Abstract
Salmonella enterica subspecies enterica serovar Typhi is a rod-shaped, Gram-negative, facultatively anaerobic bacterium. It belongs to the family Enterobacteriaceae in the class Gammaproteobacteria, and has the capability of residing in the human gallbladder by forming a biofilm and hence causing the person to become a typhoid carrier. Here we present the complete genome of Salmonella enterica subspecies enterica serotype Typhi strain P-stx-12, which was isolated from a chronic carrier in Varanasi, India. The complete genome comprises a 4,768,352 bp chromosome with a total of 98 RNA genes, 4,691 protein-coding genes and a 181,431 bp plasmid. Genome analysis revealed that the organism is closely related to Salmonella enterica serovar Typhi strain Ty2 and Salmonella enterica serovar Typhi strain CT18, although their genome structure is slightly different.
Keywords: Enterobacteriaceae, Salmonella, Typhi, Gram-negative, host-specific, pathogen, Typhoid Fever
Introduction
Salmonella enterica serovar Typhi is a particular Salmonella serovar that causes typhoid fever [1-3]. There are an estimated 20 million cases of typhoid fever and 200,000 deaths from this disease reported each year, worldwide [4,5]. S. enterica serovar Typhi belongs to the family Enterobacteriaceae. All Enterobacteriaceae ferment glucose, reduce nitrates, and are oxidatively negative [6]. In general, S. enterica serovar Typhi is motile, produces minimal H2S, and is resistant to bile acids [7]. S. enterica serovar Typhi has three types of antigens [3], namely the H antigen for motility, specific O antigen for synthesizing lipopolysaccharides and biofilm formation, and Vi antigen which is a capsular polysaccharide that acts as a major virulence factor. This Vi antigen is only specific for S. enterica serovar Typhi and is found in Salmonella Pathogenicity Island-7 [8]. In 2003, comparative genomics of S. enterica serovar Typhi strains Ty2 and CT18 was carried out by Deng et al. [9]. In that study, a half-genome interreplichore inversion in Ty2 relative to CT18 was discovered. It was reported that S. enterica serovar Typhi Ty2 does not harbor any plasmid and hence it is susceptible to antibiotics. On the other hand, S. enterica serovar Typhi CT18 carries two plasmids with one conferring multidrug resistance. We published the complete genome sequence of S. enterica serovar Typhi P-stx-12 earlier last year [10]. This sequencing project helps us to better understand the genome organization and the contribution of the virulence machinery in this pathogen. Here we present a summary of S. enterica serovar Typhi P-stx-12 and its unique features, together with the description of the complete genomic sequencing and annotation.
Classification and features
S. enterica serovar Typhi P-stx-12 was isolated from a typhoid carrier in Northern India, Uttar Pradesh, Varanasi in 2009. This serotype is known to inhabit the Peyer’s patches (lymph node) of the small intestine, liver, spleen, bone marrow, bile, and blood stream of infected humans.
Cells of S. enterica serovar Typhi P-stx-12 were Gram-negative, motile, rod-shaped, and non-spore forming. This strain grew at an optimum temperature of 35°C-37°C, but could tolerate temperatures between 7°C and 45°C. Strain P-stx-12 is a facultative anaerobe and utilizes glucose as the main carbon source. The pure isolate did not produce cytochrome oxidase but was able to reduce nitrate and break down glucose by pathways for oxidation and fermentation. This strain did not produce urease. In Triple Sugar Iron medium, there was an alkaline/acid reaction with a very small amount of H2S production. Indole was not produced in peptone water. The strain was able to ferment glucose and mannitol without production of gas; however lactose and sucrose were not fermented. The strain could be agglutinated by poly O, poly H, factors O9, H-d, and Vi antisera (data not shown).
Figure 1 shows the phylogenetic neighborhood of S. enterica serovar Typhi P-stx-12 in a 16S rRNA based tree. There were seven 16S rRNA gene copies in the genome of S. enterica serovar Typhi P-stx-12. Two out of the seven copies differed from the rest by having a single base substitution (G to A). Thus, the common gene copy was used for tree building. In relation to others in the genus Salmonella, strain P-stx-12 is closely related to S. enterica serovar Typhi strain Ty2 and S. enterica serovar Typhi strain CT18. The classification and features of this organism are summarized in Table 1.
Figure 1.
Phylogenetic tree highlighting the position of Salmonella enterica serovar Typhi strain P-stx-12 relative to other strains within the Enterobacteriaceae. Strains shown are those within the Enterobacteriaceae having corresponding GenBank accession numbers. The phylogenetic tree was constructed using Ribosomal Database Project [11] tree builder that utilizes the Weighbor weighted neighbor-joining tree building algorithm [12]. The bootstrap value was 100. Escherichia coli strain Z83205 was used as an outgroup.
