Abstract
Labrenzia alexandrii Biebl et al. 2007 is a marine member of the family Rhodobacteraceae in the order Rhodobacterales, which has thus far only partially been characterized at the genome level. The bacterium is of interest because it lives in close association with the toxic dinoflagellate Alexandrium lusitanicum. Ultrastructural analysis reveals R-bodies within the bacterial cells, which are primarily known from obligate endosymbionts that trigger “killing traits” in ciliates (Paramecium spp.). Genomic traits of L. alexandrii DFL-11T are in accordance with these findings, as they include the reb genes putatively involved in R-body synthesis. Analysis of the two extrachromosomal elements suggests a role in heavy-metal resistance and exopolysaccharide formation, respectively. The 5,461,856 bp long genome with its 5,071 protein-coding and 73 RNA genes consists of one chromosome and two plasmids, and has been sequenced in the context of the Marine Microbial Initiative.
Keywords: aerobe, motile, symbiosis, dinoflagellates, photoheterotroph, high-quality draft, Alexandrium lusitanicum, Alphaproteobacteria
Introduction
Strain DFL-11T (= DSM 17067 = NCIMB 14079) is the type strain of Labrenzia alexandrii, a marine member of the Rhodobacteraceae (Rhodobacterales, Alphaproteobacteria) [1]. Strain DFL-11T was isolated from single cells of a culture of the toxic dinoflagellate Alexandrium lusitanicum maintained at the Biological Research Institute of Helgoland, Germany [1]. L. alexandrii is the type species of the genus Labrenzia, which currently also harbors a couple of species (L. aggregata, L. alba and L. marina) that were previously classified in the genus Stappia [1]. Biebl et al. 2007 [1] did not provide a formal assignment of the genus Labrenzia to a family, but their phylogenetic analysis placed Labrenzia with high support within a clade also comprising Nesiotobacter, Pannonibacter, Pseudovibrio, Roseibium and Stappia, genera which at that time were either not formally assigned to a family or to Rhodobacteraceae [2]. Other analyses [3] indicate that the entire clade should not be placed within Rhodobacteraceae, but an alternative taxonomic arrangement has, to the best of our knowledge, not yet been published. Here we present a summary classification and a set of features for L. alexandrii DFL-11T including so far undiscovered aspects of its ultrastructure and physiology, together with the description of the high-quality permanent draft genome sequence and annotation.
This work is part of the Marine Microbial Initiative (MMI) which enabled the J. Craig Venter Institute (JCVI) to sequence the genomes of approximately 165 marine microbes with funding from the Gordon and Betty Moore Foundation. These microbes were contributed by collaborators worldwide, and represent an array of physiological diversity, including carbon fixation, photoautotrophy, photoheterotrophy, nitrification, and methanotrophy. The MMI was designed to complement other ongoing research at JCVI and elsewhere to characterize the microbial biodiversity of marine and terrestrial environments through metagenomic profiling of environmental samples.
Classification and features
16S rRNA analysis
A representative genomic 16S rRNA sequence of strain DFL-11T was compared using NCBI BLAST [4,5] using default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [6] and the relative frequencies of taxa and keywords (reduced to their stem [7]) were determined, weighted by BLAST scores. The most frequently occurring genera were Stappia (36.9%), Pannonibacter (19.6%), Pseudovibrio (18.8%), Labrenzia (10.8%) and Achromobacter (5.0%) (98 hits in total). Regarding the seven hits to sequences from other members of the genus, the average identity within HSPs was 97.3%, whereas the average coverage by HSPs was 96.4%. Among all other species, the one yielding the highest score was Stappia alba (AJ889010) (since 2007 reclassified as L. alba [1]), which corresponded to an identity of 98.2% and an HSP coverage of 99.9%. (Note that the Greengenes database uses the INSDC (= EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was AY701471 (Greengenes short name 'dinoflagellate symbiont clone GCDE08 W'), which showed an identity of 99.8% and an HSP coverage of 99.6%. The most frequently occurring keywords within the labels of all environmental samples which yielded hits were 'coral' (5.4%), 'microbi' (3.2%), 'marin' (3.0%), 'diseas' (2.8%) and 'healthi' (2.8%) (150 hits in total). The most frequently occurring keywords within the labels of those environmental samples which yielded hits of a higher score than the highest scoring species were 'coral' (11.1%), 'dinoflagel, symbiont' (5.7%), 'aquarium, caribbean, chang, dai, disease-induc, faveolata, kept, montastraea, plagu, white' (5.6%) and 'habitat, microbi, provid, threaten' (5.5%) (4 hits in total). These terms partially correspond with the known ecology of L. alexandrii.
