Abstract
Background and Purpose
Brain vascular endothelial cells express histamine H1 and H2 receptors, which regulate brain capillary permeability. We investigated whether H3 and H4 receptors are also expressed in these cells and may thus play a role in permeability regulation.
Experimental Approach
An immortalized rat brain endothelial cell line RBE4 was used to assess the presence of H3 and H4 receptors. Reverse transcription-PCR (RT-PCR) and sequencing were used to identify the receptor mRNAs. The receptors were stimulated with histamine and immepip, and specific inverse agonists/antagonists ciproxifan and JNJ 7777120 were used to block H3 and H4 receptors, respectively.
Key Results
RT-PCR of mRNA extracted from cultured immortalized RBE4 cells revealed two rat H4 receptor gene (Hrh4) transcripts, one full-length (coding sequence 1173 bp), and one with a 164 bp deletion. Also, two rat H3 receptor gene (Hrh3) isoform mRNAs were expressed in RBE4 cells, and sequencing showed they were the full-length H3 receptor and the 144 bp deletion form. Both histamine and immepip (H3 and H4 receptor agonists) activated the Erk1/2 MAPK pathway in the RBE4 cells and in vivo in brain blood vessels by activating H4 receptors, as the H4 receptor-specific inverse agonists/antagonist JNJ 7777120, but not ciproxifan, H3 receptor antagonist, dose-dependently blocked this effect in RBE4 cells.
Conclusions and Implications
Both Hrh3 and Hrh4 receptors are expressed in rat brain endothelial cells, and activation of the histamine H4 receptor activates the Erk1/2 cascade. H3 and H4 receptors in endothelial cells are potentially important for regulation of blood–brain barrier permeability, including trafficking of immunocompetent cells.
Linked Articles
This article is part of a themed issue on Histamine Pharmacology Update. To view the other articles in this issue visit http://dx.doi.org/10.1111/bph.2013.170.issue-1
Keywords: blood–brain barrier, histamine, GPCR, MAPK, Erk1/Erk2
Introduction
Histamine H3 receptors are widely expressed in the brain in histaminergic and other neurons and they regulate neurotransmitter release (Haas and Panula, 2003). The expression of the rat H4 receptor gene (Hrh4) in the brain has been difficult to establish using in situ hybridization, but antibodies made against peptides derived from the H4 receptor sequence have been used successfully to detect immunoreactivity in brain neurons (Connelly et al., 2009). Due to the difficulties in demonstrating significant expression of Hrh4 mRNA in brain neurons, we hypothesized that one of the main sites of Hrh4 mRNA expression sites in the brain could be vascular endothelial cells.
Histamine is a regulator of brain vascular permeability (Akdis and Simons, 2006), and both H1 and H2 receptors are expressed in endothelial cells (Karlstedt et al., 1999). Permeability of brain micro-vessels is important in several diseases, including multiple sclerosis (Goverman, 2009) and degenerative brain diseases, which includes Parkinson's disease (Grammas et al., 2011). In a mouse model of multiple sclerosis, experimental allergic encephalomyelitis (EAE), one of the sensitivity loci is the histamine H1 receptor. Knock-out (KO) mice lacking functional H1, H2, H3 or H4 receptors all show abnormal development of disease indicators initiated by immunological stimulation (Saligrama et al., 2012). Mice lacking H1 or H2 receptors develop less severe symptoms of disease than wild-type animals, whereas mice lacking histidine decarboxylase develop more severe symptoms, as do the mice lacking H3 or H4 receptors (Saligrama et al., 2012). Only neuronal H3 receptors have been considered as potentially responsible for the EAE pathology in H3 receptor KO mice because the H3 receptor is not expressed in lymphocytes (Teuscher et al., 2007). The pathology associated with H4 receptor KO animals has been proposed to be due to receptor expression in immunocompetent cells (del Rio et al., 2012). In this study, we investigated whether brain endothelial cells express H3 and/or H4 receptors, which could potentially be important for the entry of immunocompetent cells from the bloodstream to the brain.
As a model system, we used the immortalized rat cerebral endothelial cell line RBE4 (Durieu-Trautmann et al., 1993). These cells have been characterized in detail (Roux et al., 1994) and have been found to closely resemble brain endothelial cells in primary cultures. They express both Hrh1 and Hrh2 mRNA, which are up-regulated following exposure to dexamethasone (Karlstedt et al., 1999). These cells have been used extensively in studies on brain endothelial cells in vitro, for example for characterization of IL-15 receptor expression (Wu et al., 2010) and to study the uptake and efflux of rhodamine-123 in brain endothelial cells (Yang et al., 2012). The use of a cell line instead of primary cultures excludes the potential culture contamination with other cell types and cell remnants that could have an effect on the parameters measured. After finding both Hrh3 and Hrh4 in the immortalized rat brain endothelial cell line RBE4, we examined which of the many splice forms of these receptors are expressed and if they are functional.
