligosaccharides. Clin. Cancer Res. 15, 7593–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pakula R., Melchior A., Denys A., Vanpouille C., Mazurier J., Allain F. (2007) Syndecan-1/CD147 association is essential for cyclophilin B-induced activation of p44/42 mitogen-activated protein kinases and promotion of cell adhesion and chemotaxis. Glycobiology 17, 492–503 [DOI] [PubMed] [Google Scholar]
- 66. Zhao P., Zhang W., Wang S. J., Yu X. L., Tang J., Huang W., Li Y., Cui H. Y., Guo Y. S., Tavernier J., Zhang S. H., Jiang J. L., Chen Z. N. (2011) HAb18G/CD147 promotes cell motility by regulating annexin II-activated RhoA and Rac1 signaling pathways in hepatocellular carcinoma cells. Hepatology 54, 2012–2024 [DOI] [PubMed] [Google Scholar]
- 67. Zhu C., Pan Y., He B., Wang B., Xu Y., Qu L., Bao Q., Tian F., Wang S. (2011) Inhibition of CD147 gene expression via RNA interference reduces tumor cell invasion, tumorigenicity and increases chemosensitivity to cisplatin in laryngeal carcinoma Hep2 cells. Oncol. Rep. 25, 425–432 [DOI] [PubMed] [Google Scholar]
- 68. Staubach S., Hanisch F. G. (2011) Lipid rafts. Signaling and sorting platforms of cells and their roles in cancer. Expert Rev. Proteomics 8, 263–277 [DOI] [PubMed] [Google Scholar]
- 69. Brown R. L., Reinke L. M., Damerow M. S., Perez D., Chodosh L. A., Yang J., Cheng C. (2011) CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 121, 1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mima K., Okabe H., Ishimoto T., Hayashi H., Nakagawa S., Kuroki H., Watanabe M., Beppu T., Tamada M., Nagano O., Saya H., Baba H. (2012) CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. 72, 3414–3423 [DOI] [PubMed] [Google Scholar]
- 71. Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H., Oshima M., Ikeda T., Asaba R., Yagi H., Masuko T., Shimizu T., Ishikawa T., Kai K., Takahashi E., Imamura Y., Baba Y., Ohmura M., Suematsu M., Baba H., Saya H. (2011) CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(−) and thereby promotes tumor growth. Cancer Cell 19, 387–400 [DOI] [PubMed] [Google Scholar]
- 72. Yae T., Tsuchihashi K., Ishimoto T., Motohara T., Yoshikawa M., Yoshida G. J., Wada T., Masuko T., Mogushi K., Tanaka H., Osawa T., Kanki Y., Minami T., Aburatani H., Ohmura M., Kubo A., Suematsu M., Takahashi K., Saya H., Nagano O. (2012) Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat. Commun. 3, 883. [DOI] [PubMed] [Google Scholar]
- 73. Hsu J. M., Chen C. T., Chou C. K., Kuo H. P., Li L. Y., Lin C. Y., Lee H. J., Wang Y. N., Liu M., Liao H. W., Shi B., Lai C. C., Bedford M. T., Tsai C. H., Hung M. C. (2011) Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat. Cell Biol. 13, 174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Keilhack H., Tenev T., Nyakatura E., Godovac-Zimmermann J., Nielsen L., Seedorf K., Böhmer F. D. (1998) Phosphotyrosine 1173 mediates binding of the protein-tyrosine phosphatase SHP-1 to the epidermal growth factor receptor and attenuation of receptor signaling. J. Biol. Chem. 273, 24839–24846 [DOI] [PubMed] [Google Scholar]
- 75. VanMeter A. J., Rodriguez A. S., Bowman E. D., Jen J., Harris C. C., Deng J., Calvert V. S., Silvestri A., Fredolini C., Chandhoke V., Petricoin E. F., 3rd, Liotta L. A., Espina V. (2008) Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer. Differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Mol. Cell Proteomics 7, 1902–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yamaoka T., Frey M. R., Dise R. S., Bernard J. K., Polk D. B. (2011) Specific epidermal growth factor receptor autophosphorylation sites promote mouse colon epithelial cell chemotaxis and restitution. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G368–G376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Miller F. R., Soule H. D., Tait L., Pauley R. J., Wolman S. R., Dawson P. J., Heppner G. H. (1993) Xenograft model of progressive human proliferative breast disease. J. Natl. Cancer Inst. 85, 1725–1732 [DOI] [PubMed] [Google Scholar]
- 78. Chaffer C. L., Weinberg R. A. (2011) A perspective on cancer cell metastasis. Science 331, 1559–1564 [DOI] [PubMed] [Google Scholar]
- 79. Alexander S., Friedl P. (2012) Cancer invasion and resistance. Interconnected processes of disease progression and therapy failure. Trends Mol. Med. 18, 13–26 [DOI] [PubMed] [Google Scholar]
- 80. Slomiany M. G., Grass G. D., Robertson A. D., Yang X. Y., Maria B. L., Beeson C., Toole B. P. (2009) Hyaluronan, CD44 and emmprin regulate lactate efflux and membrane localization of monocarboxylate transporters in human breast carcinoma cells. Cancer Res. 69, 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bourguignon L. Y. (2008) Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin. Cancer Biol. 18, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hao J., Chen H., Madigan M. C., Cozzi P. J., Beretov J., Xiao W., Delprado W. J., Russell P. J., Li Y. (2010) Co-expression of CD147 (EMMPRIN), CD44v3–10, MDR1 and monocarboxylate transporters is associated with prostate cancer drug resistance and progression. Br. J. Cancer 103, 1008–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fanelli A., Grollman E. F., Wang D., Philp N. J. (2003) MCT1 and its accessory protein CD147 are differentially regulated by TSH in rat thyroid cells. Am. J. Physiol. Endocrinol. Metab. 285, E1223–E1229 [DOI] [PubMed] [Google Scholar]
- 84. Yabushita H., Shimazu M., Noguchi M., Kishida T., Narumiya H., Sawaguchi K. (2003) Vascular endothelial growth factor activating matrix metalloproteinase in ascitic fluid during peritoneal dissemination of ovarian cancer. Oncol. Rep. 10, 89–95 [PubMed] [Google Scholar]
- 85. Deora A. A., Philp N., Hu J., Bok D., Rodriguez-Boulan E. (2005) Mechanisms regulating tissue-specific polarity of monocarboxylate transporters and their chaperone CD147 in kidney and retinal epithelia. Proc. Natl. Acad. Sci. U.S.A. 102, 16245–16250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gallagher S. M., Castorino J. J., Wang D., Philp N. J. (2007) Monocarboxylate transporter 4 regulates maturation and trafficking of CD147 to the plasma membrane in the metastatic breast cancer cell line MDA-MB-231. Cancer Res. 67, 4182–4189 [DOI] [PubMed] [Google Scholar]
- 87. Chen L., Bi J., Nakai M., Bunick D., Couse J. F., Korach K. S., Nowak R. A. (2010) Expression of basigin in reproductive tissues of estrogen receptor-α or -β null mice. Reproduction 139, 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Reimers N., Zafrakas K., Assmann V., Egen C., Riethdorf L., Riethdorf S., Berger J., Ebel S., Jänicke F., Sauter G., Pantel K. (2004) Expression of extracellular matrix metalloprotease inducer on micrometastatic and primary mammary carcinoma cells. Clin. Cancer Res. 10, 3422–3428 [DOI] [PubMed] [Google Scholar]