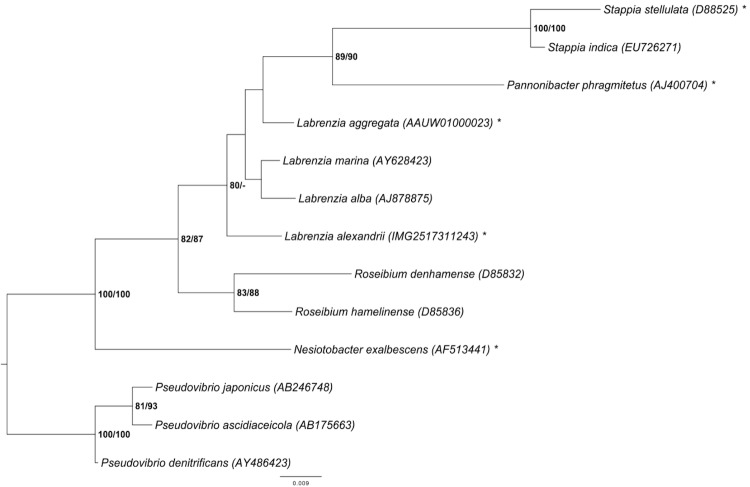
Figure 1 shows the phylogenetic neighborhood of L. alexandrii in a 16S rRNA based tree. The sequences of the three identical 16S rRNA gene copies in the genome do not differ from the previously published 16S rRNA sequence (AJ582083).
Figure 1.
Phylogenetic tree highlighting the position of L. alexandrii relative to the type strains of the species of selected genera (see [1,3] and the results of the Greengenes database search described above) within the family Rhodobacteraceae. These genera form a clade [1,3], but it might be better not to place them in this family [3]. The tree was inferred from 1,366 aligned characters [8,9] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [10] and rooted with Pseudovibrio. The branches are scaled in terms of the expected number of substitutions per site (see size bar). Numbers adjacent to the branches are support values from 1,000 ML bootstrap replicates [11] (left) and from 1,000 maximum-parsimony bootstrap replicates [12] (right) if larger than 60%. Lineages with type-strain genome sequencing projects registered in GOLD [13] are labeled with one asterisk.
Morphology and physiology
The rod-shaped cells of strain DFL-11T are 0.5 to 0.7 μm in width and 0.9 to 3.0 μm long with often unequal ends (Table 1 and Figure 2A), suggesting a polar mode of cell division which is increasingly being discovered in Alphaproteobacteria and thought to be ancient [23]. Motility is present by means of a single subpolar flagellum [1]. Star-shaped aggregated clusters occur [1]. The colonies exhibit a beige to slightly pink color [1]. Strain DFL-11T has a chemotrophic lifestyle; no fermentation occurs under aerobic or anaerobic conditions [1]. Optimal growth occurs in the presence of 1-10% NaCl and pH 7.0-8.5 at 26°C, whereas no growth occurs in the absence of NaCl or of biotin and thiamine as growth factors [1]. Several organic acids like acetate, butyrate, malate and citrate as well as glucose and fructose are metabolized, but methanol, ethanol and glycerol are not used for growth [1]. Whereas gelatin is hydrolyzed by the cells, starch is not; nitrate is not reduced [1]. The strain shows a weak resistance to potassium tellurite [1].
Table 1. Classification and general features of L. alexandrii DFL-11T according to the MIGS recommendations [14].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [15] | |
| Phylum Proteobacteria | TAS [16] | ||
| Class Alphaproteobacteria | TAS [17,18] | ||
| Order Rhodobacterales | TAS [17,19] | ||
| Family Rhodobacteraceae | TAS [17,20] | ||
| Genus Labrenzia | TAS [1] | ||
| Species Labrenzia alexandrii | TAS [1] | ||
| MIGS-7 | Subspecific genetic lineage | Strain DFL-11 | TAS [1] |
| Gram stain | Gram-negative | TAS [1] | |
| Cell shape | rod-shaped | TAS [1] | |
| Motility | motile | TAS [1] | |
| Sporulation | not reported | ||
| Temperature range | mesophile | TAS [1] | |
| Optimum temperature | 26°C | TAS [1] | |
| Salinity | 1–10 % (w/v) sea salt | TAS [1] | |
| MIGS-22 | Relationship to oxygen | aerobe | TAS [1] |
| Carbon source | acetate, butyrate and malate | TAS [1] | |
| Energy metabolism | photoheterotroph | TAS [1] | |
| MIGS-6 | Habitat | marine | TAS [1] |
| MIGS-6.2 | pH | 6–9.2 | TAS [1] |
| MIGS-15 | Biotic relationship | host-associated | TAS [1] |
| MIGS-14 | Known pathogenicity | none | TAS [1] |
| MIGS-16 | Specific host | Alexandrium lusitanicum | TAS [1] |
| MIGS-18 | Health status of host | not reported | |
| Biosafety level | 1 | TAS [21] | |
| MIGS-19 | Trophic level | not reported | |
| MIGS-23.1 | Isolation | ME207 | TAS [1] |
| MIGS-4 | Geographic location | not reported | |
| MIGS-5 | Time of sample collection | April 1, 2002 | TAS [1] |
| MIGS-4.1 | Latitude | 54.133 | TAS [1] |
| MIGS-4.2 | Longitude | 7.867 | TAS [1] |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes – TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). Evidence codes are from the Gene Ontology project [22].