Table 1. Classification and general features of S. enterica serovar Typhi P-stx-12.
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [13] | |
| Phylum Proteobacteria | TAS [14] | ||
| Class Gammaproteobacteria | TAS [15,16] | ||
| Order Enterobacteriales | TAS [17] | ||
| Family Enterobacteriaceae | TAS [18-20] | ||
| Genus Salmonella | TAS [18,21-23] | ||
| Species Salmonella enterica | TAS [23,24] | ||
| Subspecies Salmonella enterica enterica | TAS [23,24] | ||
| Gram stain | negative | TAS [6] | |
| Cell shape | Rod-shape | TAS [6] | |
| Motility | Motile | TAS [6] | |
| Sporulation | Non-spore forming | TAS [6] | |
| Temperature range | 7oC-45oC | TAS [6] | |
| Optimum temperature | 35oC-37oC | TAS [6] | |
| Carbon source | Carbohydrates (glucose) | TAS [6] | |
| Energy source | Chemoorganotrophic | TAS [6] | |
| Terminal electron receptor | Not reported | ||
| MIGS-6 | Habitat | Multi-organ pathogen that inhabits the Peyer’s patches (Lymph node) of the small intestine, liver, spleen, bone marrow, bile and blood stream of infected human. |
NAS |
| MIGS-6.3 | Salinity | Survives for days at 0.85% NaCl. | NAS |
| MIGS-22 | Oxygen | Facultative anaerobe | TAS [6] |
| MIGS-15 | Biotic relationship | Human restricted | NAS |
| MIGS-14 | Pathogenicity | Pathogenic | TAS [25,26] |
| MIGS-4 | Geographic location | Uttar Pradesh, Varanasi, India | IDA |
| MIGS-5 | Sample collection time | November 2009 | IDA |
| MIGS-4.1 | Latitude | 25° 19' 60 N | IDA |
| MIGS-4.2 | Longitude | 83° 0' 0 E | IDA |
| MIGS-4.3 | Depth | Not reported | |
| MIGS-4.4 | Altitude | 76 (meters) | IDA |
a) Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [27]. If the evidence code is IDA, then the property was directly observed for a living isolate by one of the authors or an expert mentioned in the acknowledgements.
Genome sequencing and annotation
Genome project history
S. enterica serovar Typhi P-stx-12 was selected for sequencing because it was isolated from a typhoid carrier in India, where there is a high rate of typhoid fever cases. This isolate was obtained from a 32-year old male who had been showing persistent high titers for Widal test and Vi antibody for more than one year. DNA isolation was carried out at Banaras Hindu University. This genome sequence was first published in April 2013 [10]. A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | One 454 paired-end library (4-kb insert size), one Illumina library |
| MIGS-29 | Sequencing platforms | Illumina GA IIx, 454 GS FLX Titanium |
| MIGS-31.2 | Fold coverage | 100× Illumina, 18× pyrosequencing |
| MIGS-30 | Assemblers | Newbler, Burrows-Wheeler Alignment |
| MIGS-32 | Gene calling method | Glimmer, RNAmmer, tRNAscan-SE |
| Genbank ID | CP003278 (Chromosome) CP003279 (Plasmid) |
|
| Genbank Date of Release | February 1, 2012 | |
| Project relevance | Biotechnology, Pathway, Pathogenic |
Growth conditions and DNA isolation
The stool specimen of strain P-stx-12 was collected from a known chronic typhoid carrier patient. For the isolation of the bacterium, 5 gm of freshly passed unpreserved stool was sieved through a gauze piece to remove the coarse particles. The filtrate was centrifuged at 4,000 rpm for 5 min. The pellet was washed twice with Phosphate Buffered Saline, pH 7.2 and suspended in selenite F broth (50 ml) for enrichment with some modified technique (under process of patenting). After overnight incubation, the broth was examined for turbidity and subcultured on deoxycholate citrate agar and MacConkey agar.
Extraction of genomic DNA was carried out using a Phenol-Chloroform and Proteinase K method with some modification [28]. The DNA preparation was checked by PCR amplification of the flagellin (fliC) gene of S. enterica serovar Typhi [29,30] and 16S rRNA gene [31].
