Background: The nature of proteoglycans that function as co-receptors for Sonic Hedgehog (Shh) is not known.
Results: Glypican 5 and 2-O-sulfo-iduronic acid are expressed in neural precursors, adjacent to primary cilia. Elimination of these components reduce Shh signaling.
Conclusion: Glypican 5 bearing 2-O-sulfo-iduronic acid at the nonreducing ends is a Shh co-receptor.
Significance: Attributes of Shh co-receptors have been identified.
Keywords: Cerebellum, Hedgehog, Neurodevelopment, Proliferation, Proteoglycan
Abstract
Sonic Hedgehog (Shh) signaling is crucial for growth, cell fate determination, and axonal guidance in the developing nervous system. Although the receptors Patched (Ptch1) and Smoothened (Smo) are required for Shh signaling, a number of distinct co-receptors contribute to these critical responses to Shh. Several membrane-embedded proteins such as Boc, Cdo, and Gas1 bind Shh and promote signaling. In addition, heparan sulfate proteoglycans (HSPGs) have also been implicated in the initiation of Shh responses. However, the attributes of HSPGs that function as co-receptors for Shh have not yet been defined. Here, we identify HSPGs containing a glypican 5 core protein and 2-O-sulfo-iduronic acid residues at the nonreducing ends of the glycans as co-receptors for Shh. These HSPG co-receptors are expressed by cerebellar granule cell precursors and promote Shh binding and signaling. At the subcellular level, these HSPG co-receptors are located adjacent to the primary cilia that act as Shh signaling organelles. Thus, Shh binds to HSPG co-receptors containing a glypican 5 core and 2-O-sulfo-iduronic acid to promote neural precursor proliferation.
Introduction
Hedgehog (Hh)4 proteins are critical, evolutionarily conserved morphogens that pattern developing tissues and promote growth of stem/precursor cells (1). Biochemical and genetic studies identified Patched (Ptch1) and Smoothened (Smo) as core components of canonical Hh receptors and demonstrated that Hh ligands directly bind Ptch1 to initiate signaling (2, 3). Subsequently, several additional Hh-interacting proteins have been identified, including the Hedgehog-interacting protein, CAM-regulated/down-regulated by oncogenes (CDO), brother of CDO (BOC), and growth arrest-specific 1 (GAS1). In vertebrates, CDO, BOC, and GAS1 are receptors that positively regulate multiple functions of Hh ligands, including cell fate specification, progenitor proliferation, and maintenance (4–8), whereas Hedgehog-interacting protein negatively regulates such functions (9, 10). In Drosophila and vertebrates, proteoglycan core components and glycan-synthesizing enzymes are critical for Hh responses (11–14), and Hh interacts with proteoglycans to promote Hh responses (15–17), suggesting that heparan sulfate proteoglycans (HSPGs) can also function as co-receptors for Hh ligands.
Shh is the Hh ligand most widely expressed in the developing mammalian nervous system, and this ligand is critical for normal development. Shh binds several receptors, including Ptch1, Ptch2, CDO, BOC, Gas1, and HSPGs. Although the proteins that function as receptors for Shh are well defined, the nature of the cell surface proteoglycan receptors that bind Shh and contribute to intracellular signaling is not yet understood. Proteoglycans contain long, linear, negatively charged glycosaminoglycan chains attached to a variety of core proteins. Several core proteins, including perlecan, glypican 3, and glypican 5 in vertebrates and Dally and Dally-like protein in invertebrates, have been implicated in Hh interactions (14, 18–21). Intriguingly, glypican 5 has been implicated in positive effects, whereas glypican 3 has been associated with negative effects on Shh signals (19, 21, 22). The glycosaminoglycans attached to these core proteins are necessary for binding to Shh, but specific features of the glycans that are responsible for binding to Shh are not known.
Heparan sulfates, the glycosaminoglycans that interact with Shh (12, 19, 23, 24), are composed of repeating disaccharide units of N-acetyl-d-glucosamine (GlcNAc) and d-glucuronic acid (GlcUA) linked via a β1–4-glycosidic bond. Individual monosaccharides are modified in specific patterns as follows: GlcNAc can be deacetylated and sulfated at the N-, 3-O-, and 6-O-positions, whereas glucuronic acid can be epimerized to l-iduronic acid and sulfated at the 2-O-position (25). Thus, proteoglycans exhibit immense structural variation.
Here, we identify features of proteoglycan receptors that bind Shh and are needed for neural precursor cell proliferation. Consistent with previous studies in tumor cells, glypican 5 is a core protein of these receptors (14, 19, 26, 27). Importantly, sulfation of iduronic acid residues at the nonreducing ends of glycosaminoglycan chains attached to glypican 5 promotes binding to Shh and Shh-dependent proliferation. Thus, HSPGs that contain glypican 5 and 2-O-sulfo-iduronic acid function as Shh co-receptors. We find that these receptors are expressed by proliferating cerebellar granule cells; at the subcellular level, these receptors are adjacent to primary cilia, organelles critical for Shh signaling. Together, these data indicate that proteoglycans, with specific core components and particular glycan modifications, are critical for binding Shh and regulating proliferation.
EXPERIMENTAL PROCEDURES
Microarray Fabrication
Synthetic glycans 1–13 were prepared as described (28, 29). Glycan 14 was prepared by functionalization of the deaminated 5-kDa heparin with 1,11-diamino-3,6,9-trioxaundecane by reductive amination. Glycans were dissolved in sodium phosphate buffer (50 mm, pH 8.5) and printed robotically using a piezoelectric spotting device (S11, Scienion, Berlin, Germany) onto N-hydroxysuccinimide-activated CodeLink slides in 75% relative humidity at 23 °C. All samples were printed at four different concentrations (1, 0.25, 0.063, and 0.016 mm) in replicates of 10. Prior to the experiment, slides were washed three times with water to remove noncovalently attached carbohydrates from the surface. Remaining succinimidyl groups were quenched in 100 mm ethanolamine in sodium phosphate buffer (pH 9, 50 mm) for 1 h at 50 °C. Slides were rinsed three times with water and dried by centrifugation.
Microarray Binding Assay
Arrayed slides were blocked for 1 h with 100 μl of 1% (w/v) BSA and 0.01% (v/v) Tween 20 in Tris-buffered saline, TBS (20 mm Tris/HCl, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2), then washed three times with TBS, and dried by centrifugation (5 min, 200 × g). Protein incubation was performed with 100 μl with 20 μg of protein in TBS with 0.01% (v/v) Tween 20. Inhibition solution contained 2.5 mm deaminated heparin. Slides were incubated with protein solutions for 1.5 h at room temperature, washed three times with TBS, and dried by centrifugation. For detection, 100 μl of a fluorescent-labeled antibody solution was applied (1 μl of Alexa Fluor 594 goat anti-mouse IgG in TBS containing 1% (w/v) BSA and 0.01% (v/v) Tween 20) for 1 h in the dark. Slides were washed three times with TBS and dried by centrifugation.
Microarray Detection and Signal Processing
Slides were scanned with an LS400 scanner and analyzed by Array-Pro Analyzer. Quantification of Alexa Fluor 594 binding signals was carried out using GenePix PrO 7 software (Molecular Devices). For data analysis, the mean intensity of each spot was used. Signal background of each slide was subtracted from the hybridization signal. An average of 10 spots on the same array at 250 μm glycan concentration was determined.
SPR Immobilization Procedure
Synthetic glycans 1–6 and 11 were covalently bound to the sensor surface using primary amine coupling. HBS-N containing 0.005% (v/v) surfactant P20 was employed as running buffer. The carboxymethylated dextran matrix (CM5 chip) was activated at a flow rate of 10 μl/min using an 8-min injection pulse of an aqueous solution containing N-hydroxysuccinimide (0.05 m) and N-ethyl-N′-(dimethylaminopropyl) carbodiimide (0.2 m). A solution of glycan (50 μg/ml) containing 1 mm hexadecyltrimethylammonium chloride was flowed over the activated surface for 8 min at 5 μl/min. The remaining reactive groups on the chip surface were quenched by injection of 1 m ethanolamine hydrochloride, pH 8.5, for 7.5 min at 10 μl/min. For each immobilized synthetic glycan, a parallel flow cell was coupled with heparan sulfate/heparin 12 as reference. The following binding levels were established (in response units): glycan 1, 1100; glycan 2, 614; glycan 3, 383; glycan 4, 300; glycan 5, 604; glycan 6, 702; 596; glycan 11, 557; HS/heparin 12, ±300.
SPR Binding and Inhibition Assay
HBS-N containing 0.005% (v/v) surfactant P20 was used as running buffer. Protein solutions containing 0.2 μm protein in HBS-N buffer were injected into the analyte and the reference flow cell for 30 s at 20 μl/min. After sample injection, running buffer was flowed over the sensor surface for 2 min to enable dissociation. Chip surface was regenerated for the next sample by injection of the following solutions for 1 min at 80 μl/min: 0.1% (w/v) SDS, 0.085% (v/v) H3PO4, 1 m NaCl, and 0.1% (v/v) HCl. Response was calculated as the difference in response units between analyte and reference flow cell and monitored as a function of time (sensorgram).