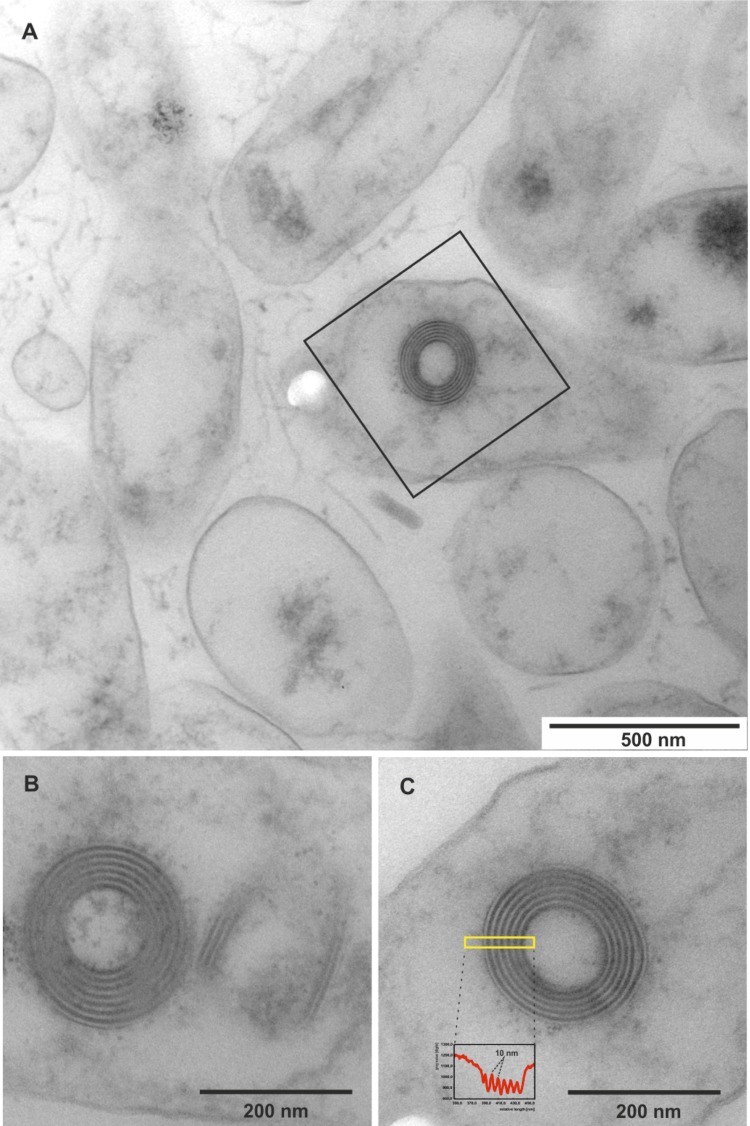
Figure 2.
Ultrastructure of L. alexandrii DFL-11T and its R-bodies. (A) Survey view of the cells from the near-surface position of a colony. Many bacterial remnants are visible, one of which contains an R-body; such bodies are shown enlarged in (B) and (C). (B) A pair of R-bodies, oriented at right angle towards each other, one as a cross-section and the other one cut oblique-longitudinally. The bipartite, black-white organization of the spiral layers is shown, and the averaged intensity profile (C, inset) of the boxed area shows a regular spacing of 10 nm.
The utilization of carbon compounds by L. alexandrii DSM 17067T was also determined for this study using PM01 microplates in an OmniLog phenotyping device (BIOLOG Inc., Hayward, CA, USA). The microplates were inoculated at 28°C with a cell suspension at a cell density of approximately 85% Turbidity and dye D. Further additives were artificial sea salts, vitamins, trace elements and NaHC03. The exported measurement data were further analyzed with the opm package for R [24], using its functionality for statistically estimating parameters from the respiration curves such as the maximum height, and automatically translating these values into negative, ambiguous, and positive reactions. The strain was studied in six independent biological replicates, and reactions with a distinct behavior between the repetitions were regarded as ambiguous and are not listed below.
L. alexandrii DSM 17067T was positive for glycerol, D-xylose, D-mannitol, L-glutamic acid, D,L-malic acid, D-ribose, D-fructose, D-glucose, α-keto-glutaric acid, α-keto-butyric acid, uridine, L-glutamine, α-hydroxy-butyric acid, myo-inositol, fumaric acid, propionic acid, glycolic acid, inosine, tricarballylic acid, L-threonine, D-malic acid, L-malic acid and pyruvic acid. The strain was negative for D-saccharic acid, D-galactose, D-alanine, D-trehalose, dulcitol, D-serine, L-fucose, D-glucuronic acid, D-gluconic acid, D,L-α-glycerol-phosphate, sodium formate, D-glucose-6-phosphate, D-galactonic acid-γ-lactone, tween 20, L-rhamnose, D-maltose, L-asparagine, D-aspartic acid, D-glucosaminic acid, 1,2-propanediol, tween 40, α-methyl-D-galactoside, α-D-lactose, lactulose, sucrose, m-tartaric acid, α-D-glucose-1-phosphate, D-fructose-6-phosphate, tween 80, α-hydroxy-glutaric acid-γ-lactone, β-methyl-D-glucoside, adonitol, maltotriose, 2'-deoxy-adenosine, adenosine, gly-asp, D-threonine, bromo-succinic acid, mucic acid, D-cellobiose, glycyl-L-glutamic acid, L-alanyl-glycine, acetoacetic acid, N-acetyl-β-D-mannosamine, methyl pyruvate, tyramine, D-psicose, glucuronamide, L-galactonic acid-γ-lactone, D-galacturonic acid and β-phenylethylamine.