Genome sequencing and assembly
Whole-genome sequencing was performed with a combined strategy of 454 and Illumina sequencing technologies. A 4-kb paired-end library was constructed according to the manufacturer’s instructions (454). A total of 242,499 reads were generated using the GS FLX Titanium system, giving ~18× coverage of the genome. Initial assembly of 97.09% of the reads using the Newbler assembler (Roche) resulted in ~200 large contigs within 11 scaffolds. A total of ~500 Mb of 3-kb mate-pair sequencing data were generated to reach a depth of 100× coverage with an Illumina GA IIx. These sequences were mapped to the scaffolds using the Burrows-Wheeler Alignment (BWA) tool [32]. A majority of the gaps within the scaffolds were filled by local assembly of 454 and Illumina reads. The remaining gaps were filled by sequencing the PCR products of the gaps using an ABI 3730xl capillary sequencer. The putative sequencing errors were verified by the coverage of 454 and Illumina reads.
Genome annotation
Annotation of the S. enterica serovar Typhi P-stx-12 genome was done using a combination of ISGA (Integrative Services for Genomic Analysis) [33] and the DIYA (Do-It-Yourself Annotator) pipeline [34], which comprises of Glimmer [35], tRNAscan-SE [36], RNAmmer [37], BLAST [38], and Asgard [39]. RPS-BLAST searches against the Clusters of Orthologous Groups (COG) database enabled assignment of COG functional categories to the ORFs. CLC Genomics Workbench was used to further improve and check the annotation results. Frameshifts and partial gene fragments that indicate potential pseudogenes were identified by the NCBI Submission Check tool and manually verified. Protein coding genes were searched against the NCBI RefSeq database using BLASTP [40]. Clustered Regularly Interspersed Short Palindromic Repeats (CRISPR) regions were identified using the CRISPR Finder program [41]. PHAST (PHAge Search Tool) [42] was used to search for prophage sequences within the genome. Potential genomic islands were identified using the IslandViewer web server [43]. Comparison between different S. enterica serovar Typhi strains was done using progressiveMauve [44].
Genome properties
The complete genome of S. enterica serovar Typhi P-stx-12 contains a single circular chromosome of 4,768,352 bp with a GC content of 52.1%, and a circular plasmid of 181,431 bp with a GC content of 46.4% (Figure 2 and Figure 3). The chromosome consists of 4,885 predicted genes, of which there are 4,691 protein-coding genes, 22 rRNA genes, and 76 tRNA genes. Specific COGs were assigned to 75.34% of the genes in the chromosome, and 25% of these genes were also assigned with enzyme classification numbers which were involved in 268 metabolic pathways. The properties and statistics of the genome are summarized in Tables 3 and 4. The plasmid harbors 234 protein-coding genes, with 187 annotated as hypothetical proteins with unknown function. The remaining genes were grouped into specific COGs, the majority of which fell into the category of information storage and processing with respect to replication, recombination and repair.
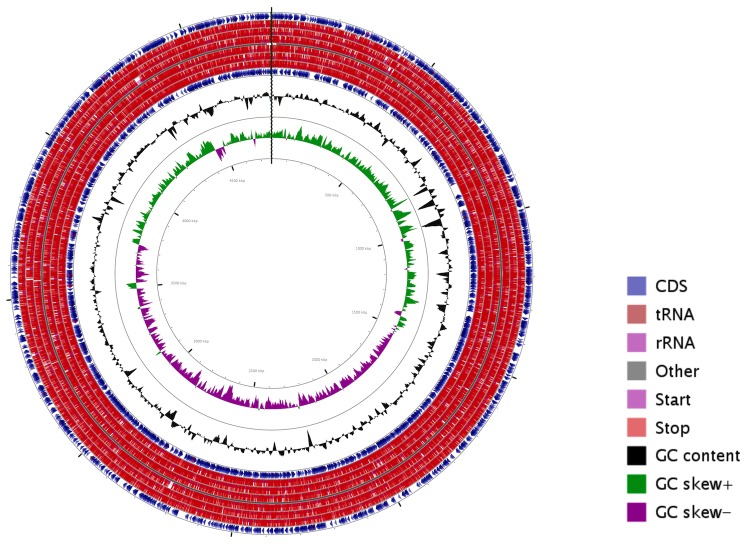
Figure 2.
Circular map of the Salmonella enterica serovar Typhi P-stx-12 chromosome. From the inside to outside, the first and second circles show GC skew and G+C content respectively. The third circle shows the CDS, tRNA and rRNA in the reverse strand; the fourth circle shows the CDS, tRNA, rRNA in the forward strand. This figure was generated by CGView [45].