Methods
Cell cultures
The cultured RBE4 cells used in this study were immortalized rat brain microvessel endothelial cells (Durieu-Trautmann et al., 1993; Roux et al., 1994). Basic cell culture conditions were as stated in Karlstedt et al. (1999). The cells were originally a kind gift from Dr PO Couraud, CNRS, Paris. Briefly, prior to ligand treatment, cell cultures were incubated for 10–12 h in the regular culture media with no serum (necessary for cell synchronization, and also for avoiding any natural histamine receptor ligands in the media). All ligand treatments were performed for 10 min at 37°C. Ligand treatments were ended by aspiration of the culture media followed by a quick wash with PBS (pH 7.4) at 37°C. Cells were immediately fixed in concentrated methanol at −20°C for 30 min, followed by a transfer to PBS (pH 7.4) at +4°C until used for immunocytochemical staining. The cells used for RNA isolation were harvested after treatment by scraping them off culture plates, collected in tubes, centrifuged to a pellet and eventually frozen at −80°C.
Reverse transcription-PCR (RT-PCR)
RT-PCR amplifications were performed on total RNA extracts from harvested RBE4 cells and different rat tissues according to the manufacturer's instructions (RNeasy kit, Qiagen, Hilden, Germany). cDNA synthesis was performed as instructed in ‘Superscript first-strand synthesis system for RT-PCR’ (Invitrogen, Carlsbad, CA, USA). For PCR amplification 23-mer oligonucleotides, partly covering the beginning and the end of the full-length coding sequence (CDS), were constructed for rat histamine Hrh3 and Hrh4 receptors. As a negative control (contr/RT) for cDNA synthesis and PCR amplification, we included a sample with no tissue/cells in the RNA isolation procedure. This control sample was used in parallel with all amplifications.
PCR products were eventually cloned into proper vectors and sequenced (Applied Biosystems Big Dye Terminator [v1.1] kit [Foster City, CA, USA] for PCR and Applied Biosystems ABI Prism 3130xl 16 capillary Genetic Analyzer for analysing).
In vivo experiments
Male Wistar rats (16-weeks-old, 300 g) were anaesthetized with sodium pentobarbital (Mebunat, Orion, Finland) and perfused through the heart ventricle. The total number of rats used was 7. Control rats were first perfused with 200 mL PBS (pH 7.4) at room temperature (RT) and then with 200 mL ice-cold 4% paraformaldehyde (pFA) in PB (pH 7.4). Histamine-perfused brains were first perfused with 200 mL PBS (pH 7.4) at RT, then with 200 mL 10 μM histamine in PBS (pH 7.4), and finally with 200 mL ice-cold 4% pFA in PB (pH 7.4). Histamine receptor antagonist-treated rats were first perfused with 200 mL PBS (pH 7.4) at RT, then with 200 mL of 10 μM JNJ 7777120 in PBS (pH 7.4) and then with 200 mL 10 μM histamine in PBS (pH 7.4). Finally, these rats were perfused with 200 mL ice-cold 4% pFA in PB (pH 7.4). pFA-fixed brains were immediately removed from the skull and for cryoprotection, all brains were kept in 20% sucrose in 0.1 M PB (pH 7.4) at 4°C until they sank. Brains were finally frozen on dry ice, embedded and kept at −70°C until used. Immunostainings for MAPK (Erk1/2) were carried out on 20 μm thick cryosections mounted on superfrost plus slides (Thermo Scientific, Braunschweig, Germany). The principles of the Finnish Act on the Use of Animals for Experimental Purposes were followed and all protocols were approved by the Animal Experiment Committee of the State Provincial Office of Southern Finland. All studies involving animals are reported in accordance with the ARRIVE guidelines for reporting experiments involving animals (Kilkenny et al., 2010; McGrath et al., 2010).
Immunocytochemistry
Coverslips with methanol fixed RBE4 cells were washed briefly in PBS (pH 7.4) containing 0.05% Triton X-100 (PBS-T), followed by incubation with phospho-p44/42 MAPK antibody (#9101, Cell Signaling, Danvers, MA, USA) diluted (1:200) in PBS-T containing 1% normal serum for 1 h at RT. After being washed in PBS-T, the cells were incubated for 1 h at RT with Alexa fluor-568-conjugated goat anti rabbit antibody (Molecular Probes, Eugene, OR, USA) diluted 1:500 in PBS-T containing 1% normal serum. After a final wash in PBS, the coverslips were embedded in glycerol on glass slides and examined with a fluorescence microscope (Olympus AX70 Provis, Olympus, Tokyo, Japan).