In an electron microscopic survey colonies of strain DFL-11T, grown on half-strength MB (Roth CP73.1) agar plates, were fixed with 2.5% glutardialdehyde, 10 mM Hepes, pH 7.1, and embedded in Spurr's epoxide resin as described in detail elsewhere [25]. Ultrathin sections (90 nm) were analyzed in the elastic bright-field mode with an energy-filter transmission electron microscope (TEM) (Libra 120 plus; Zeiss, Oberkochen), and micrographs were recorded with a 2k × 2k cooled CCD-camera (SharpEye; Tröndle, Moorenweis, Germany) at a magnification range of 4000 × to 25000 ×.
TEM analysis showed that individual cells of strain DFL-11T, assembled in clusters, contained refractile inclusion bodies, known as R-bodies [26,27], when plate-grown bacteria were embedded as microcolonies of different growth states. R-bodies are highly insoluble protein ribbons coiled to form a hollow cylinder within the cytoplasma of the bacterial cells [26,27]. In strain DFL-11T these unusual structures were generally observed in cell remnants, which contained only small amounts of cytoplasmic material (Figure 2A). They were built mainly as five- to six-layered spirals and often had a loose electron-dense, amorphous matrix. In concentric cross- or longitudinal sections the individual layers appeared to be composed of an electron-dense dark and an electron-translucent bright layer; each doublet was found to have an average thickness of 10.1 nm (standard deviation: 0.7 nm; N = 16), ranging from minimal 8.7 nm to maximum 11.9 nm. The overall diameter of the R-bodies ranged from 183 nm to 242 nm, which is in good accordance with the dimensions of furled R-body ribbons reviewed in [27].
To date only a few bacterial species are known to produce R-bodies [26,27]. They were first described in members of the genus 'Caedibacter'. These bacteria live as obligate endosymbionts in Paramecium species and confer the so-called “killer trait” to their hosts: “killer-phenotype” paramecia release 'Caedibacter' cells via their cytopyge into the environment and these kill sensitive paramecia (i.e. 'Caedibacter'-free ciliates) after being ingested. The toxic effect of 'Caedibacter' is strictly correlated with R-body synthesis. Once incorporated into sensitive paramecia, the R-body extrudes in a telescopic fashion, thereby disrupting the bacterial cell. Cellular components are subsequently released into the cytoplasma of Paramecium, finally causing the ciliate’s death. It has been proposed that a lethal toxin is involved in this process, but it has not been identified so far [28]. Interestingly, a phylogenetic study based on comparative 16S rRNA gene sequencing revealed that 'Caedibacter' is a polyphyletic assemblage, comprising Gammaproteobacteria related to Francisella tularensis as well as Alphaproteobacteria affiliated with Rickettsiales (including the obligate Paramecium endosymbiont 'Holospora') [29]. In addition to the obligate endosymbionts, some free-living bacteria, i.e. Hydrogenophaga taeniospiralis, Acidovorax avenae subsp. avenae (both Burkholderiales), Rhodospirillum centenum, an anoxygenic phototrophic alphaproteobacterium, and Marinomonas mediterranea, a marine gammaproteobacterium, were observed to produce R-bodies [30].
Chemotaxonomy
Ubiquinone 10 was found as the single respiratory lipoquinone, which is a common feature in most Alphaproteobacteria. The spectrum of polar lipids consists of phosphatidylglycerol, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholin, phosphatidylmono-methylethanolamine, sulphoquinovosyldiacylglyceride, as well as an unidentified aminolipid [1]. In the fatty acids spectrum is dominated by C18 : 1ω7 (71%) and complemented by C20 : 1ω7 (9.1%), C18 : 0 (6.5%), 11-methyl C18:1ω6t (3.7%) and some hydroxy fatty acids C14:0 3-OH (3.4%) and C16:0 3-OH (1.5%) as well as traces of C18 : 1ω9 and cyclo C21:0 [1]. The presence of photosynthetic pigments was tested in [1] and the absorption spectrum of the acetone/methanol extract showed that bacteriochlorophyll a was present at low concentrations. Another peak at 420 and 550 nm indicated the presence of an additional photosynthetic pigment, most probably a yet unidentified carotinoid.