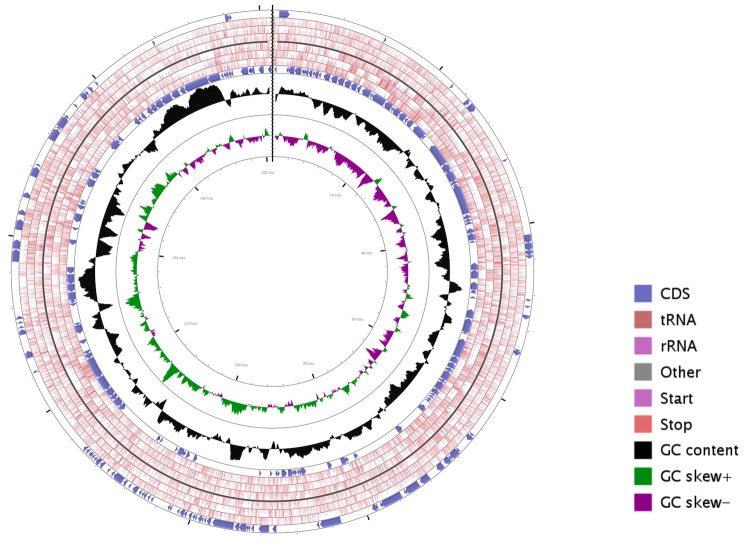
Figure 3.
Circular map of the Salmonella enterica serovar Typhi P-stx-12 plasmid. From the inside to outside, the first and second circles show GC skew and G+C content respectively. The third circle shows the CDS, tRNA and rRNA in the reverse strand; the fourth circle shows the CDS, tRNA, rRNA in the forward strand. This figure was generated by CGView [45].
Table 3. Genome statistics.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 4,768,352 | 100.00 |
| DNA coding region (bp) | 4,018,014 | 84.26 |
| DNA G+C content (bp) | 2,484,311 | 52.10 |
| Total genesb | 4,885 | 100.00 |
| RNA genes | 98 | 2.00 |
| Protein-coding genes | 4,691 | 96.03 |
| Pseudogenes | 96 | 1.97 |
| Genes in paralog clusters | 623 | 13.28 |
| Genes assigned to COGs | 3,534 | 75.34 |
| Genes with signal peptides | 388 | 8.27 |
| Genes with transmembrane helices | 1,096 | 23.36 |
| CRISPR repeat | 1 |
a) The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
b) Also includes 96 pseudogenes
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 182 | 3.9 | Translation |
| A | 1 | 0.0 | RNA processing and modification |
| K | 307 | 6.5 | Transcription |
| L | 191 | 4.1 | Replication, recombination and repair |
| B | 0 | 0.0 | Chromatin structure and dynamics |
| D | 33 | 0.7 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 48 | 1.0 | Defense mechanisms |
| T | 181 | 3.9 | Signal transduction mechanisms |
| M | 246 | 5.2 | Cell wall/membrane biogenesis |
| N | 122 | 2.6 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 1 | 0.0 | Extracellular structures |
| U | 139 | 3.0 | Intracellular trafficking and secretion |
| O | 156 | 3.3 | Posttranslational modification, protein turnover, chaperones |
| C | 262 | 5.6 | Energy production and conversion |
| G | 352 | 7.5 | Carbohydrate transport and metabolism |
| E | 349 | 7.4 | Amino acid transport and metabolism |
| F | 89 | 1.9 | Nucleotide transport and metabolism |
| H | 176 | 3.8 | Coenzyme transport and metabolism |
| I | 85 | 1.8 | Lipid transport and metabolism |
| P | 193 | 4.1 | Inorganic ion transport and metabolism |
| Q | 65 | 1.4 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 421 | 9.0 | General function prediction only |
| S | 355 | 0.1 | Function unknown |
| - | 1157 | 24.7 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Paralog clusters
In order to identify paralog families, BLASTP was used to calculate all possible protein homologs in the S. enterica serovar Typhi P-stx-12 genome. Homologs that had at least 30% shared amino acid similarity were selected. Paralog pairs were imported into the S. enterica serovar Typhi P-stx-12 database in Pathway Studio as a new type of interaction called “Paralog” [46]. Protein functional families were identified as clusters in the global Paralog network using the direct force layout algorithm. The biological function was assigned to each paralog cluster based on the functional annotation of the protein (Figure 4). The major paralog clusters identified include ATPase components that are mainly involved in transport systems, transcriptional regulator, transcriptional repressor, transposases, major facilitator superfamily permeases, response-regulator containing CheY-like receiver domain and an HTH DNA binding domain, P-pilus assembly proteins, multidrug efflux system proteins, and fimbrial-like adhesins.
Figure 4.
Paralog network of functional families in the S. enterica serovar Typhi P-stx-12 genome.