Generation of Shh and ShhAla Ligand
Plasmid containing full-length Shh was a generous gift of P. Chuang. Mutations to generate the ShhAla allele were introduced by QuikChange (Stratagene). Primer sequences were designed to introduce the desired amino acid changes (R34A and K38A). Sense and antisense mutagenesis primer sequences are as follows: 5′-GGCCTGGCAGAGGGTTTGGAAAGGCGCGCCACCCCGCAAAGCTGAC-3′ and 5′-TCAGCTTTGCGGGGTGGCGCGCCTTTCCAAACCCTCTGCCAGGCC-3′. Plasmids containing sequences for alkaline phosphatase-tagged N-terminal wild type or ShhAla were described previously (12).
Plasmids containing full-length Shh and ShhAla were transfected into HEK cells, and plasmids containing N-Shh:AP and N-ShhAla:AP were transfected into HEK or COS7 cells, using Lipofectamine2000 (Invitrogen). The 24-h serum-free conditioned media were collected 60 h post-transfection and concentrated 10-fold using AmiconUltra concentration devices with a molecular weight cutoff of 10,000 (dually lipidated product) or 30,000 (AP-tagged ligand) (Millipore). Sample concentrations were determined by Western blot in comparison with Shh protein standards using anti-Shh antibody as follows: Shh N-19; Santa Cruz Biotechnology, sc-1194, or by alkaline phosphatase activity (12). Parallel preparations from mock-transfected HEK cells were generated and used as vehicle controls to provide the data for a Shh concentration of 0 ng/ml.
Heparin Plate Binding Assay
Heparin-coated plates (Lifespan Technologies) were washed with 100 μl/well binding buffer (20 mm Tris/HCl, pH 7.4, 150 mm sodium chloride, 2 mm calcium chloride, 2 mm magnesium chloride, .01% Tween 20) and blocked with 100 μl/well of 1% BSA in binding buffer for 1 h at RT. Blocking solution was removed and 100 μl of Shh ligands were added (3.2 ng/μl) to each well and incubated for 1 h at RT. Wells were washed (100 μl/well, three times for 5 min) with binding buffer and then washed (100 μl/well, one time for 5 min) with alkaline phosphatase assay buffer (1160 μl of diethanolamine, 8290 μl of water, 500 μl of 5 mg/ml BSA, 50 μl of 50 mm magnesium chloride). 100 μl of substrate buffer (1160 μl of diethanolamine, 7790 μl of water, 500 μl of 5 mg/ml BSA, 500 μl of 120 mm 4-nitrophenyl phosphate disodium salt hexahydrate, 50 μl of 50 mm magnesium chloride) was added to each well and developed for 90–120 min at 37 °C. Absorbance was read at 405 nm.
For enzymatic treatment, 0.04 μg/μl of iduronate 2-sulfatase (IDS (Elaprase) Shire Pharmaceuticals) or vehicle in 50 μl of binding buffer, pH 4.5, were added to wells for 2 h at 37 °C. Plates were processed as described above. For competition studies, complex glycans isolated from the urine of patients with mucopolysaccharidosis type II were added together with ligand (100 μl total volume) to the wells for 1 h (RT).
Primary Granule Cell Precursor (GCP) Cultures
Cerebella from P6 mice were dissected; meninges were removed, and tissue was incubated in 20 units/ml papain with 100 units/ml DNase (Worthington) for 30 min at 37 °C. Tissue digestion was quenched with ovomucoid protease inhibitor solution and GCP media (1:1). (The ovomucoid protease inhibitor solution contained the following: ovomucoid protease inhibitor (Worthington, 3.75 mg/ml) with BSA (3.75 mg/ml) and DNase (250 units/ml) (Worthington); GCP media contained the following: B-27 (1×, Invitrogen, 17504044), 20 mm NaCl, 100 units/ml penicillin, 100 μg/ml streptomycin (Invitrogen, 15140122) in Neurobasal (Invitrogen, 21103049).). Cells were washed and dissociated by serial trituration in DMEM/F-12 (1:1). After panning, dissociated cells were counted and plated in GCP media in 12-well plates (2 × 106 cells/well) coated with 0.0015% poly-l-ornithine (Sigma, P4957) and 10 μg/ml laminin (Invitrogen, 23017015). Recombinant enzyme (IDS (3.5 μg/ml)) or vehicle control were added at the time of plating, and cells were incubated for 1 h at 37 °C. 1 h after plating, 300 ng/ml of Shh, ShhAla, or vehicle was added. Volumes of Shh in concentrated conditioned media were matched. RNA was harvested 6 h after stimulation.
Quantitative RT-PCR
RNA was purified from GCP cultures using RNAqueous-4-PCR (Ambion). RNA samples were treated with DNase I (Ambion). cDNA was generated using the high capacity cDNA archive kit (Applied Biosystems). TaqMan quantitative PCR was performed on an Applied Biosystems 7700 quantitative PCR instrument with Sequence Detector Software. The following primer and probe mixes from Applied Biosystems were used: GAPDH, Ptch1, Gli1, Gli2, Gli3, Mycn (N-Myc), Cyclin D1, and Cyclin D2.
Samples were run in triplicate, and the mean of the triplicate values was taken as the transcript level. Transcript levels (S) were normalized to GAPDH transcript levels (G) (31) in the same cDNA sample. Each figure represents the mean of 3–6 independent experiments. Significance was calculated by two-tailed z test.
In Vitro Proliferation Assay
GCPs were prepared as stated above. IDS (3.5 μg/ml) or vehicle was added at the time of plating, and cells were incubated for 1 h at 37 °C. 1 h after plating, 300 ng/ml prepared Shh, ShhAla, or vehicle was added. 18–24 h after plating, cells were gently resuspended in Ca2+ and Mg2+-free PBS, triturated until a single cell suspension, fixed for 30 min in cold 70% ethanol (4 °C), and rinsed in PBS, 0.5% BSA. Cells were centrifuged and then resuspended in 500 μl of PI/RNase Staining Buffer (BD Biosciences, 550825) for 1 h at 37 °C. Percentage of cells in S-phase was assessed by FACS analysis. Values were normalized to cultures stimulated with vehicle.
Immunocytochemistry
6 × 105 dissociated P6 GCPs were plated onto poly-l-ornithine- and laminin-coated 12-mm glass coverslips (Bellco). Cells were stimulated with Smoothened agonist (300 nm, Alexis Biochemicals, 270–426-M001) for 24 h and fixed with 4% paraformaldehyde. Cold EtOH was applied for 5 min, and cells were rinsed with PBS. For visualization of glycosaminoglycan sulfations using phage display antibodies, nonspecific antibody binding was blocked by incubating cells in PBS/BSA (0.1% (w/v)) for 20 min (RT). The periplasmic fraction, containing the single chain antibody, was diluted in PBS/BSA and then applied at 1:2.5 (AO4B08V, MW3G3V3, or HS4E4V) for 1.5 h at RT. Cultures were rinsed with PBS (three times for 10 min) and then incubated for 1 h (RT) with a mouse anti-vesicular stomatitis virus glycoprotein-Cy3 antibody diluted in PBS/BSA (Sigma, C7706, clone P5D4, 1:250). Cultures were rinsed with PBS (two times for 10 min) and then fixed with 4% paraformaldehyde (30 min, 4 °C). After this initial staining and PBS rinses (three times for 10 min), cells were permeabilized and blocked for 60 min (RT) in PBS/NGS/BSA/Triton (10% normal goat serum, 1% BSA, 0.1% Triton in PBS) and then incubated in primary antibodies (diluted in block) overnight at 4 °C. The following primary antibodies were used: rabbit anti-adenylyl cyclase III (C-20, Santa Cruz Biotechnology, sc-588, lot J1007, 1:400) and/or rabbit anti-γ-tubulin (Sigma, T5192, 1:400). Staining was visualized with Alexa Fluor 488-labeled secondary antibodies (Invitrogen) and DAPI counterstain. Cultures were mounted in Prolong Gold antifade reagent (Invitrogen P36934).
For visualizing Shh binding, GCP cultures were prepared as above and then treated with or without IDS for 1 h. Cells were then stimulated with full-length wild type Shh, ShhAla (150, 300, and 450 ng/ml), an equivalent volume of vehicle, or no addition for 6 h. Cells were fixed and immunostained with rabbit anti-Shh (Abcam) and Alexa® 546-conjugated goat anti-rabbit IgG (Molecular Probes) and DAPI. Images were taken with Nikon Eclipse E800 microscope equipped with a Spot Camera under the control of the imaging software (Nikon NIS-Element BR 2.30, SP4 (Build 387), Roper Scientific version 5.03). Fluorescence intensity was calculated using ImageJ software (ImageJ 1.46j; National Institutes of Health) to measure the mean fluorescent intensity (normalized to background) associated with cultured cells in each condition. Approximately 50–70 cells in 4–6 fields were measured for each condition.
Immunohistochemistry
The antibodies used are as follows: rabbit anti-Shh (Abcam, ab73958, 1:175), mouse anti-glypican 5 (R&D Systems, MAB2607, 1:50), and goat anti-glypican 3 (Santa Cruz Biotechnology, sc-34096, 1:100). After hydration with PBS (15 min), nonspecific antibody binding was blocked by incubating slides in 5% NGS, 1% BSA in PBS for 1 h. Primary antibodies in block were applied overnight at 4 °C. Staining was visualized with Alexa Fluor 546- or 488-labeled secondary antibodies (Invitrogen, 1:400) and DAPI counterstain.