Genome sequencing and annotation
Genome project history
The genome was sequenced within the MMI supported by the Gordon and Betty Moore Foundation. Initial Sequencing was performed by the JCVI (Rockville, MD, USA) and a high-quality draft sequence was deposited at INSDC. The number of scaffolds and contigs was reduced and the assembly improved by a subsequent round of manual gap closure at HZI/DSMZ. A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High quality draft |
| MIGS-28 | Libraries used | Two genomic libraries: 40kb fosmid library and 3 kB pUC18 plasmid library |
| MIGS-29 | Sequencing platforms | ABI3730 |
| MIGS-31.2 | Sequencing coverage | 9.1 × Sanger |
| MIGS-30 | Assemblers | Consed 20.0 |
| MIGS-31.3 | Contig count | 6 |
| MIGS-32 | Gene calling method | Genemark 4.6b, tRNAScan-SE-1.23, Infernal 0.81 |
| INSDC ID | Final ID pending; previous version ACCU00000000 | |
| Genbank Date of Release | N/A | |
| GOLD ID | Gi01459 | |
| NCBI project ID | 19367 | |
| Database: IMG | 2517287006 | |
| MIGS-13 | Source Material Identifier | DSM 17067 |
| Project relevance | Environmental, Marine Microbial Initiative |
Growth conditions and DNA extractions
A culture of DSM 17067 was grown for two to three days on a LB & sea-salt agar plate, containing (l-1) 10 g tryptone, 5 g yeast extract, 10 g NaCl, 17 g sea salt (Sigma-Aldrich S9883) and 15 g agar. A single colony was used to inoculate LB & sea-salt liquid medium and the culture was incubated at 28°C on a shaking platform. The genomic DNA was isolated using the Qiagen Genomic 500 DNA Kit (Qiagen 10262) as indicated by the manufacturer. DNA quality and quantity were in accordance with the instructions of the genome sequencing center.
Genome sequencing and assembly
The genome was sequenced with the Sanger technology using a combination of two libraries. All general aspects of library construction and sequencing performed at the JCVI can be found on the JCVI website. Base calling of the sequences were performed with the phredPhrap script using default settings. The reads were assembled using the phred/phrap/consed pipeline [31]. The last gaps were closed by adding new reads produced by recombinant PCR and PCR primer walks. In total 21 reads were required for gap closure and improvement of low quality regions. The final consensus sequence was built from 60,668 Sanger reads (9.1 × coverage).
Genome annotation
Gene prediction was carried out using GeneMark as part of the genome annotation pipeline in the Integrated Microbial Genomes Expert Review (IMG-ER) system [32]. To identify coding genes, Prodigal [33] was used, while ribosomal RNA genes within the genome were identified using the tool RNAmmer [34]. Other non-coding genes were predicted using Infernal [35]. Manual functional annotation was performed within the IMG platform [32] and the Artemis Genome Browser [36].
Genome properties
The genome statistics are provided in Table 3 and Figures 3a, 3b and 3c. The genome consists of a 5,299,280 bp long chromosome and two plasmids with 68,647 bp and 93,929 bp length, respectively, with a G+C content of 56.4%. Of the 5,144 genes predicted, 5,071 were protein-coding genes, and 73 RNAs; pseudogenes were not identified. The majority of the protein-coding genes (81.0%) were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 5,461,856 | 100.00 |
| DNA coding region (bp) | 4,871,168 | 89.19 |
| DNA G+C content (bp) | 3,080,828 | 56.41 |
| Number of replicons | 3 | |
| Extrachromosomal elements | 2 | |
| Total genes | 5,144 | 100.00 |
| RNA genes | 73 | 1.42 |
| rRNA operons | 3 | |
| tRNA genes | 52 | 1.01 |
| Protein-coding genes | 5,071 | 98.58 |
| Pseudo genes | 0 | |
| Genes with function prediction | 4,168 | 81.03 |
| Genes in paralog clusters | 1,866 | 36.28 |
| Genes assigned to COGs | 4,140 | 80.48 |
| Genes assigned Pfam domains | 4,203 | 81.71 |
| Genes with signal peptides | 1,147 | 22.30 |
| Genes with transmembrane helices | 1,264 | 24.57 |
| CRISPR repeats | 0 |
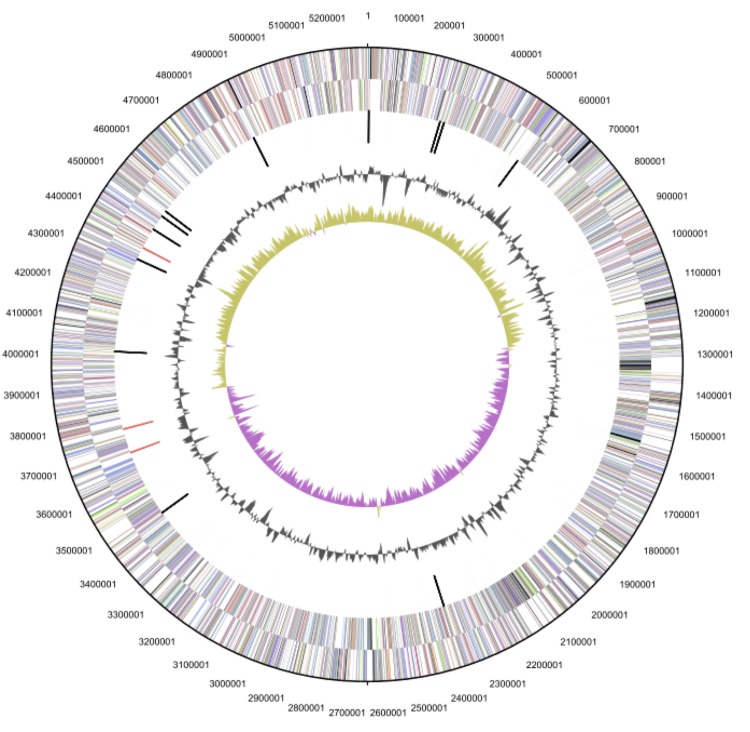
Figure 3a.