Insights into the genome
Comparisons with other fully sequenced S. enterica serovar Typhi genomes
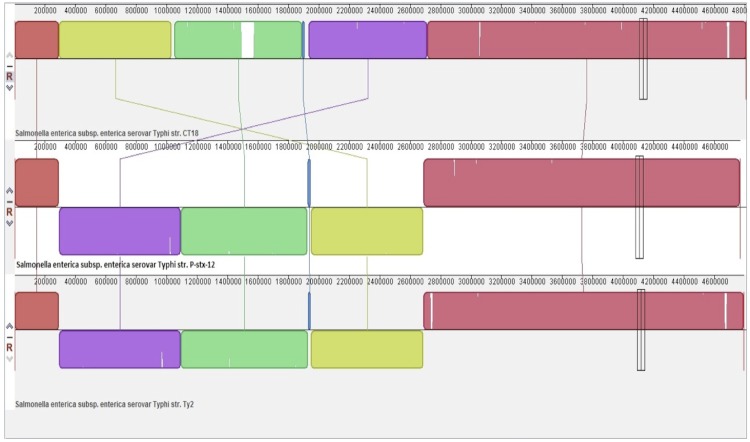
The genome of S. enterica serovar Typhi P-stx-12 was compared with the other two published S. enterica serovar Typhi genomes, CT18 (isolated from Vietnam) and Ty2 (isolated from Russia). Comparison between these three genomes revealed that the coding genes of S. enterica serovar Typhi P-stx-12 were 84% similar to those of CT18 [47] and Ty2 [9]. The genome organization of these three strains is shown in Figure 5. The location of the genes in strains P-stx-12 and Ty2 are identical. Both have three blocks of genes that are inverted from strain CT18. Our observations are in agreement with the work of Deng et al. [9], where they discovered that half of the Ty2 genome was inverted relative to the CT18 genome. Nevertheless, most of the genes have the same function, indicating that these are the possible housekeeping genes which maintain the survival of this pathogen. Besides that, this P-stx-12 strain has one plasmid which shares 169 orthologous CDSs with pHCM1, the plasmid belonging to CT18 (Genbank accession number AL513383). pHCM1 is a conjugative plasmid which encodes resistance to antimicrobial agents and heavy metals; similar to IncHI plasmid R27. This further supports the hypothesis that the presence of a plasmid signifies a dynamic link between resistance and pathogenicity. Indeed, it was reported that the stable maintenance of IncHI1 plasmids in S. enterica serovar Typhi occurred throughout the development of antibiotic resistance in S. enterica serovar Typhi [48]. It is worth noting that the plasmid of P-stx-12 carries genes encoding the tetracycline resistance protein and tetracycline repressor protein TetR, possibly conferring drug resistance to this strain. This resistance protein is also found in strain CT18. On the other hand, the number of pseudogenes in this genome appears to be only 96, which is less than those in S. enterica serovar Typhi CT18 and S. enterica serovar Typhi Ty2 (> 200).
Figure 5.
Alignment of the S. enterica serovar Typhi CT18, S. enterica serovar Typhi P-stx-12, and S. enterica serovar Typhi Ty2 genomes using progressive Mauve [44]. Colored blocks in the first genome are connected by lines to similar colored blocks in the second and third genomes. Inverted regions in S. enterica serovar Typhi P-stx-12 and S. enterica serovar Typhi Ty2 are presented as blocks below the center line of the genome. Lines indicate regions in each genome that are homologous.
Genomic Islands (GIs) and Salmonella Pathogenicity Island (SPIs)
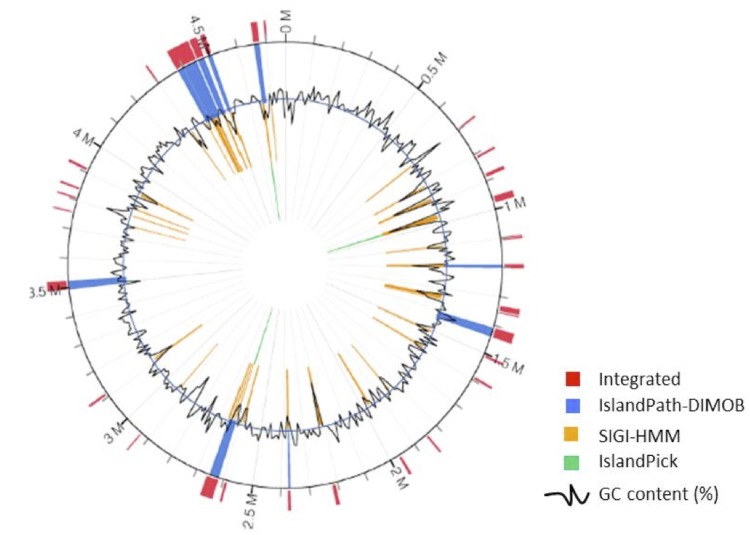
There are 31 possible genomic islands (GIs) as predicted by IslandViewer (Figure 6). Analysis of these GIs revealed that most of the genes within the islands encode for hypothetical proteins. Eight Salmonella Pathogenicity Islands (SPI-11, SPI-2, SPI-16, SPI-6, SPI-8, SPI-4, SPI-7 and SPI-10) were found to be embedded in these GIs, whereas the rest of the SPIs spanned between the GIs. Interestingly, the proteins found in SPI-8 are located next to the proteins of SPI-13, which is not classified as one of the predicted GIs. Three GIs within the coordinate 4,376,723 to 4,508,803 make up the total region for SPI-7.