Immunostaining for phospho-p44/42 MAPK in 20 μm thick rat brain cryosections was carried out using biotin-streptavidin method followed by visualization peroxidase activity with diaminobenzidine (DAB) and H2O2 (ABC kit, Vector laboratories, Burlingame, CA, USA). Sections were incubated with phospho-p44/42 MAPK primary antibody (#9101, Cell Signaling) diluted (1:200) in PBS-T containing 2% normal serum for 24–48 h at +4°C. For signal detection, a biotinylated anti-rabbit IgG secondary antibody diluted 1:700 in PBS-T containing 2% normal serum was applied for 2 h at RT. After DAB treatment, sections were dried for 12–24 h in RT and then mounted with DPX mountant (BDH, Poole, UK).
Western blotting
For Western blotting, endothelial cells were harvested in SDS-buffer (62,5 mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 50 mM DTT, 0.01% brom phenol blue).
Western blotting was performed according to instructions from the provider of the primary antibodies (Cell Signaling). Proteins were separated on 10% SDS polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad, 162-0115, Hercules, CA, USA) using a semi-dry transfer cell. Membranes were blocked for 1–2 h at RT with 5% non-fat dry milk in Tris-buffered saline (TBS) plus 0.1% Tween and immunoblotted with rabbit phospho-p44/42 MAPK antibody (#9101 Cell Signaling) or p44/42 MAPK antibody (#9102 Cell Signaling). Primary antibodies (diluted 1:1000 in TBS, 0.1% Tween-20 with 5% non-fat dry milk) were incubated for 12–24 h at +4°C in a shaking bath, followed by incubation with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Bio-Rad, 170-6515, Hercules, CA, USA, diluted 1:4000 in TBS, 0.1% Tween-20 with 5% non-fat dry milk) incubated for 1–2 h at RT in a shaking bath. For reblotting filters were washed according to instructions (ReBlot Plus Mild, Millipore, Temecula, CA, USA). All blots were developed with the ECL-Plus detection system (Amersham Pharmacia, Cambridge, UK).
Quantification
Quantification of Western blot films was done by digitizing the X-ray film images with a computer-based MCID image analysis system (Imaging Research, St Catherines, Canada). The result presented is as a ratio between phosphorylated Erk1/2 and total pool of Erk1/2-protein in each sample.
Statistical analysis
Data are expressed as mean or mean ± SD. Treatment effects over the time course were compared using Student's paired t-test. Statistical significance was taken as P < 0.05.
Materials
Histamine hydrochloride and ciproxyfan hydrochloride were from Sigma-Aldrich (St Louis, MO, USA). JNJ 7777120 was obtained from Johnson & Johnson Pharmaceutical Research & Development, L.L.C. (San Diego, CA, U.S.A.).
The nomenclature used for the receptors conforms to BJP's Guide to Receptors and Channels (Alexander et al., 2011).
Results
Histamine H4 and H3 receptors are expressed in rat cultured brain endothelial (RBE4) cells
Histamine Hrh4 is known to be highly expressed in the spleen. Using RT-PCR, we verified this and by using PCR primers, partly flanking the coding region, we found two different transcripts. RT-PCR on mRNA extracted from cultured RBE4 cells revealed identical transcripts, one being the full-length Hrh4 (CDS 1173 bp, as in NCBI Reference Sequence: NM_131909.1) and the other one an identical transcript with the full-length transcript but with a 164 bp deletion (Figure 1). The RBE4 cells showed this expression pattern after incubation in both the normal culture medium (10% serum) and after 12 h in the starvation medium (regular culture media with serum omitted). The shorter transcript was differentially spliced with the second exon excluded. A schematic picture (Figure 2A) shows the Hrh4 with the second exon marked in green, the putative stop codon for the truncated (and frame shifted) transcript is indicated in red. Part of the transcripts are aligned in Figure 2B with the deleted area marked in green (second exon) and putative stop codon marked in red.
Figure 1.
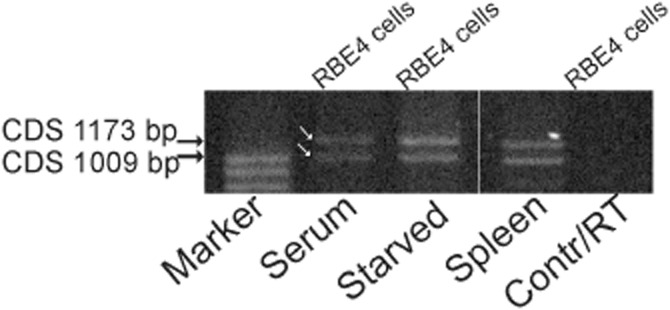
RT-PCR amplification of two variants of the histamine H4 receptor messages in RBE4 cells, as well as in a rat spleen sample. Culturing RBE4 cells in regular medium supplemented with 10% calf serum revealed the same two H4 receptor transcripts that are seen in cells after 12 h of starvation (no calf serum supplement). White arrows indicate the transcripts with 1073 bp and 1009 bp CDS, respectively. The contr/RT lane indicates the negative control sample with mRNA omitted from the amplification reaction.