Graphical map of the chromosome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
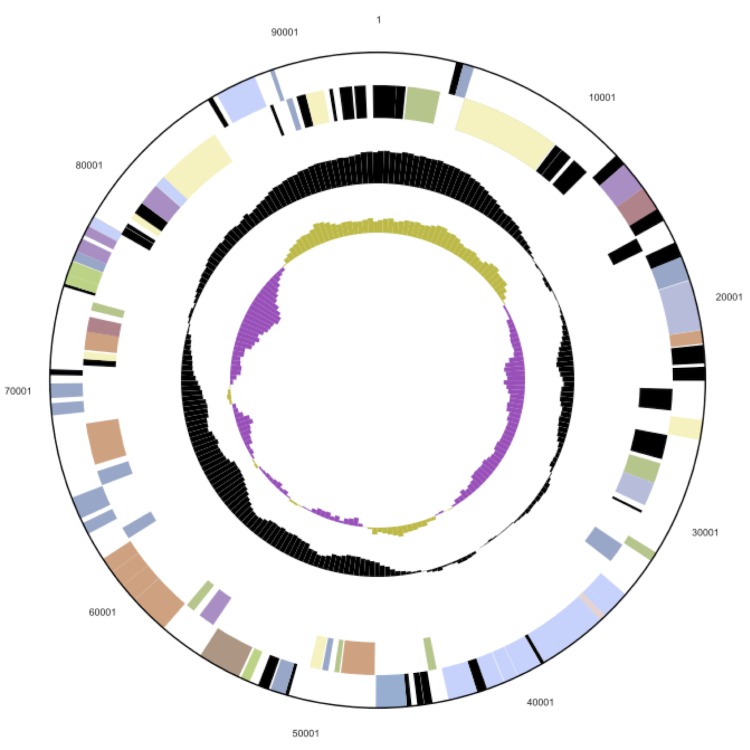
Figure 3b.
The larger of the two plasmids (LADFL_5, not drawn to scale with the chromosome). From outside to the center: Genes on forward strand (color by COG categories), genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Figure 3c.
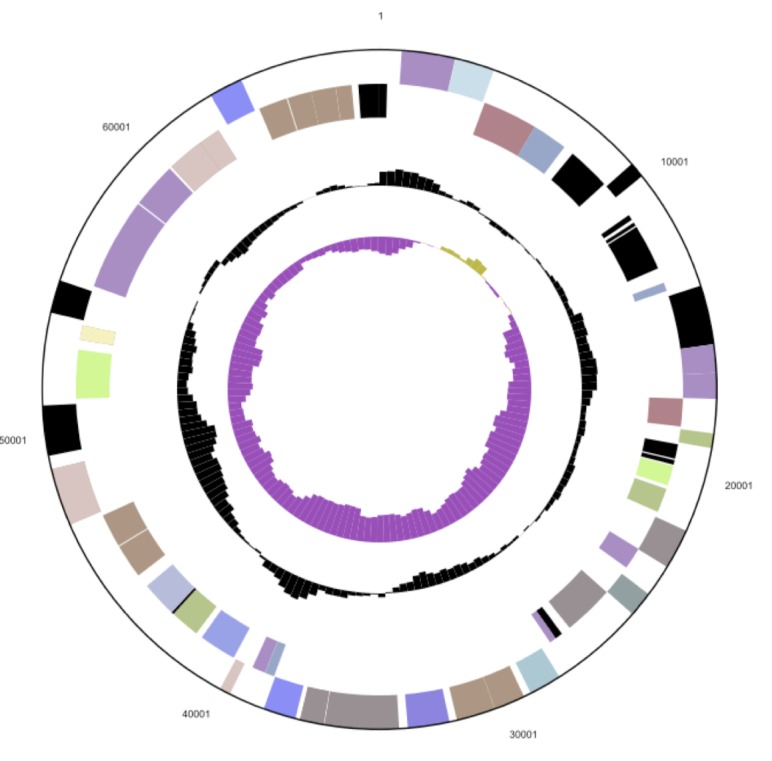
The smaller of the two plasmids (LADFL_6, not drawn to scale with the chromosome). From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 179 | 3.88 | Translation, ribosomal structure and biogenesis |
| A | 2 | 0.04 | RNA processing and modification |
| K | 363 | 7.86 | Transcription |
| L | 133 | 2.88 | Replication, recombination and repair |
| B | 3 | 0.06 | Chromatin structure and dynamics |
| D | 38 | 0.82 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0 | Nuclear structure |
| V | 51 | 1.10 | Defense mechanisms |
| T | 330 | 7.15 | Signal transduction mechanisms |
| M | 223 | 4.83 | Cell wall/membrane/envelope biogenesis |
| N | 115 | 2.49 | Cell motility |
| Z | 2 | 0.04 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 91 | 1.97 | Intracellular trafficking, secretion, and vesicular transport |
| O | 167 | 3.62 | Posttranslational modification, protein turnover, chaperones |
| C | 260 | 5.63 | Energy production and conversion |
| G | 276 | 5.98 | Carbohydrate transport and metabolism |
| E | 518 | 11.22 | Amino acid transport and metabolism |
| F | 91 | 1.97 | Nucleotide transport and metabolism |
| H | 174 | 3.77 | Coenzyme transport and metabolism |
| I | 186 | 4.03 | Lipid transport and metabolism |
| P | 232 | 5.02 | Inorganic ion transport and metabolism |
| Q | 136 | 2.95 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 582 | 12.61 | General function prediction only |
| S | 465 | 10.07 | Function unknown |
| - | 1,004 | 19.52 | Not in COGs |
Insights into the genome
R-body genes
In 'Caedibacter taeniospiralis', three genes (rebA, rebB and rebC) were identified to determine the R-body production. They are clustered on large plasmids, ranging from 41-49 kb, and encompass 345 bp, 318 bp and 171 bp (accession number U04524), respectively. The corresponding proteins RebA (114 aa, 18 kDa), RebB (105 aa, 13 kDa) and RebC (56aa, 10 kDa) are necessary to assemble R-bodies through polymerization processes [37]. Furthermore, a putative forth gene rebD (249 bp; RepD 82aa) is located between rebB and rebC and might be involved in R-body formation.