For staining with phage display antibodies, 12-μm cryostat sections were hydrated with PBS for 15 min. Nonspecific antibody binding was blocked by incubating slides in PBS/BSA (0.1% (w/v)) for 20 min (RT). The periplasmic fraction, containing the single chain antibody, was diluted in PBS/BSA and then applied at 1:2.5 (AO4B08V or HS4E4V) for 1.5 h at RT. Sections were rinsed with PBS (three times for 10 min) and then incubated for 1 h (RT) with a mouse anti-VSV glycoprotein-Cy3 antibody diluted in PBS/BSA (Sigma, C7706, clone P5D4, 1:250). Sections were rinsed with PBS (two times for 10 min), counterstained with DAPI, rinsed with PBS (three times for 10 min), dried with ethanol, and coverslipped (Immunomount, Thermo Electron Corp., 9990402). For co-staining using phage display antibodies, staining with the phage display antibody was done subsequent to staining with the other primary antibody.
For PI-PLC treatment, 12-μm sagittal cryosections from P6 BALB/c pups were treated with PBS or 500 milliunits/ml of phosphatidylinositol-specific phospholipase C (PI-PLC) (Sigma, P5542) in PBS, first for 1 h at 37 °C and then overnight at 4 °C. After PBS washes (three times for 5 min), nonspecific binding was blocked with PBS/BSA (1% (w/v)) for 1 h at RT.
Organotypic Slice Culture
Cerebella from P6 C57BL/6, Shh+/+, ShhCtl/Ctl, or ShhAla/Ala littermates were washed in cold dissection buffer (Hanks' balanced salt solution, pH 7.4, containing 36 mm glucose, 15 mm HEPES, and 24 mm KCl), embedded in 2–4% low melting point agarose in dissection buffer, and 100–200-μm thick midsagittal slices were cut with a vibrating microtome (VT1000S; Leica, Wetzlar, Germany) and transferred to porous membrane inserts (Millipore) in GCP media containing IDS (3.5 μg/ml) or vehicle control and were maintained in a 37 °C, 5% CO2 humidified incubator for 16 h. Slices were fixed (30 min, 4 °C) in 4% paraformaldehyde. Organotypic slice cultures were immunostained using anti-Shh at 1:50. Quantification of the EGL and the upper molecular layer (ML) was done by averaging the mean fluorescence in each area obtained using ImageJ, and the average mean fluorescent intensity of EGL was compared with that of the ML.
Shh Quantification
Organotypic slice cultures were established, and tissue was lysed in Tris buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% (v/v) Nonidet P-40, 0.25% (w/v) sodium deoxycholate, 1 mm DTT, 10 mm NaF, 1 mm sodium orthovanadate, 1 mm PMSF, protease inhibitors) for 2 h at 4 °C. Suspensions were centrifuged for 1 h at 13,000 rpm at 4 °C. Samples were subjected to SDS-PAGE and blotted with anti-Shh (Cell Signaling, 2287) or anti-pan-actin (Cell Signaling, 4968).
Proliferation in Organotypic Slices
Culture procedure above was used; for the last 2 h of enzyme or vehicle treatment, slices were incubated with 30 μg/ml of BrdU. After fixation, slices were permeabilized with 0.4% Triton X-100 and 5% NGS in PBS for 20 min, treated with 2 n HCl for 30 min at 37 °C, followed by 0.1 m sodium borate two times for 10 min and a PBS rinse. Slices were blocked for 1 h in blocking solution (5% NGS, 0.1% Triton X-100 in PBS) and then incubated in anti-BrdU (BD Biosciences 555627, 1:50) in blocking solution overnight at 4 °C. Staining was visualized with anti-mouse Alexa Fluor 546-labeled secondary antibody and DAPI counterstain. Images were taken on Zeiss LSM 510 META confocal microscope with a Plan-Apochromat ×63/1.4 oil objective. Optical sections of 1–4 μm were taken through the slice, and a maximum intensity projection was generated.
For shRNA Knockdown Experiments
GCPs were grown as neurospheres and infected with lentivirus constructs. GCPs were infected with control GFP shRNA or glypican 5 shRNA with Polybrene (8 μg/ml) and SAG (300 nm) for 24 h. After infection, media were changed to remove the Polybrene for 3 days, and then cells were treated as stated above for Shh binding quantification, phage display immunohistochemistry, or quantitative RT-PCR.
Statistical Analysis
Significance was assessed using two-tailed Student's t test or two-tailed z test for normalized data, with a Bonferroni correction for multiple comparisons.
RESULTS
Glypican 5 Is the Core Protein of the Shh Proteoglycan Receptor
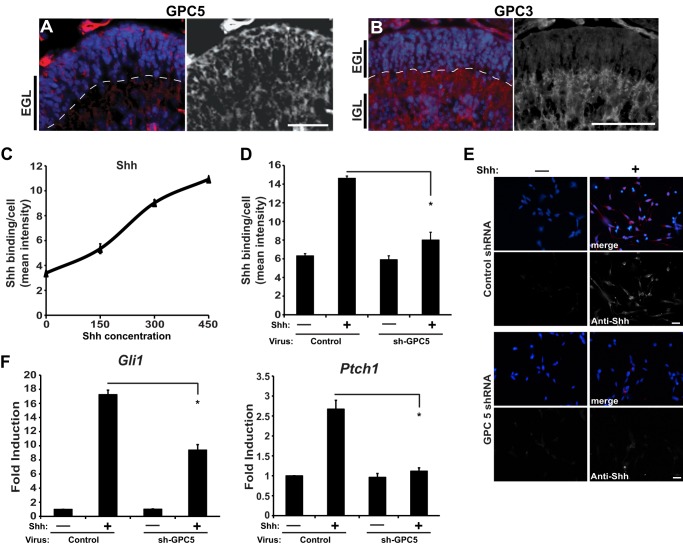
Several proteoglycan core components have been implicated in binding Shh and mediating biological responses, including perlecan, glypican 3, and glypican 5 (18, 19, 32). As shown in Fig. 1, glypicans 3 and 5 are both expressed in the developing cerebellum. However, glypican 5 is enriched in granule cell precursors located in the EGL, the mitogenic niche of the cerebellum (Fig. 1A), whereas glypican 3 is predominantly expressed by post-mitotic cells within the internal granule cell layer (IGL) (Fig. 1B). Although other glypicans, perlecans, and syndecans are expressed in the cerebellum (data not shown), glypican 5 is appropriately expressed to promote Shh-proliferative responses, suggesting that it might serve as a core component of the Shh proteoglycan receptor involved in neural precursor proliferation.
FIGURE 1.
Glypican 5 is expressed in the cerebellar EGL and promotes Shh binding and signaling. Immunohistochemistry of glypican 5 (A) and glypican 3 (B) in P6 wild type mouse cerebellum is shown. Glypican 5 is expressed in the outer EGL, whereas glypican 3 is in the IGL. (EGL, external granule cell layer, dashed line delineates edge of EGL; IGL, internal granule cell layer). Glypican staining is shown in red and DAPI staining in blue. C, mean fluorescent intensity of Shh staining. GCPs were stimulated for 6 h with Shh at four concentrations (0, 150, 300, and 450 ng/ml). D, shRNA-mediated knockdown of glypican 5 reduces Shh binding. E, Shh immunostaining is shown in red (merge) or white; nuclei are counterstained with DAPI (blue). Scale bar, 32 μm. F, shRNA-mediated knockdown of glypican 5 decreases Gli1 and Ptch1 mRNA in P6 GCPs stimulated for 6 h with Shh (*, p < 0.05).
As shown in Fig. 1C, exogenous full-length Shh protein binds to cultured cerebellar GCPs across a physiological range of concentrations. When glypican 5 expression is reduced by shRNA-mediated knockdown in GCPs, the ability of these cells to bind Shh is substantially impaired (Fig. 1, D and E). Induction of the Shh target genes, Gli1 and Ptch1, provide a specific indicator of Shh response. Knockdown of glypican 5 decreases Shh-dependent induction of these target genes in GCPs (Fig. 1F). Together, these data indicate that glypican 5 is needed for Shh binding and Shh responses and demonstrate that this membrane-attached protein functions as a core component of a Shh co-receptor on neural precursors.
Shh Interacts with Glycans Containing 2-O-Sulfo-iduronic Acid
Although glypican 5 provides the protein core of an Shh co-receptor, it is the attached glycan chains that are responsible for direct binding to Shh (19). To determine whether there is specificity in the glycan components of the Shh receptor, we used glycan microarrays containing a library of synthetic glycans that mimic the structures found in heparan sulfate proteoglycans (Fig. 2A). To focus on biologically relevant binding, we examined binding of wild type Shh and Shh with mutations in the Cardin-Weintraub motif (ShhAla) (12). The ShhAla mutant (R34A and K38A) exhibits decreased binding to HSPGs without defects in affinity for Ptc receptors (12). Among the synthetic glycans bearing a single sugar residue (structures 11–13), we find that glycans with sulfation at the 2-O-position of iduronic acid bind to Shh tagged with alkaline phosphatase (Fig. 2B). These data suggest that 2-O-sulfation of iduronic acid is important for Shh binding. Similarly, binding of Shh to more complex structures in the microarray depends on the presence of 2-O-sulfo-iduronic acid; complex structures containing 2-O-sulfo-iduronic acid (1, 2, 5, 6, 7, 10, 14) bind Shh, regardless of the sulfation status of other positions (Fig. 2B). These data suggest that Shh interacts with specific glycans that contain 2-O-sulfated iduronic acid.
FIGURE 2.