Figure 2.
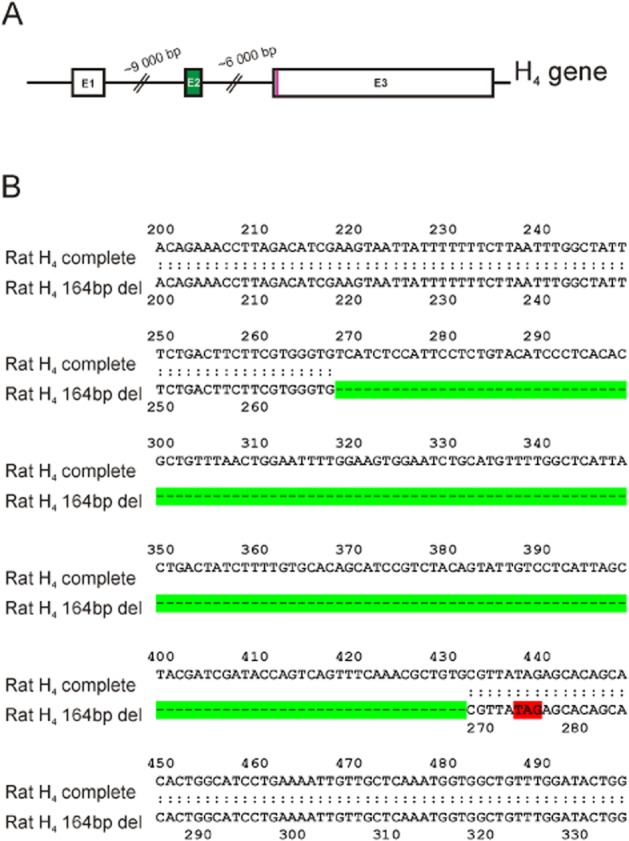
A schematic drawing of the tree exon rat histamine H4 receptor gene and the partial sequence of the two isolated H4 isoforms. (A) The E1, E2 and E3 exons are indicated and the deleted region in the shorter isoform is marked in green. (B) A part of the cloned sequences are aligned and identical accept for the 164 bp deletion in the shorter cDNA (gap marked in green). The potential new stop codon for the frame shifted shorter message is marked in red.
We have previously shown that the RBE4 cells express histamine receptor genes Hrh1 and Hrh2 (Karlstedt et al., 1999), but the existence of Hrh3 was not demonstrated. In the present study, Hrh3 mRNA was found to be expressed in RBE4 cells during normal cell culture conditions as shown in Figure 3. RT-PCR amplification on RBE4 cells revealed the distinct expression of both histamine H3A (the full-length isoform) and H3C (the 144 bp deletion form) gene isoforms (Figure 3).
Figure 3.
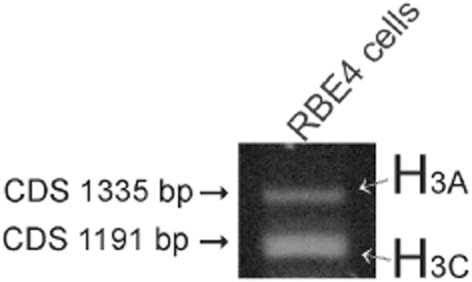
RT-PCR amplification of the histamine H3 receptor messages in RBE4 cells. The presence of the H3A (full-length transcript) and H3C (144 bp deletion) isoforms are verified by cloning and sequencing.
All these RT-PCR results were verified by subsequent cloning and sequencing (data not shown).
Histamine H4 receptor-mediated activation of the MAPK (Erk1/2) pathway in RBE4 cells
Histamine was shown to activate the Erk1/2 MAPK pathway in RBE4 cells. Histamine treatment (10 nM) resulted in an elevated level of MAPK phosphorylation as visualized by Western blotting (Figure 4) and immunohistochemical staining (Figure 5A). The MAPK activation was dose-dependently blocked by the histamine H4 receptor antagonist JNJ 7777120. A Western blot quantification of representative, in parallel, treated cell samples is shown in Figure 5B. The level of phosphorylation is shown as the ratio between phosphorylated Erk1/2 and total pool of Erk1/2-protein for each sample. Immepip-treated (10 μM) RBE4 cells revealed, similar to those treated with histamine, an increase in Erk1/2 MAPK phosphorylation (Figure 5C). This phosphorylation was also dose-dependently blocked by the histamine H4 receptor antagonist JNJ 7777120. The inserts in Figure 5B and C show the Erk1/2 phosphorylated bands on RBE4 cells at baseline and after the indicated treatments.
Figure 4.