Figure 6.
Genomic islands as predicted using IslandViewer. Predicted genomic islands are colored within the circular image based on the tool IslandPath-DIMOB, SIGI-HMM, IslandPick, and an integration of the three tools.
A comparison between the SPIs found in strains CT18 and P-stx-12 revealed that the location of several SPIs in both genomes is different (Figure 7). Indeed, the orientation for SPI-6, SPI-16, SPI-5, SPI-18, SPI-2, SPI-11, SPI-12, and SPI-17 was inverted in both genomes. These SPIs fall within the inverted genomic regions shown in Figure 5.
Figure 7.
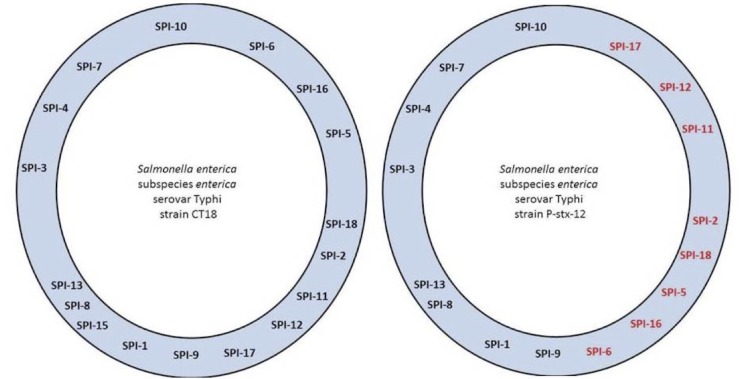
Distribution of SPIs in S. enterica serovar Typhi CT18 and S. enterica serovar Typhi P-stx-12.
Prophage Regions
Prophage are one of the diverse mobile genetic elements that are acquired through horizontal gene transfer. These prophage genes are involved in lysogenic conversion. PHAST (PHAge Search Tool) was used to identify the prophage regions of S. enterica serovar Typhi P-stx-12. Based on the analysis, five predicted prophage regions (three intact, two partial) were identified in the genome. The three intact prophage regions have the size of 44.2 kb, 50.8kb, and 68.2 kb, respectively. These regions consist of a total of 165 coding sequences for the phages phage_Gifsy_2 and Enterobacteria_phage_Fels2. In comparison, S. enterica serovar Typhi CT18 and S. enterica serovar Typhi Ty2 each have eight predicted prophage regions. Out of the eight regions, only four intact regions (247 proteins) were found in S. enterica serovar Typhi CT18 whereas three intact regions (170 proteins) were found in S. enterica serovar Typhi Ty2. The phage regions of S. enterica serovar Typhi P-stx-12 are the same types as those found in S. enterica serovar Typhi Ty2, while S. enterica serovar Typhi CT18 carries an additional phage region of the Enterobacteria_phage_SfV type. A summary of the prophage regions in each genome is shown in Table 5.
Table 5. Prophage regions identified in S. enterica serovar Typhi P-stx-12, S. enterica serovar Typhi CT18, and S. enterica serovar Typhi Ty2.