Based on high sequence similarities to the C. taeniospiralis R-body protein RebB, three homologues (ladfl_0000085, ladfl_0000090 and ladfl_0000091) were detected on the chromosome of strain DFL-11T. Their amino acid sequence length is 122 aa, 109 aa and 76 aa, respectively, which is in accordance with R-body proteins found in C. taeniospiralis 47, and they were all assigned to the Pfam family RebB (PF11747). The chromosomal arrangement of R-body genes in strain DFL-11T is not contiguous; ladfl_0000085 is separated from ladfl_0000090 and ladfl_0000091 by four hypothetical genes (ladfl_0000086 -ladfl_0000089). Interestingly, a putative alternative sigma-factor of the ECF subfamily (ladfl_0000084, upstream of ladfl_0000085) flanks the R-body gene cluster, indicating that reb gene expression in strain DFL-11T is regulated by extracytoplasmic stimuli. Gene arrangements orthologous to the L. alexandrii DFL-11T reb gene cluster were found in the alphaproteobacteria Roseibium sp. TrichSKD4 (NZ_GL47637) and Polymorphum gilvum (NC_015259), organisms which are closely related to L. alexandrii [38].
Plasmids
Genome sequencing of L. alexandrii DSM 17067T reveals the presence of two RepABC-type plasmids designated LADFL_5 and LADFL_6 with sizes of 93,929 bp and 68,647 bp, respectively. This outcome is in agreement with a previous study about the genome organization of different marine Alphaproteobacteria including DFL-11T [39]. Pulsed-field gel electrophoresis (PFGE) showed faint bands with estimated sizes of 88 kb and 65 kb, and their circular conformation has been documented by comparative analyses with distinct PFGE parameters. An additional linear fragment of about 35 kb, which has not been recovered by genome sequencing, may represent a prophage (see below) whose excision from the genome depends on the cultivation conditions. Both plasmids represent RepABC-type replicons with the partitioning genes repA and repB as well as the replicase repC that are located in a typical operon [40]. Phylogenetic analyses of the replicases provides the basis for the classification of alphaproteobacterial plasmids [41]. The respective phylogeny of both RepC sequences from L. alexandrii DSM 17067T (ladfl_05027, ladfl_05140) documents a close affiliation with rhizobial genes to an exclusion of sequences from Rhodobacterales that are located in distinct subtrees (data not shown [42] ). Both plasmids seem to be equipped with characteristic post segregational killing systems consisting of a toxin/antitoxin operon that prevent plasmid loss (ladfl_05100/ladfl_05101, ladfl_05128/ladfl_05129 [43] ).