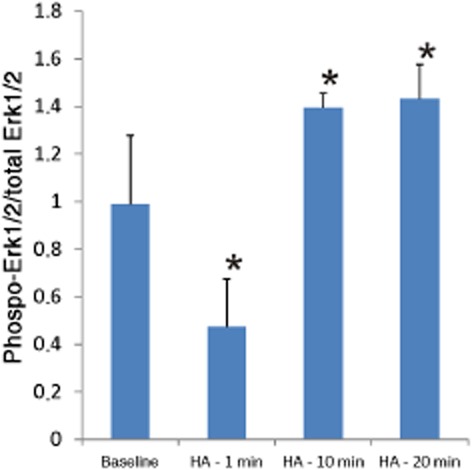
Time dependence of 10 nM histamine (HA)-induced MAPK (Erk1/2, p44/p42) phosphorylation in RBE4 cells. Sample n ≥ 3 per time point, *P < 0.05.
Figure 5.
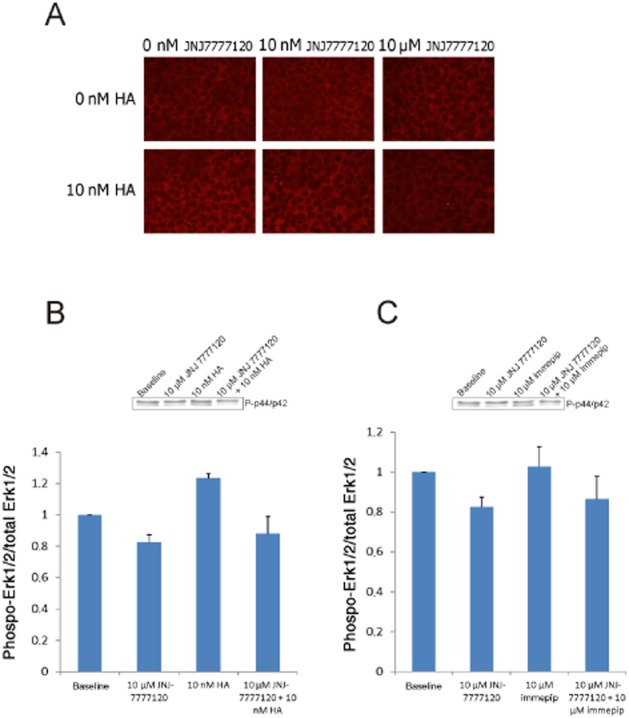
Histamine (HA)- and immepip-induced MAPK (Erk1/2, p44/p42) phosphorylation in RBE4 cells. (A) Immunostaining detecting phosphorylated MAPK in cultured and differentially treated RBE4 cells. (B) Representative quantification of MAPK phosphorylation in HA and histamine H4 receptor antagonist (JNJ 7777120) treated RBE4 cells. (C) Representative quantification of MAPK phosphorylation in immepip and JNJ 7777120-treated RBE4 cells. The quantifications were done on Western blot films (see inserts), and results are indicated as the ratio between phosphorylated and non-phosphorylated MAPK. Untreated cells show a weak but consistent level of phosphorylated MAPK (base line), the degree of background phosphorylation was moderately affected by JNJ 7777120. Histamine (10 nM) as well as immepip (10 μM) increased the level of phosphorylation, and this activation of MAPK was inhibited by JNJ 7777120. In (B) and (C) n = 2, SD indicated.
Histamine H3 receptor does not activate the MAPK (Erk1/2) pathway in RBE4 cells
Histamine-mediated activation of the Erk1/2 MAPK pathway in RBE4 cells was not blocked by the histamine H3 receptor inverse agonist/antagonist, ciproxifan (10 μM). Immepip treatment (10 nM–10 μM) increased the level of MAPK phosphorylation in RBE4 cells, but pretreatment with ciproxifan had no inhibitory effect on the phosphorylation in these cells (Figure 6).
Figure 6.
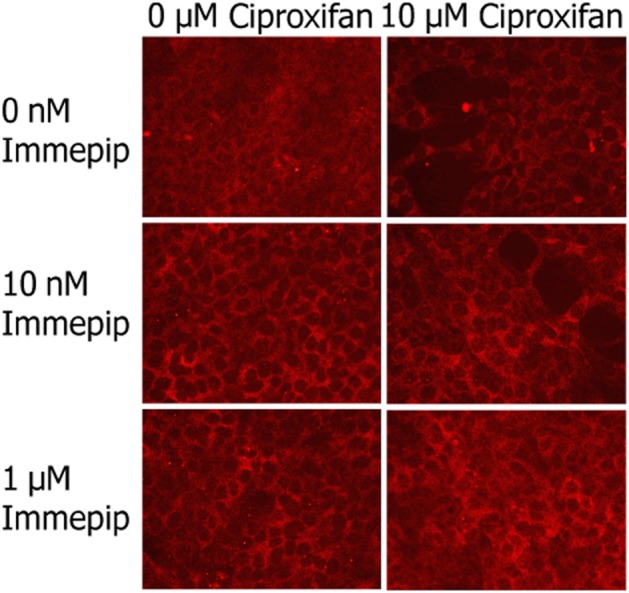
Immepip-induced Erk1/2 phosporylation in RBE4 cells is not affected by the histamine H3 receptor antagonist ciproxifan.