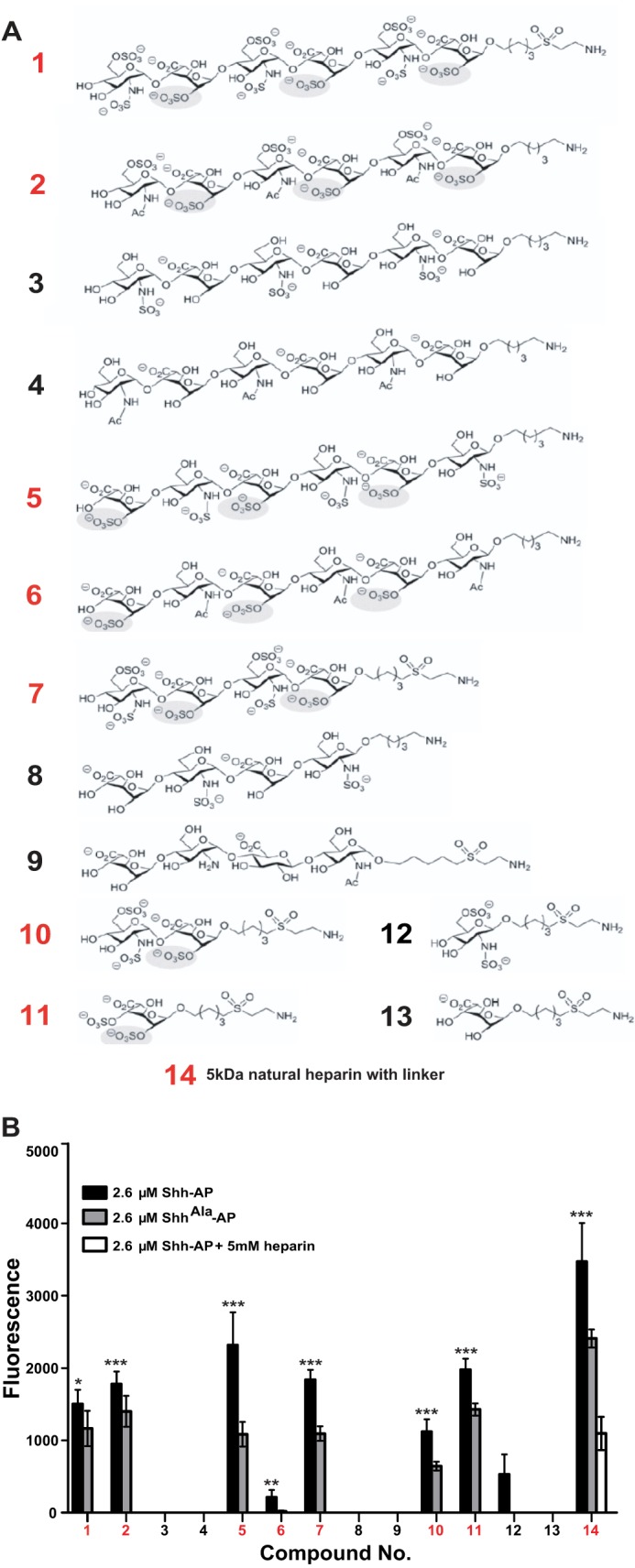
Shh binding to glycan microarray. A, synthetic glycans tested in the microarray. B, glycan microarray binding results. Glycans that bind Shh-AP more than ShhAla-AP are characterized by the presence of 2-O-sulfated iduronic acid (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
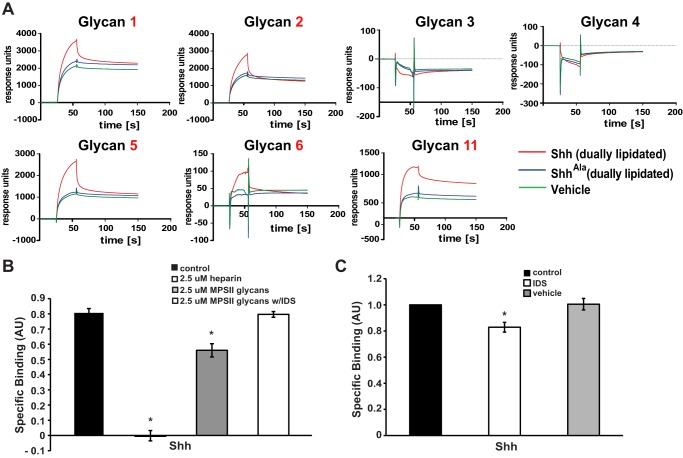
Although glycan microarrays allow rapid screening of binding to several distinct disaccharide motifs, they do not provide detailed real time information on binding. Furthermore, there is variation in the extent to which different glycans are successfully immobilized on the array, making quantification more difficult. Therefore, we carried out binding studies using surface plasmon resonance (SPR) to monitor binding of Shh to glycans. In these studies, we used mature Shh ligand that is dually lipidated and shed from producing cells (31) rather than the Shh-alkaline phosphatase fusion constructs used for high throughput rapid screening in array analysis. The AP-tagged Shh and the biologically mature Shh ligand show equivalent heparan sulfate proteoglycan binding (data not shown). In agreement with the microarray results, all glycan structures containing a sulfate group at the 2-O-position of iduronic acid bind Shh (1, 2, 5, 6, 7, 10, 11, 14), whereas structures desulfated at this position do not (Fig. 3A). In contrast, ShhAla shows little binding to structures sulfated at the 2-O-position of iduronic acid (Fig. 3A). Notably, although glycan 6, which contains 2-O-sulfated iduronic acid, showed low but detectable levels of Shh binding in the array, specific binding was readily apparent by SPR. Thus, the comparatively low level of binding seen in the array may reflect issues in immobilization of glycan 6. Together, these array studies and SPR analyses demonstrate that sulfation at the 2-O-position of iduronic acid is a critical feature of glycans that bind the Cardin-Weintraub motif of Shh.
FIGURE 3.
2-O-Sulfo-iduronic acid promotes Shh binding. A, surface plasmon resonance results for the interaction of dually lipidated Shh and ShhAla with synthetic glycans 1–6 and 11. For each synthetic glycan, the following samples were analyzed: 0.2 μm Shh, 0.2 μm ShhAla, and vehicle. B, competition assay with complex glycans from urine of patients with MPS II. Glycans containing 2-O-sulfo-iduronic acid at the nonreducing end (MPS II glycans) efficiently compete for binding to Shh, whereas glycans lacking 2-O-sulfo-iduronic acid at the nonreducing end (MPS II glycans + IDS) do not (*, p < 0.05, n = 3). Specific binding equals bound Shh-AP minus bound ShhAla-AP. C, pretreatment of heparin-coated plates with IDS alters specific binding of Shh to heparin (*, p < 0.05, n = 5).
Nonreducing Ends of Glycans Are Critical for Shh Binding
We used competition binding assays to further examine the role of glycans containing 2-O-sulfated iduronic acid in Shh binding. Shh binding to heparin plates can be inhibited with excess heparin in a dose-dependent fashion (23). We evaluated the effects of heparin and biological heparan sulfate glycosaminoglycans at 2.5 μm, a concentration where heparin sulfates abrogate specific binding to heparin plates (Fig. 3B). Complex glycans isolated from the urine of patients with an IDS deficiency (mucopolysaccharidosis type II) uniformly contain 2-O-sulfated iduronic acid at the nonreducing ends (30). As shown in Fig. 3B, these heparan sulfate glycans compete with heparin attached to the plates for Shh binding. In contrast, when these same glycans are digested with IDS to desulfate the 2-O-position of the nonreducing end iduronic acid, they no longer compete for binding (Fig. 3B). Thus, glycans with 2-O-sulfo-iduronic acid at the nonreducing end mediate binding to Shh.
To determine whether 2-O-sulfation found on the terminal disaccharide promotes binding to Shh, we used IDS in concert with the heparin plate binding assay (23) to further probe the specificity of Shh-glycan interactions. When the heparin plate is pretreated with IDS, which selectively removes a single 2-O-sulfate from iduronic acid at the nonreducing end of glycan chains (30), binding of Shh is reduced (Fig. 3C). Together, these data indicate that Shh binds to glycans wherein 2-O-sulfated iduronic acid is present at the nonreducing end.