| Organism | Region length | # CDS | Region Position | Phage | % GC |
|---|---|---|---|---|---|
| S. enterica serovar Typhi P-stx-12 | 44.2 kb | 59 | 1924908-1969179 | Phage_Gifsy_2 | 50.35 |
| 50.8 kb | 49 | 3478537-3529349 | Enterobacteria_phage_Fels2 | 52.21 | |
| 68.2 kb | 57 | 4424417-4492645 | Enterobacteria_phage_Fels2 | 50.07 | |
| S. enterica serovar Typhi CT18 | 44.3 kb | 64 | 1008698-1053060 | Phage_Gifsy_2 | 50.19 |
| 59.7 kb | 74 | 1879760-1939495 | Enterobacteria_phage_SfV | 48.12 | |
| 50.8 kb | 49 | 3504242-3555052 | Enterobacteria_phage_Fels2 | 52.21 | |
| 48.1 kb | 60 | 4459144-4507270 | Enterobacteria_phage_Fels2 | 51.15 | |
| S. enterica serovar Typhi Ty2 | 44.2 kb | 63 | 1928058-1972330 | Phage_Gifsy_2 | 50.35 |
| 50.8 kb | 48 | 3489900-3540712 | Enterobacteria_phage_Fels2 | 52.20 | |
| 45.1 kb | 59 | 4446021-4491188 | Enterobacteria_phage_Fels2 | 51.36 |
CRISPR Region
By using the CRISPR Finder tool, one CRISPR repeat region with a length of 394 bp was identified in the S. enterica serovar Typhi P-stx-12 genome. The CRISPR region starts at the position 2,900,675 and ends at the position 2,901,069 with 6 spacers in between. The confirmed CRISPR has the following direct repeat consensus sequence: CGGTTTATCCCCGCTGGCGCGGGGAACAC. Strains CT18 and Ty2 also have a single CRISPR repeat region with the lengths of 385 bp and 394 bp, respectively. The location for the CRISPR region of all three strains falls within the region of 2.9 Mbp on the chromosome. All the strains have 6 spacers and share the common direct repeat consensus sequence. It is worth noting that the CRISPR region, including the length and the spacer sequence, of S. enterica serovar Typhi P-stx-12 is exactly identical to S. enterica serovar Typhi Ty2. It suggests a strong evidence of their evolutionary relevance and shows that the CRISPR region in S. enterica serovar Typhi is conserved. As CRISPRs function as a prokaryotic immune system and confer resistance towards plasmids and phages (thus interfering with the spread of antibiotic resistance and virulence factors), it is reasonable to find only one CRISPR with very few spacers in this pathogen as compared to other bacterial strains that are not pathogenic [49].
Acknowledgement
This work was supported by APEX funding (Malaysia Ministry of Higher Education) to the Centre for Chemical Biology, Universiti Sains Malaysia; and the contributions of the Council of Scientific and Industrial Research (CSIR), New Delhi, India.
Abbreviations:
- NCBI- National Center for Biotechnology Information
RDP- Ribosomal Database Project
References
- 1.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med 2002; 347:1770-1782 10.1056/NEJMra020201 [DOI] [PubMed] [Google Scholar]
- 2.Hornick RB, Greisman SE, Woodward TE, DuPont HL, Dawkins AT, Snyder MJ. Typhoid fever: pathogenesis and immunologic control. N Engl J Med 1970; 283:686-691 10.1056/NEJM197009242831306 [DOI] [PubMed] [Google Scholar]
- 3.Baker S, Dougan G. The genome of Salmonella enterica serovar Typhi. Clin Infect Dis 2007; 45(Suppl 1):S29-S33 10.1086/518143 [DOI] [PubMed] [Google Scholar]
- 4.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ 2004; 82:346-353 [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang XL, Jeza VT, Pan Q. Salmonella typhi: from a human pathogen to a vaccine vector. Cell Mol Immunol 2008; 5:91-97 10.1038/cmi.2008.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holt JG, Kreig NR, Sneath PHA, Staley JT, Williams ST. Bergey's manual of determinative bacteriology. Baltimore: Williams & Wilkins; 1994. [Google Scholar]
- 7.Crawford RW, Gibson DL, Kay WW, Gunn JS. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immun 2008; 76:5341-5349 10.1128/IAI.00786-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seth-Smith HM. SPI-7: Salmonella's Vi-encoding Pathogenicity Island. J Infect Dev Ctries 2008; 2:267-271 10.3855/jidc.220 [DOI] [PubMed] [Google Scholar]
- 9.Deng W, Liou SR, Plunkett G, III, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J Bacteriol 2003; 185:2330-2337 10.1128/JB.185.7.2330-2337.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong SY, Pratap CB, Wan X, Hou S, Abdul Rahman AY, Saito JA, Nath G, Alam M. Complete genome sequence of Salmonella enterica subsp. enterica serovar Typhi P-stx-12. J Bacteriol 2012; 194:2115-2116 10.1128/JB.00121-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009; 37(Database issue):D141-D145 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruno WJ, Socci ND, Halpern AL. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol Evol 2000; 17:189-197 10.1093/oxfordjournals.molbev.a026231 [DOI] [PubMed] [Google Scholar]
- 13.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. Second Edition ed. Volume 2, Part B. New York: Springer; 2005. p 1. [Google Scholar]
- 15.Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235-2238 10.1099/ijs.0.64108-0 [DOI] [PubMed] [Google Scholar]