In vivo activation of the MAPK (Erk1/2) pathway in rat brain blood vessels
Endothelial cells in rat brain blood vessels in vivo have a phenotype that varies depending on the brain region studied. Immunohistological staining to reveal phosphorylated MAPK showed a low level (or none) of basal MAPK activity in microvessels of, for example, the forebrain and thalamus (data not shown). In the pons and the brainstem regions, basal level of staining for phosphorylated MAPK in the capillary endothelium was locally very distinct (Figure 7A). Furthermore, brain perfusion with histamine (10 μM) increased the phospho-MAPK staining in capillary endothelial cells in some regions, for example, in the dorsomedial tegmental area (Figure 7B). Pre-perfusion of the brain with the H4 receptor antagonist JNJ 7777120 resulted in a partial block of the histamine-induced MAPK phosphorylation (Figure 7C).
Figure 7.
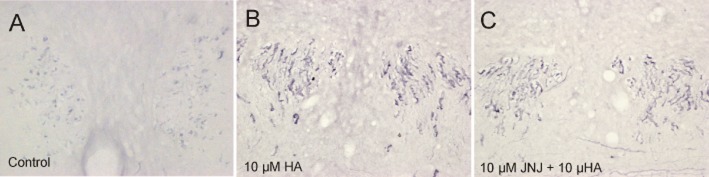
MAPK (Erk1/2, p44/p42) phosphorylation in rat brain endothelial cells in vivo. (A) Modest phospho-MAPK immunoreactivity in the brainstem of untreated control rats. (B) MAPK phosphorylation in brain capillaries in dorsomedial tegmental area after perfusion with 10 μM histamine (HA). (C) Preperfusion with 10 μM JNJ 4444120, a histamine H4 receptor antagonist, down-regulated locally the histamine-induced MAPK phosphorylation.
Discussion
Histamine H4 receptor expression has, in the past, been considered to be largely restricted to haematopoietic cells such as mast cells, eosinophils and T cells (Zampeli and Tiligada, 2009). Stimulation of this receptor was thought to mediate chemotaxis (Buckland et al., 2003; Ling et al., 2004) and modulate cell migration (Gschwandtner et al., 2010). Recently, it has been suggested that the H4 receptor also acts as a modulator of anti-inflammatory responses within the CNS (Passani and Ballerini, 2012). Histamine H4 receptor-KO mice develop more severe EAE upon myelin oligodendrocyte glycoprotein administration, therefore, it was speculated that H4 receptors are involved in T cell recruitment and regulation of T cell chemotaxis and suppressor activity, thereby having an effect on the anti-inflammatory response (del Rio et al., 2012).
Histamine is thought to be a major player in leukocyte trafficking through activation of histamine H1 receptors, but recently the histamine H4 receptor has been shown to be a key player as well (Buckland et al., 2003; Ling et al., 2004). Published data show that histamine receptors are expressed and activated in different inflammatory cells, but we found in this study that histamine H4 receptors are also expressed and activated in blood vessel endothelial cells. To the best of our knowledge, this is the first time this has been demonstrated and indicates that histamine could have an important function in, for example, vascular permeability not only through stimulation of histamine H1 receptors but also through H4 receptor activation.
The main bulk of ligand treatments in the present work were performed for 10 min. This was done based on time scale experiments where 10–20 min incubations resulted in a significant increase in phosphorylation compared to the baseline levels (Figure 4). The significant drop in phosphorylation at 1 min of ligand incubation must be due to the present of an unknown inhibitory second messenger signalling pathway. According to a recent publication (Rosethorne and Charlton, 2011) based on an H4 receptor expression system in a transfected cell line, histamine should increase the MAPK phosphorylation levels after between 1 and 5 min of incubation. The same article also showed an increase in MAPK phosphorylation through a β–arrestin-dependent pathway at 20–30 min. As the RBE4 cells did not exhibit the early 1 min histamine effects, and to minimize the risk of a β–arrestin-dependent effect, the time window for ligand treatment in this work was chosen to be 10 min.