Specific Proteoglycans Are Co-receptors for Shh
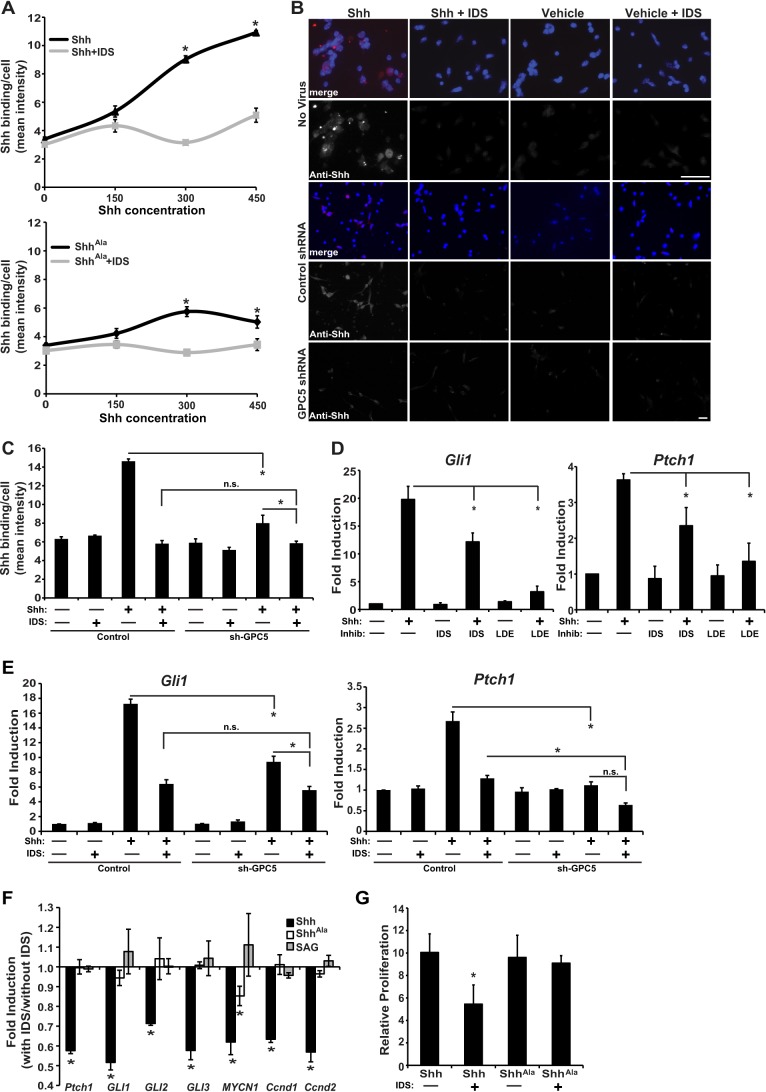
To consider whether 2-O-sulfo-iduronic acid at the nonreducing ends of glycans is important for Shh binding to proteoglycans in a biological context, we treated cerebellar GCPs with IDS. As shown, IDS treatment reduces the amount of Shh that binds to precursor cells across a range of Shh stimulation (Fig. 4, A and B). ShhAla binds less well to cells than does wild type Shh, due to impaired binding to HSPGs (12, 23, 24). We find that IDS has a smaller effect on ShhAla binding. Thus, 2-O-sulfo-iduronic acid at the nonreducing end of glycans is a critical feature of the proteoglycan receptor for Shh expressed by cerebellar GCPs. Furthermore, the Cardin-Weintraub domain of Shh, which is mutated in ShhAla, promotes binding to glycans that contain 2-O-sulfo-iduronic acid at the nonreducing end.
FIGURE 4.
Sulfation of nonreducing end 2-O-iduronic acid enables Shh responses. A, mean fluorescent intensity of Shh staining associated with GCPs pretreated with IDS and then stimulated for 6 h with Shh or ShhAla at 0, 150, 300, and 450 ng/ml (*, p < 0.05). Data in A include datasets from Fig. 1A. B, IDS pretreatment reduces Shh binding. Shh immunostaining is shown in red (merge) or white; nuclei are counterstained with DAPI (blue). Scale bar, 32 μm. Knockdown of glypican 5 also reduces Shh binding. C, mean fluorescent intensity of Shh bound to GCPs pretreated with control shRNA or shRNA to glypican 5. Data in C include datasets from Fig. 1D. D, mRNA in P6 GCPs stimulated for 6 h with Shh or vehicle control; pretreating cells with IDS attenuates Shh-dependent increase in Gli1 and Ptch1 mRNA by 40%. Pretreating with a Smo inhibitor (NVP-LDE225, 500 μm) is shown for comparison (*, p < 0.05, n = 3). E, shRNA-mediated knockdown of glypican 5 reduces Shh-dependent Gli1 and Ptch1 induction, and subsequent treatment with IDS has little additional effect. Data in E include datasets from Fig. 1F. IDS pretreatment selectively modulates Shh-induced transcription (F) and proliferation (G) but does not affect ShhAla-induced or SAG-induced response (*, p < 0.05, n = 4). n.s., not significant.
To determine whether the newly identified glycan epitopes are attached to glypican 5 core proteins, we reduced expression of glypican 5 in GCPs using shRNA, and we then treated the cells with IDS. As shown in Fig. 4, B and C, treatment with IDS and shRNA for glypican 5 exhibit a nonadditive effect on binding of Shh. The overlapping effects of glypican 5 knockdown and IDS treatment suggest that HSPGs that function as Shh co-receptors contain a glypican 5 core protein and attached glycan chains that have 2-O-sulfo-iduronic acid at the nonreducing end.
If these newly identified glycan structures are indeed essential components of the proteoglycan co-receptor for Shh, they should be needed for biological responses to Shh, as well as for Shh binding. In the developing cerebellum, Shh promotes granule cell precursor proliferation (35) and regulates expression of a gene program that includes Gli1 and Ptch1, as well as Gli 2–3, Mycn (MYCN1), Cyclin D1 (Ccnd1), and Cyclin D2 (Ccnd2) (36–38). In ShhAla/Ala mice, where Shh interactions with glycans are specifically abrogated due to mutations of the Cardin-Weintraub motif, cerebellar precursor cell proliferation is reduced and the transcriptional program is altered (23). We isolated precursor cells from postnatal day 6 wild type mice, treated the cells with IDS to remove 2-O-sulfate from nonreducing end iduronic acid residues, and then stimulated the cells with Shh. Treatment with IDS, which decreases Shh binding to glycans, reduces Shh-dependent induction of Gli1 and Ptch1 by more than 40%. For comparison, pretreatment with an antagonist of the canonical Shh receptor Smoothened, NVP-LDE225, decreases Gli1 and Ptch1 induction in this assay by 80 and 60%, respectively (Fig. 4D). As shown in Fig. 4E, IDS treatment or genetic knockdown of glypican 5 each reduce Shh-dependent changes in gene expression. Furthermore, IDS and shRNA for glypican 5 exhibit a nonadditive effect on Shh-induced changes in gene expression. Taken together, these data indicate that HSPGs that function as Shh co-receptors to mediate changes in gene expression contain a glypican 5 core component and attached glycan chains with 2-O-sulfo-iduronic acid at the nonreducing ends.
To determine whether the biological effects of IDS and glypican 5 knockdown on gene induction are due to reduced binding of Shh that is mediated by the Cardin-Weintraub motif, we again pretreated cerebellar cultures with IDS, then stimulated with Shh, ShhAla, or chlorobenzothiophene-containing Shh-pathway agonist (SAG) that directly binds and activates Smoothened (39). Cleavage of the 2-O-sulfate by IDS impacts Gli1 and Ptch1 induction in response to Shh but does not prevent responses to ShhAla or to SAG (Fig. 4F). Thus, this co-receptor promotes binding of Shh and initiation of a biological response by interacting with the Cardin-Weintraub motif of Shh, and it does not affect signaling steps subsequent to Smo activation.
Proliferation of neural stem/precursor cells is the biological response that is most dependent on Shh-proteoglycan interactions and is altered by the ShhAla mutation in vivo (23). As shown, Shh-dependent proliferation is reduced in cerebellar precursor cell cultures treated with IDS, whereas proliferation in response to ShhAla is not affected by IDS (Fig. 4G). Taken together, these data indicate that specialized proteoglycan receptors, characterized by a glypican 5 core and 2-O-sulfated iduronic acid at the nonreducing end, bind Shh via the Cardin-Weintraub motif and promote Shh-dependent transcription and proliferation.
Shh Proteoglycan Receptors Are in the Cerebellar EGL
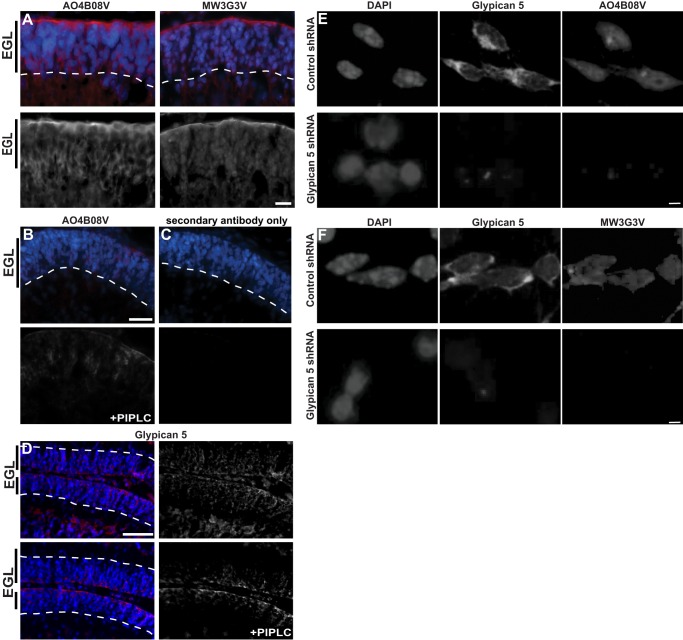
To examine the tissue localization of the endogenous proteoglycan co-receptors that bind Shh, we used a set of well characterized phage display antibodies that recognize particular glycan epitopes (40, 41). Specificity of the phage display antibodies is based on the pattern of disaccharide sulfation and acetylation and is also affected by chain length. Two of these antibodies, AO4B08V and MW3G3V, depend on 2-O-sulfo-iduronic acid for binding (42, 43). We found that AO4B08V and MW3G3V immunostain precursor cells in the EGL of the P6 cerebellum (Fig. 5A). Thus, glycans containing 2-O-sulfo-iduronic acid can be visualized on precursor cells of the EGL in vivo.
FIGURE 5.