- 16.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. Second Edition ed. Volume 2, Part B. New York: Springer; 2005. p 1. [Google Scholar]
- 17.Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey's Manual of Systematic Bacteriology. Second Edition ed. Volume 1. New York: Springer; 2001. p 155-166. [Google Scholar]
- 18.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 19.Rahn O. New principles for the classification of bacteria. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg 1937; 96:273-286 [Google Scholar]
- 20.Judicial Commission of the International Committee on Bacteriological Nomenclature Conservation of the family name Enterobacteriaceae, of the name of the type genus, and designation of the type species. Opinion No. 15. Int Bull Bacteriol Nomencl Taxon 1958; 8:73-74 [Google Scholar]
- 21.Lignieres J. Maladies du porc. Bulletin of the Society for Central Medical Veterinarians 1900; 18:389-431 [Google Scholar]
- 22.Le Minor L, Rohde R. Genus IV. Salmonella Lignieres 1900, 389. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 298-318. [Google Scholar]
- 23.Judicial Commission of the International Committee on Systematics of Prokaryotes The type species of the genus Salmonella Lignieres 1900 is Salmonella enterica (ex Kauffmann and Edwards 1952) Le Minor and Popoff 1987, with the type strain LT2T, and conservation of the epithet enterica in Salmonella enterica over all earlier epithets that may be applied to this species. Opinion 80. Int J Syst Evol Microbiol 2005; 55:519-520 10.1099/ijs.0.63579-0 [DOI] [PubMed] [Google Scholar]
- 24.Le Minor L, Popoff MY. Request for an opinion. Designation of Salmonella enterica sp. nov., nom. rev., as the type and only species of the genus Salmonella. Int J Syst Bacteriol 1987; 37:465-468 10.1099/00207713-37-4-465 [DOI] [Google Scholar]
- 25.Pegues DA, Ohl ME, Miller SI. Salmonella species, including Salmonella Typhi. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and Practice of Infectious Disease. Philadelphia: Churchill-Livingstone; 2004. p 2636-2653. [Google Scholar]
- 26.WHO. Background document: the diagnosis, treatment and prevention of typhoid fever. http://www.who.int/vaccines-documents WHO; 2003.
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Third Edition. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 29.Song JH, Cho H, Park MY, Na DS, Moon HB, Pai CH. Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. J Clin Microbiol 1993; 31:1439-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frankel G. Detection of Salmonella typhi by PCR. J Clin Microbiol 1994; 32:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll NM, Jaeger EE, Choudhury S, Dunlop AA, Matheson MM, Adamson P, Okhravi N, Lightman S. Detection of and discrimination between Gram-positive and Gram-negative bacteria in intraocular samples by using nested PCR. J Clin Microbiol 2000; 38:1753-1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754-1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hemmerich C, Buechlein A, Podicheti R, Revanna KV, Dong Q. An Ergatis-based prokaryotic genome annotation web server. Bioinformatics 2010; 26:1122-1124 10.1093/bioinformatics/btq090 [DOI] [PubMed] [Google Scholar]
- 34.Stewart AC, Osborne B, Read TD. DIYA: a bacterial annotation pipeline for any genomics lab. Bioinformatics 2009; 25:962-963 10.1093/bioinformatics/btp097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007; 23:673-679 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410 [DOI] [PubMed] [Google Scholar]
- 39.Alves JM, Buck GA. Automated system for gene annotation and metabolic pathway reconstruction using general sequence databases. Chem Biodivers 2007; 4:2593-2602 10.1002/cbdv.200790212 [DOI] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 2007; 35(Web Server issue):W52-57. [DOI] [PMC free article] [PubMed]
- 42.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res 2011; 39(Web Server issue):W347-352. [DOI] [PMC free article] [PubMed]
- 43.Langille MG, Brinkman FS. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 2009; 25:664-665 10.1093/bioinformatics/btp030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010; 5:e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics 2005; 21:537-539 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- 46.Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio-the analysis and navigation of molecular networks. Bioinformatics 2003; 19:2155-2157 10.1093/bioinformatics/btg290 [DOI] [PubMed] [Google Scholar]
- 47.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 2001; 413:848-852 10.1038/35101607 [DOI] [PubMed] [Google Scholar]
- 48.Phan MD, Wain J. IncHI plasmids, a dynamic link between resistance and pathogenicity. J Infect Dev Ctries 2008; 2:272-278 [DOI] [PubMed] [Google Scholar]
- 49.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in Bacteria and Archaea. Nat Rev Genet 2010; 11:181-190 10.1038/nrg2749 [DOI] [PMC free article] [PubMed] [Google Scholar]