In the present study we showed that histamine and immepip (an H3 and H4 receptor agonist) can activate the Erk1/2 MAPK pathway both in cultured brain microvessel endothelial cells and in vivo in brain blood vessels through H4 receptor activation. The H4 receptor-specific inverse agonist/antagonist JNJ 7777120 dose-dependently inhibited this effect, whereas the selective H3 receptor inverse agonist/antagonist ciproxifan did not block the immepip-induced effects. This led us to speculate that the activation seen is predominantly conveyed through H4 receptors. JNJ 7777120 appeared to inhibit completely the histamine-induced Erk1/2 MAPK phosphorylation indicating that other Erk1/2 MAPK-coupled receptors (e.g. the H1 receptor) are not involved. Interestingly, we showed that both the full-length Hrh3 (H3A receptor gene) and the H3C (the 144 bp deletion form) receptor gene isoform are expressed in cultured cells. Further studies are needed to determine if the lack of obvious H3 receptor-mediated effects is due to lack of functional endothelial receptors or if the endothelial H3 receptor might be un-coupled from the Erk1/2 MAPK pathway. In a recent paper by Gbahou et al. (2012), it was shown that one of the rat short H3 receptor isoforms (H3[413]; also known as H3B in Drutel et al., 2001) can act as an autoreceptor. This could indicate that H3 receptor-mediated effects and second messenger coupling can be cell-specific and differentially regulated, as the different histamine receptor isoforms seem to have a regulatory effect on each other. In brain blood vessels, it is possible that the H3C receptor has a regulatory effect on the availability of the full-length receptor cell membrane. The two Hrh4 transcripts, shown in this study to be expressed in endothelial cells, resemble the human full-length receptor and the truncated human H4 receptor, H4(67) (van Rijn et al., 2008). It has been shown that the H4(67) isoform does not bind histamine, but it has a dominant negative effect on the surface expression of the full-length H4 receptor (van Rijn et al., 2008). Future studies are needed to establish whether this receptor isoform is present in rat endothelial cells and regulates the expression of histamine receptors on the endothelial surface.
H4 receptors have recently also been shown to be functionally expressed in the CNS (Connelly et al., 2009; Strakhova et al., 2009), indicating that the receptor could have a direct synaptic function that is poorly understood. However, the presence of H4 receptor mRNA in significant neuronal populations in the brain has not yet been demonstrated, and the specificity of the antibodies needs be verified by mRNA colocalization studies. We propose that the vascular endothelium is a major site of H4 receptor expression in the brain and this receptor has the potential to be an important factor, which may affect vascular permeability in the brain.
Acknowledgments
We thank Johnson & Johnson Pharmaceutical Research & Development, L.L.C. (San Diego, CA, U.S.A.) for JNJ 7777120. The study was supported by the Academy of Finland, the Sigrid Juselius Foundation, Magnus Ehrnrooths Foundation, Swedish Cultural Foundation in Finland, and EU COST BM806.
Glossary
- CDS
coding sequence
- EAE
experimental allergic encephalomyelitis
- Hrh3
rat H3 receptor gene
- Hrh4
rat H4 receptor gene
- KO
knock-out
- pFA
paraformaldehyde
- RT
room temperature
Conflict of interest
P. P. received payments for lectures from Abbott Laboratories. The other authors declare no conflicts of interest.
References
- Akdis CA, Simons FE. Histamine receptors are hot in immunopharmacology. Eur J Pharmacol. 2006;533:69–76. doi: 10.1016/j.ejphar.2005.12.044. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Mathie A, Peter JA. Guide to Receptors and Channels (GRAC), 5th edition. Br J Pharmacol. 2011;164(Suppl. S1):S1–S324. doi: 10.1111/j.1476-5381.2011.01649_1.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland KF, Williams TJ, Conroy DM. Histamine induces cytoskeletal changes in human eosinophils via the H(4) receptor. Br J Pharmacol. 2003;140:1117–1127. doi: 10.1038/sj.bjp.0705530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly WM, Shenton FC, Lethbridge N, Leurs R, Waldvogel HJ, Faull RL, et al. The histamine H4 receptor is functionally expressed on neurons in the mammalian CNS. Br J Pharmacol. 2009;157:55–63. doi: 10.1111/j.1476-5381.2009.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drutel G, Peitsaro N, Karlstedt K, Wieland K, Smit MJ, Timmerman H, et al. Identification of rat H3 receptor isoforms with different brain expression and signaling properties. Mol Pharmacol. 2001;59:1–8. [PubMed] [Google Scholar]
- Durieu-Trautmann O, Federici C, Creminon C, Foignant-Chaverot N, Roux F, Claire M, et al. Nitric oxide and endothelin secretion by brain microvessel endothelial cells: regulation by cyclic nucleotides. J Cell Physiol. 1993;155:104–111. doi: 10.1002/jcp.1041550114. [DOI] [PubMed] [Google Scholar]