2-O-Sulfo-iduronic acid attached to glypican 5 in the EGL. A, phage display antibodies that recognize specific glycan epitopes were used to immunostain P6 cerebellum. AO4BO8V and MW3G3V, which preferentially bind glycans containing 2-O-sulfo-iduronic acid, stain the proliferative zone of the EGL. B, P6 cerebellar sections were treated with PI-PLC and then stained for AO4B08V (red on top and gray on bottom) or for glypican 5 (D, red on left and gray on right). PI-PLC, which cleaves glycosylphosphatidylinositol-linked components, reduces stain for AO4BO8V and glypican 5 to levels comparable with secondary only antibody control (C). Scale bar, A–C, 12.5 μm; D, 50 μm. shRNA-mediated knockdown of glypican 5 (middle panel) reduces immunostaining with AO4B08V (E) or MW3G3V (F) (last panel); nuclei are counterstained with DAPI (1st panel). Scale bar, 10 μm.
To determine whether the 2-O-sulfo-iduronic acid-containing glycans in the EGL that are recognized by the phage display antibodies are attached to glypican 5, we treated cerebellar slices with PI-PLC to cleave the glycosylphosphatidylinositol that links glypican 5 to the cell surface. Treatment with PI-PLC abrogates immunostaining with AO4B08V (Fig. 5B) and immunostaining with antibodies to glypican 5 (Fig. 5D) to a level comparable with control staining with secondary antibody only (Fig. 5C). As PI-PLC releases all glycosylphosphatidylinositol-linked components, including other glypicans, we asked whether immunostaining with specific phage display antibodies is affected by more selective interventions. As shown, after shRNA-mediated knockdown of glypican 5 in dissociated GCPs, immunostaining with antibodies to glypican 5 and with AO4B08V (Fig. 5E) and MW3G3V (Fig. 5F) is greatly reduced. Thus, glypican 5 constitutes a protein core of proteoglycans that contain 2-O-sulfo-iduronic acid.
Shh Co-receptor Is Localized at the Base of Primary Cilia
To understand the role of the newly identified glycan receptor motif at the cellular level, we immunostained precursor cells with AO4B08V, a phage display antibody that recognizes 2-O-sulfo-iduronate residues, and we examined the subcellular localization of staining. The basal bodies of GCPs are readily visualized with antibodies to γ-tubulin or genetic labeling of centrin (44), although the primary cilia can be detected using antibodies to adenylate cyclase III (45, 46). We found that AO4B08V immunostaining is located adjacent to the basal bodies of primary cilia in control precursor cells or in precursor cells treated with an activator of the Shh pathway, SAG (Fig. 6A). Similar results are seen with MW3G3V (Fig. 6B), a second antibody that preferentially recognizes 2-O-sulfo-iduronic acid residues (42, 43), and with antibodies to glypican 5 (Fig. 6C). The glycan epitopes surround the small primary cilia, visualized with antibodies to adenylate cyclase III (Fig. 6D). Thus, the proteoglycan receptors encircle the primary cilia that are a major site of Shh signaling. However, unlike Ptch1 and Smo (47–49), the proteoglycan receptors do not relocalize following pathway stimulation. Therefore, the proteoglycan co-receptors are appropriately situated to concentrate Shh signaling at the base of the primary cilia in a sustained fashion.
FIGURE 6.
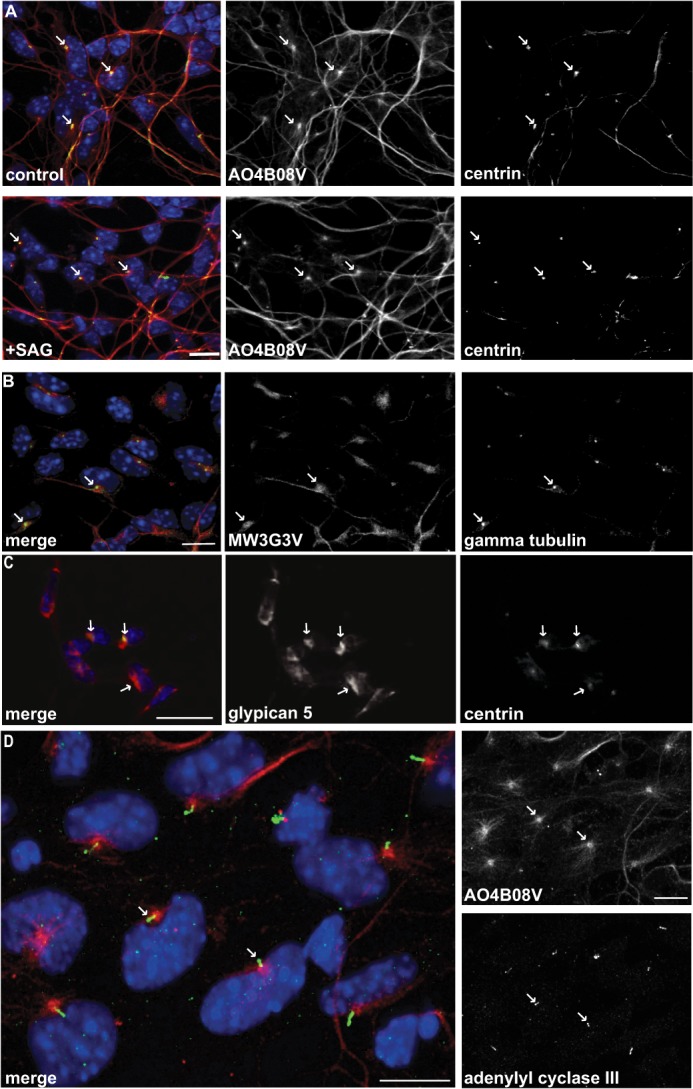
Shh co-receptor proteoglycans surround primary cilia. Glycan epitopes recognized by AO4B08V (A) or MW3G3V (B) are localized near basal bodies, both in control and SAG-stimulated GCPs. Basal bodies are visualized by GFP-tagged centrin or immunostaining with antibodies to γ-tubulin. C, glypican 5 staining shown in red (merge) or white is localized primarily around the basal bodies as visualized by GFP-tagged centrin. Scale bar, 25 μm. D, these glycan epitopes are adjacent to the primary cilia and visualized with antibodies to adenylate cyclase III. Scale bar, 10 μm; arrows indicate basal bodies.
Shh Co-receptors Promote Precursor Proliferation in the EGL
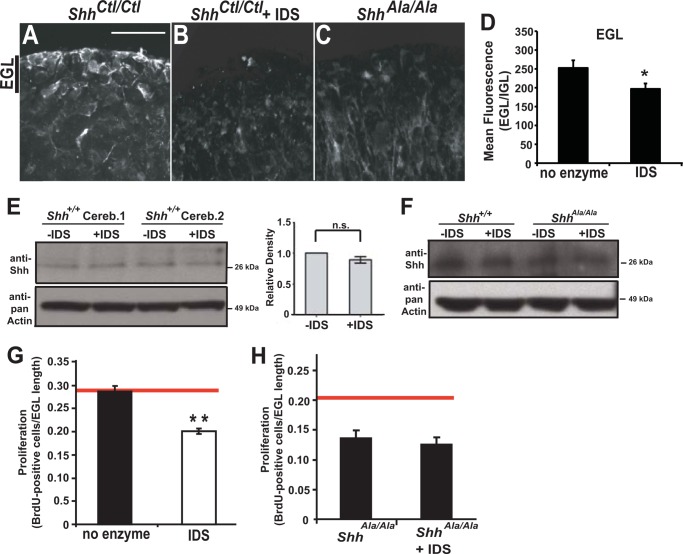
As shown above, proteoglycans with glypican 5 and with 2-O-sulfo-iduronic acid residues are expressed by precursors in the EGL. To determine whether these endogenous proteoglycans enable binding and concentration of Shh in this mitogenic niche, we treated cerebellar organotypic slices with IDS. IDS removes 2-O-sulfate from nonreducing end iduronic acid residues and reduces endogenous Shh immunostaining in the EGL (Fig. 7, A, B and D). Notably, the Shh-immunostaining pattern in IDS-treated ShhCtl/Ctl slices resembles the pattern seen in untreated ShhAla/Ala slices (Fig. 7, B and C) (23). Although IDS treatment or the ShhAla mutation alters the distribution pattern of Shh, these two interventions do not affect the overall level of Shh protein in the cerebellum (Fig. 7, E and F).
FIGURE 7.
Glycans are needed for the EGL mitogenic niche. A, Shh is localized in the cerebellar EGL. IDS treatment of slice cultures of P6 cerebellum from ShhCtl/Ctl mice (B) alters Shh immunostaining pattern (white), reduces the localization of Shh to this mitogenic niche, and resembles the Shh staining pattern seen in untreated ShhAla/Ala mice (C). Scale bar, 25 μm. D, quantification of Shh immunostaining. Mean signal intensity in the EGL normalized to IGL is reduced by IDS (*, p < 0.05). E and F, Western blot analysis reveals no significant difference in overall Shh levels in wild type slice cultures, wild type cultures treated with IDS, or in ShhAla/Ala. (Cereb, cerebellum; −IDS, slices without prior treatment; +IDS, slices treated with 3.5 μg/ml IDS, n = 3, n.s., not significant.) G, treating P6 cerebellar slices with IDS decreases proliferation in the EGL (*, p < 0.05; **, p < 1 × 10−6, n = 31–38). H, GCP proliferation in explant cultures from ShhAla/Ala mice was not affected by recombinant IDS (n = 4–9). Proliferation in the EGL of ShhAla/Ala slices is reduced compared with ShhCtl/Ctl slices (proliferation level in ShhCtl/Ctl slices designated by red line, p = 0.001).
Importantly, treatment of organotypic slice cultures with IDS not only alters the Shh immunostaining pattern (Fig. 7, A and B) but also reduces precursor cell proliferation in the EGL (Fig. 7G). Although functions of many growth factors can be influenced by sulfation motifs of proteoglycans (50, 51), our data indicate that changes in proliferation after IDS treatment are due to specific modulation of Shh binding to its proteoglycan receptors, as proliferation of precursor cells in ShhAla/Ala slice cultures is not altered by IDS (Fig. 7H). Thus, specialized HSPG co-receptors promote Shh-dependent proliferation of neural precursor cells within the cerebellar mitogenic niche.
DISCUSSION
Here, we identify proteoglycan receptors that promote Shh-dependent signaling, and we show that they are appropriately placed at both the tissue level and the subcellular level to mediate responses to Shh. We find that glypican 5, with attached glycosaminoglycan chains containing 2-O-sulfo-iduronic acid, is enriched in cerebellar precursor cells that require Shh for proliferation. At the subcellular level, these proteoglycan receptors are localized adjacent to primary cilia, which function as critical organelles for Shh signal transduction. We demonstrate that Shh binding and Shh responses are attenuated by genetic manipulations that reduce expression of glypican 5 or by enzymatic treatments that desulfate iduronic acid residues at the nonreducing ends of glycan chains. Together, these data indicate that proteoglycans containing a glypican 5 core and 2-O-sulfo-iduronic acid function as Shh co-receptors to promote neural precursor proliferation.
The ubiquitous distribution and structural complexity of glycans allow for participation in a variety of fundamental biological processes (52). Here, we used a genetic approach in combination with chemical synthesis of defined glycans (29) to identify motifs involved in Shh-HSPG interactions. Short synthetic glycans used for microarrays and for surface plasmon resonance binding studies, together with a comparison between the wild type Shh and a mutant isoform with impaired glycan binding (ShhAla), led to identification of glycan determinants critical for biologically relevant interactions with Shh.
We complemented chemical approaches with enzymatic methods that modify glycans on Shh-responsive cells, and we then assessed changes in Shh binding and function. The enzyme IDS selectively cleaves sulfate from the 2-O-position of iduronic acid situated at the nonreducing end of a glycosaminoglycan chain. We find that IDS attenuates Shh binding to precursor cells and reduces responses to Shh, including target gene induction and cell proliferation. Thus, proteoglycan receptors for Shh contain 2-O-sulfated iduronic acid at the nonreducing end of the glycosaminoglycan chains. Although sulfo-iduronic acid residues are unusual at the nonreducing end of uncleaved heparan sulfate chains (53), frame shifts in heparan sulfate biosynthetic enzymes may allow formation of such epitopes (54). Alternatively, cleavage of the glycan chain by a heparanase could give rise to a specialized Shh-binding epitope. It is notable that the nonreducing end of glycans, which appears to be the sites for Shh binding, are remarkably regulated during proteoglycan synthesis (54, 55).
Although treatment with IDS alters binding and biological responses to wild type Shh, IDS does not alter binding and biological responses of a mutant isoform ShhAla, providing validation that basic amino acids in the Shh Cardin-Weintraub motif are important for binding to proteoglycan co-receptors (12, 23, 24). Intriguingly, in Shh, a histidine and a proline residue are in the middle of the highly basic Cardin-Weintraub motif (amino acids 33–39, KRRHPKK); this may engender a structural bend so that Shh “caps” the end of glycan chains where a 2-O-sulfate is present (55). Detailed structural analyses will be necessary to test this possibility.
Our studies identify glypican 5 as a core component of the heparan sulfate proteoglycans that function as Shh co-receptors in neural precursors. Previous studies indicated that glypican 5 can also function as a core component in rhabdomyosarcoma tumor cells; however, the nature of the attached glycosaminoglycan chains that directly interact with Shh in rhabdomyosarcoma was not identified (19). Here, we show that glycan epitopes containing 2-O-sulfo-iduronic acid are attached to glypican 5 proteins (Figs. 5 and 6), and interventions that affect either the protein core or the sulfate modification reduce Shh binding and Shh responses. Following genetic knockdown of glypican 5, treatment with IDS has little additional effect on Shh binding or signaling (Fig. 4), providing further evidence that glycosaminoglycan chains containing 2-O-sulfo-iduronic acid and attached to glypican 5 mediate Shh responses. Although treatment with IDS does not fully abrogate Shh binding or Shh-dependent responses, the effects of IDS phenocopy those observed in ShhAla/Ala mutant mice, where cerebellar size is reduced, and are similar in magnitude to the effect of mutating either Gas1 or Boc alone (4), indicating that these effects are both statistically and biologically significant.
Multiple components of Shh signaling exhibit a distinctive subcellular localization, as they are found at or near the primary cilia (47, 48, 56). We find that proteoglycans containing a glypican 5 core and glycan chains with 2-O-sulfo-iduronic acid are localized near the base of the primary cilia both in unstimulated and in Shh-stimulated neural precursors. In unstimulated cells, the Ptch receptor is also near the base of the cilia (47). After binding of Shh, Ptch moves away from the cilia and Smo receptors accumulate in the primary cilia and initiate a signal (47). Thus, in contrast to Ptch and Smo, proteoglycan receptors remain at the base of the cilia throughout stimulation. The unvarying subcellular localization of proteoglycan receptors may enable continuous recruitment and concentration of Shh near the primary cilia on precursor cells within a mitogenic niche. In this way, the proteoglycan receptor may alter the kinetics or intensity of Shh signaling (23) and thereby augment the biological response (57). The localization of proteoglycan receptors adjacent to primary cilia is consistent with evidence that these co-receptors contribute to transcriptional and proliferative responses to Shh and may play a lesser role in functions independent of the primary cilia such as chemotaxis and neurite outgrowth (33).
The studies reported here identify critical attributes of proteoglycans that function as Shh co-receptors in neural precursor cells. A glypican 5 core component, with attached glycosaminoglycan chains containing 2-O-sulfo-iduronic acid at the nonreducing end, binds Shh and promotes intracellular signaling. Localization of these proteoglycan co-receptors adjacent to the primary cilia is likely to be important for their role in Shh signaling and proliferative responses. As the Shh pathway is critical for normal brain development and for tumor growth, identification of this co-receptor provides new therapeutic targets for developmental disorders and for Shh-dependent cancers.
Acknowledgments
We thank Jennifer Kalscheuer, Jose Alfaro, and Emily Chadwick for excellent assistance; T. Nielsen for MPS II (Lysosomal Diseases Research Unit, SA Pathology, Adelaide, Australia) urine oligosaccharides; S. Josiah and Brian Felice (Shire Human Genetics Therapies, Inc.) for iduronate 2-sulfatase (IDS Elaprase®; idursulfase); and J. Zaia, C. Stiles, M. Greenberg, and the Segal laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Director's Pioneer Award (Grant DP1 0D000839; to R. A. S.). This work was also supported by a Quan Fellowship (to R. M. W.), the Swiss National Science Foundation (to M. L. H.), ETH Zurich (to P. H. S.), the Max-Planck Society (to P. H. S.), and an ERC advanced grant (to P. H. S. and Harvard NeuroDiscovery Center).
This article was selected as a Paper of the Week.