- Gbahou F, Rouleau A, Arrang JM. The histamine autoreceptor is a short isoform of the H(3) receptor. Br J Pharmacol. 2012;166:1860–1871. doi: 10.1111/j.1476-5381.2012.01913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goverman J. Autoimmune T cell responses in the central nervous system. Nat Rev Immunol. 2009;9:393–407. doi: 10.1038/nri2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammas P, Martinez J, Miller B. Cerebral microvascular endothelium and the pathogenesis of neurodegenerative diseases. Expert Rev Mol Med. 2011;13:e19. doi: 10.1017/S1462399411001918. [DOI] [PubMed] [Google Scholar]
- Gschwandtner M, Rossbach K, Dijkstra D, Baumer W, Kietzmann M, Stark H, et al. Murine and human Langerhans cells express a functional histamine H4 receptor: modulation of cell migration and function. Allergy. 2010;65:840–849. doi: 10.1111/j.1398-9995.2009.02279.x. [DOI] [PubMed] [Google Scholar]
- Haas H, Panula P. The role of histamine and the tuberomamillary nucleus in the nervous system. Nat Rev Neurosci. 2003;4:121–130. doi: 10.1038/nrn1034. [DOI] [PubMed] [Google Scholar]
- Karlstedt K, Sallmen T, Eriksson KS, Lintunen M, Couraud PO, Joo F, et al. Lack of histamine synthesis and down-regulation of H1 and H2 receptor mRNA levels by dexamethasone in cerebral endothelial cells. J Cereb Blood Flow Metab. 1999;19:321–330. doi: 10.1097/00004647-199903000-00010. [DOI] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. NC3Rs Reporting Guidelines Working Group. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P, Ngo K, Nguyen S, Thurmond RL, Edwards JP, Karlsson L, et al. Histamine H4 receptor mediates eosinophil chemotaxis with cell shape change and adhesion molecule upregulation. Br J Pharmacol. 2004;142:161–171. doi: 10.1038/sj.bjp.0705729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Drummond G, McLachlan E, Kilkenny C, Wainwright C. Guidelines for reporting experiments involving animals: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1573–1576. doi: 10.1111/j.1476-5381.2010.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passani MB, Ballerini C. Histamine and neuroinflammation: insights from murine experimental autoimmune encephalomyelitis. Front Syst Neurosci. 2012;6:32. doi: 10.3389/fnsys.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn RM, van Marle A, Chazot PL, Langemeijer E, Qin Y, Shenton FC, et al. Cloning and characterization of dominant negative splice variants of the human histamine H4 receptor. Biochem J. 2008;414:121–131. doi: 10.1042/BJ20071583. [DOI] [PubMed] [Google Scholar]
- del Rio R, Noubade R, Saligrama N, Wall EH, Krementsov DN, Poynter ME, et al. Histamine H4 receptor optimizes T regulatory cell frequency and facilitates anti-inflammatory responses within the central nervous system. J Immunol. 2012;188:541–547. doi: 10.4049/jimmunol.1101498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosethorne EM, Charlton SJ. Agonist-biased signaling at the histamine H4 receptor: JNJ7777120 recruits beta-arrestin without activating G proteins. Mol Pharmacol. 2011;79:749–757. doi: 10.1124/mol.110.068395. [DOI] [PubMed] [Google Scholar]
- Roux F, Durieu-Trautmann O, Chaverot N, Claire M, Mailly P, Bourre JM, et al. Regulation of gamma-glutamyl transpeptidase and alkaline phosphatase activities in immortalized rat brain microvessel endothelial cells. J Cell Physiol. 1994;159:101–113. doi: 10.1002/jcp.1041590114. [DOI] [PubMed] [Google Scholar]
- Saligrama N, Noubade R, Case LK, del Rio R, Teuscher C. Combinatorial roles for histamine H1-H2 and H3-H4 receptors in autoimmune inflammatory disease of the central nervous system. Eur J Immunol. 2012;42:1536–1546. doi: 10.1002/eji.201141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakhova MI, Nikkel AL, Manelli AM, Hsieh GC, Esbenshade TA, Brioni JD, et al. Localization of histamine H4 receptors in the central nervous system of human and rat. Brain Res. 2009;1250:41–48. doi: 10.1016/j.brainres.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Teuscher C, Subramanian M, Noubade R, Gao JF, Offner H, Zachary JF, et al. Central histamine H3 receptor signaling negatively regulates susceptibility to autoimmune inflammatory disease of the CNS. Proc Natl Acad Sci U S A. 2007;104:10146–10151. doi: 10.1073/pnas.0702291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Pan W, Stone KP, Zhang Y, Hsuchou H, Kastin AJ. Expression and signaling of novel IL15Ralpha splicing variants in cerebral endothelial cells of the blood-brain barrier. J Neurochem. 2010;114:122–129. doi: 10.1111/j.1471-4159.2010.06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Fawcett JP, Ostergaard J, Zhang H, Tucker IG. Mechanistic studies of the effect of bile salts on rhodamine 123 uptake into RBE4 cells. Mol Pharm. 2012;9:29–36. doi: 10.1021/mp200201y. [DOI] [PubMed] [Google Scholar]
- Zampeli E, Tiligada E. The role of histamine H4 receptor in immune and inflammatory disorders. Br J Pharmacol. 2009;157:24–33. doi: 10.1111/j.1476-5381.2009.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