- Hh
- Hedgehog
- Shh
- Sonic Hedgehog
- Ptch1
- Patched
- Smo
- Smoothened
- HSPG
- heparan sulfate proteoglycan
- IDS
- iduronate 2-sulfatase
- GCP
- granule cell precursor
- PI-PLC
- phosphatidylinositol-specific phospholipase C
- EGL
- external granule cell layer
- IGL
- internal granule cell layer
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Fuccillo M., Joyner A. L., Fishell G. (2006) Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 772–783 [DOI] [PubMed] [Google Scholar]
- 2. Marigo V., Davey R. A., Zuo Y., Cunningham J. M., Tabin C. J. (1996) Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176–179 [DOI] [PubMed] [Google Scholar]
- 3. Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H., Noll M., Hooper J. E., de Sauvage F., Rosenthal A. (1996) The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129–134 [DOI] [PubMed] [Google Scholar]
- 4. Izzi L., Lévesque M., Morin S., Laniel D., Wilkes B. C., Mille F., Krauss R. S., McMahon A. P., Allen B. L., Charron F. (2011) Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev. Cell 20, 788–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allen B. L., Song J. Y., Izzi L., Althaus I. W., Kang J. S., Charron F., Krauss R. S., McMahon A. P. (2011) Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev. Cell 20, 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tenzen T., Allen B. L., Cole F., Kang J. S., Krauss R. S., McMahon A. P. (2006) The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev. Cell 10, 647–656 [DOI] [PubMed] [Google Scholar]
- 7. Zhang W., Kang J. S., Cole F., Yi M. J., Krauss R. S. (2006) Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev. Cell 10, 657–665 [DOI] [PubMed] [Google Scholar]
- 8. Allen B. L., Tenzen T., McMahon A. P. (2007) The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beachy P. A., Hymowitz S. G., Lazarus R. A., Leahy D. J., Siebold C. (2010) Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 24, 2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chuang P. T., McMahon A. P. (1999) Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397, 617–621 [DOI] [PubMed] [Google Scholar]
- 11. Bellaiche Y., The I., Perrimon N. (1998) Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394, 85–88 [DOI] [PubMed] [Google Scholar]
- 12. Rubin J. B., Choi Y., Segal R. A. (2002) Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development 129, 2223–2232 [DOI] [PubMed] [Google Scholar]
- 13. Koziel L., Kunath M., Kelly O. G., Vortkamp A. (2004) Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev. Cell 6, 801–813 [DOI] [PubMed] [Google Scholar]
- 14. Han C., Belenkaya T. Y., Wang B., Lin X. (2004) Drosophila glypicans control the cell-to-cell movement of Hedgehog by a dynamin-independent process. Development 131, 601–611 [DOI] [PubMed] [Google Scholar]
- 15. The I., Bellaiche Y., Perrimon N. (1999) Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell 4, 633–639 [DOI] [PubMed] [Google Scholar]
- 16. Yan D., Wu Y., Yang Y., Belenkaya T. Y., Tang X., Lin X. (2010) The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development 137, 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gallet A., Staccini-Lavenant L., Thérond P. P. (2008) Cellular trafficking of the glypican Dally-like is required for full-strength Hedgehog signaling and wingless transcytosis. Dev. Cell 14, 712–725 [DOI] [PubMed] [Google Scholar]
- 18. Park Y., Rangel C., Reynolds M. M., Caldwell M. C., Johns M., Nayak M., Welsh C. J., McDermott S., Datta S. (2003) Drosophila perlecan modulates FGF and hedgehog signals to activate neural stem cell division. Dev. Biol. 253, 247–257 [DOI] [PubMed] [Google Scholar]
- 19. Li F., Shi W., Capurro M., Filmus J. (2011) Glypican-5 stimulates rhabdomyosarcoma cell proliferation by activating Hedgehog signaling. J. Cell Biol. 192, 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim M. S., Saunders A. M., Hamaoka B. Y., Beachy P. A., Leahy D. J. (2011) Structure of the protein core of the glypican Dally-like and localization of a region important for hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 13112–13117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Capurro M. I., Xu P., Shi W., Li F., Jia A., Filmus J. (2008) Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev. Cell 14, 700–711 [DOI] [PubMed] [Google Scholar]
- 22. Yan D., Lin X. (2008) Opposing roles for glypicans in Hedgehog signalling. Nat. Cell Biol. 10, 761–763 [DOI] [PubMed] [Google Scholar]
- 23. Chan J. A., Balasubramanian S., Witt R. M., Nazemi K. J., Choi Y., Pazyra-Murphy M. F., Walsh C. O., Thompson M., Segal R. A. (2009) Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nat. Neurosci. 12, 409–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Farshi P., Ohlig S., Pickhinke U., Höing S., Jochmann K., Lawrence R., Dreier R., Dierker T., Grobe K. (2011) Dual roles of the Cardin-Weintraub motif in multimeric Sonic hedgehog. J. Biol. Chem. 286, 23608–23619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turnbull J., Powell A., Guimond S. (2001) Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 11, 75–82 [DOI] [PubMed] [Google Scholar]
- 26. Lum L., Yao S., Mozer B., Rovescalli A., Von Kessler D., Nirenberg M., Beachy P. A. (2003) Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039–2045 [DOI] [PubMed] [Google Scholar]
- 27. Ayers K. L., Gallet A., Staccini-Lavenant L., Thérond P. P. (2010) The long-range activity of Hedgehog is regulated in the apical extracellular space by the glypican Dally and the hydrolase Notum. Dev. Cell 18, 605–620 [DOI] [PubMed] [Google Scholar]
- 28. de Paz J. L., Noti C., Seeberger P. H. (2006) Microarrays of synthetic heparin oligosaccharides. J. Am. Chem. Soc. 128, 2766–2767 [DOI] [PubMed] [Google Scholar]
- 29. Noti C., de Paz J. L., Polito L., Seeberger P. H. (2006) Preparation and use of microarrays containing synthetic heparin oligosaccharides for the rapid analysis of heparin-protein interactions. Chemistry 12, 8664–8686 [DOI] [PubMed] [Google Scholar]
- 30. Fuller M., Chau A., Nowak R. C., Hopwood J. J., Meikle P. J. (2006) A defect in exodegradative pathways provides insight into endodegradation of heparan and dermatan sulfates. Glycobiology 16, 318–325 [DOI] [PubMed] [Google Scholar]
- 31. Cummings R. D. (2009) The repertoire of glycan determinants in the human glycome. Mol. Biosyst. 5, 1087–1104 [DOI] [PubMed] [Google Scholar]
- 32. Ho M., Kim H. (2011) Glypican-3: a new target for cancer immunotherapy. Eur. J. Cancer 47, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bijlsma M. F., Damhofer H., Roelink H. (2012) Hedgehog-stimulated chemotaxis is mediated by smoothened located outside the primary cilium. Sci. Signal. 5, ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deleted in proof [Google Scholar]
- 35. Wechsler-Reya R. J., Scott M. P. (1999) Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22, 103–114 [DOI] [PubMed] [Google Scholar]
- 36. Molofsky A. V., Pardal R., Iwashita T., Park I. K., Clarke M. F., Morrison S. J. (2003) Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425, 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kenney A. M., Cole M. D., Rowitch D. H. (2003) Nmyc up-regulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130, 15–28 [DOI] [PubMed] [Google Scholar]
- 38. Kenney A. M., Rowitch D. H. (2000) Sonic hedgehog promotes G1 cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 20, 9055–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen J. K., Taipale J., Young K. E., Maiti T., Beachy P. A. (2002) Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. U.S.A. 99, 14071–14076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Kuppevelt T. H., Dennissen M. A., van Venrooij W. J., Hoet R. M., Veerkamp J. H. (1998) Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J. Biol. Chem. 273, 12960–12966 [DOI] [PubMed] [Google Scholar]
- 41. Jenniskens G. J., Oosterhof A., Brandwijk R., Veerkamp J. H., van Kuppevelt T. H. (2000) Heparan sulfate heterogeneity in skeletal muscle basal lamina: demonstration by phage display-derived antibodies. J. Neurosci. 20, 4099–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. ten Dam G. B., van de Westerlo E. M., Smetsers T. F., Willemse M., van Muijen G. N., Merry C. L., Gallagher J. T., Kim Y. S., van Kuppevelt T. H. (2004) Detection of 2-O-sulfated iduronate and N-acetylglucosamine units in heparan sulfate by an antibody selected against acharan sulfate (IdoA2S-GlcNAc)n. J. Biol. Chem. 279, 38346–38352 [DOI] [PubMed] [Google Scholar]
- 43. Thompson S. M., Fernig D. G., Jesudason E. C., Losty P. D., van de Westerlo E. M., van Kuppevelt T. H., Turnbull J. E. (2009) Heparan sulfate phage display antibodies identify distinct epitopes with complex binding characteristics: insights into protein binding specificities. J. Biol. Chem. 284, 35621–35631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Higginbotham H., Bielas S., Tanaka T., Gleeson J. G. (2004) Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res. 13, 155–164 [DOI] [PubMed] [Google Scholar]
- 45. Berbari N. F., Bishop G. A., Askwith C. C., Lewis J. S., Mykytyn K. (2007) Hippocampal neurons possess primary cilia in culture. J. Neurosci. Res. 85, 1095–1100 [DOI] [PubMed] [Google Scholar]
- 46. Berbari N. F., O'Connor A. K., Haycraft C. J., Yoder B. K. (2009) The primary cilium as a complex signaling center. Curr. Biol. 19, R526–R535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rohatgi R., Milenkovic L., Scott M. P. (2007) Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–376 [DOI] [PubMed] [Google Scholar]
- 48. Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005) Vertebrate smoothened functions at the primary cilium. Nature 437, 1018–1021 [DOI] [PubMed] [Google Scholar]
- 49. Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87 [DOI] [PubMed] [Google Scholar]
- 50. Kamimura K., Maeda N., Nakato H. (2011) In vivo manipulation of heparan sulfate structure and its effect on Drosophila development. Glycobiology 21, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pye D. A., Vives R. R., Turnbull J. E., Hyde P., Gallagher J. T. (1998) Heparan sulfate oligosaccharides require 6-O-sulfation for promotion of basic fibroblast growth factor mitogenic activity. J. Biol. Chem. 273, 22936–22942 [DOI] [PubMed] [Google Scholar]
- 52. Lowe J. B., Marth J. D. (2003) A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 72, 643–691 [DOI] [PubMed] [Google Scholar]
- 53. Jacobsson I., Lindahl U., Jensen J. W., Rodén L., Prihar H., Feingold D. S. (1984) Biosynthesis of heparin. Substrate specificity of heparosan N-sulfate d-glucuronosyl 5-epimerase. J. Biol. Chem. 259, 1056–1063 [PubMed] [Google Scholar]
- 54. Rudd T. R., Yates E. A. (2012) A highly efficient tree structure for the biosynthesis of heparan sulfate accounts for the commonly observed disaccharides and suggests a mechanism for domain synthesis. Mol. Biosyst. 8, 1499–1506 [DOI] [PubMed] [Google Scholar]
- 55. Shih P. C., Yang M. S., Lin S. C., Ho Y., Hsiao J. C., Wang D. R., Yu S. S., Chang W., Tzou D. L. (2009) A turn-like structure “KKPE” segment mediates the specific binding of viral protein A27 to heparin and heparan sulfate on cell surfaces. J. Biol. Chem. 284, 36535–36546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scholey J. M., Anderson K. V. (2006) Intraflagellar transport and cilium-based signaling. Cell 125, 439–442 [DOI] [PubMed] [Google Scholar]
- 57. Dessaud E., Yang L. L., Hill K., Cox B., Ulloa F., Ribeiro A., Mynett A., Novitch B. G., Briscoe J. (2007) Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature 450, 717–720 [DOI] [PubMed] [Google Scholar]