Plasmid LADFL_5 contains several genes that are related to heavy-metal resistance [44] and eight of them are related to the COG category “Inorganic ion transport and metabolism” (see also Table 4). This set includes the mer-operon composed of merR, merT, merF and mercuric reductase MerA, which are part of the Gram-negatives' mercury-resistance system [45]. This plasmid also harbors a predicted P-type ATPase translocating heavy-metal ions and components of a Cd2+, Zn2+ or Co2+ efflux system. The resistance to a wide pallet of heavy-metal ions may enable the strain to dwell in polluted environments [44]. The second conspicuous trait of LADFL_5 is the presence of a complete type-IV secretion system (T4SS [46] ). The virB operon (ladfl_05033 to ladfl_05043) is required for the formation of a functional transmembrane channel and pilus formation. Moreover, the virD gene cluster including the characteristic DNA relaxase (ladfl_05091) and the coupling protein VirD4 (ladfl_05093) indicates that the T4SS machinery represents a functional conjugation system. The lysozyme TraH_2 (ladfl_05088), which is required for the degradation of the peptidoglycan cell wall and transmembrane channel formation, is annotated as specific protein of Rhizobiales, an affiliation that is in agreement with the outcome of the phylogenetic RepC analysis [42].
Plasmid LADFL_6 is dominated by more than a dozen genes that are involved in sugar metabolism. It contains the complete operon for the conversion of glucose-1-phosphate into dTDP-L-rhamnose (rmlC, rmlD, rmlA, rmlB) that is a common component of the cell wall and capsule of many pathogenic bacteria [47]. Three glycosyltransferases, some components of an ABC-type polysaccharide transport system as well as a sugar transferase for lipopolysaccharide synthesis and a lipid A core O-antigen ligase (ladfl_05144, ladfl_05145) are indicative for a functional role of the plasmid for exopolysaccharide formation. Extracellular polysaccharids of the Sym plasmid are required for root hair attachment in Rhizobium leguminosarum [48] and the plasmid LADFL_6 may also be required for biofilm generation. This prediction is compatible with the origin of strain DFL-11T that has been isolated from the dinoflagellate A. lusitanicum [1].
Acknowledgements
This work was conducted as part of the Marine Microbial Initiative supported by the Gordon and Betty Moore Foundation. Additional support via the German Research Foundation (DFG) SFB/TRR 51 is gratefully acknowledged. We also thank the European Commission which supported phenotyping via the Microme project 222886 within the Framework 7 program.
References
- 1.Biebl H, Pukall R, Lünsdorf H, Schulz S, Allgaier M, Tindall BJ, Wagner-Döbler I. Description of Labrenzia alexandrii gen. nov., sp. nov., a novel alphaproteobacterium containing bacteriochlorophyll a, and a proposal for reclassification of Stappia aggregata as Labrenzia aggregata comb. nov., of Stappia marina as Labrenzia marina comb. nov. and of Stappia alba as Labrenzia alba comb. nov., and emended descriptions of the genera Pannonibacter, Stappia and Roseibium, and of the species Roseibium denhamense and Roseibium hamelinense. Int J Syst Evol Microbiol 2007; 57:1095-1107 10.1099/ijs.0.64821-0 [DOI] [PubMed] [Google Scholar]
- 2.Garrity GM, Bell JA, Lilburn T. Order III. Rhodobacteraceae fam. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 161. [Google Scholar]
- 3.Lee KB, Liu CT, Anzai Y, Kim H, Aono T, Oyaizu H. The hierarchical system of the ‘Alphaproteobacteria’: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol 2005; 55:1907-1919 10.1099/ijs.0.63663-0 [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410 [DOI] [PubMed] [Google Scholar]
- 5.Korf I, Yandell M, Bedell J. BLAST, O'Reilly, Sebastopol, 2003. [Google Scholar]
- 6.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137.
- 8.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 9.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 10.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 11.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [Google Scholar]
- 12.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland, 2002. [Google Scholar]
- 13.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2012; 40:D571-D579 10.1093/nar/gkr1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 17.Validation List No 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 18.Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 1. [Google Scholar]
- 19.Garrity GM, Bell JA, Lilburn T. Order III. Rhodobacterales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 161. [Google Scholar]
- 20.Garrity GM, Bell JA, Lilburn T. Family I. Rhodobacteraceae fam. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 161. [Google Scholar]
- 21.BAuA. 2010, Classification of Bacteria and Archaea in risk groups. http://www.baua.de TRBA 466, p. 209.
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown PJB, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA 2012; 109:1697-1701 10.1073/pnas.1114476109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaas LAI, Sikorski J, Michael V, Göker M, Klenk HP. Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 2012; 7:e34846 10.1371/journal.pone.0034846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yakimov MM, Golyshin PN, Lang S, Moore ERB, Abraham WR, Lünsdorf H, Timmis KN. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int J Syst Bacteriol 1998; 48:339-348 10.1099/00207713-48-2-339 [DOI] [PubMed] [Google Scholar]
- 26.Lalucat J, Meyer O, Mayer F, Parés R, Schlegel HG. R-Bodies in newly isolated free-living hydrogen-oxidizing bacteria. Arch Microbiol 1979; 121:9-15 10.1007/BF00409199 [DOI] [Google Scholar]
- 27.Pond FR, Gibson I, Lalucat J, Quackenbush RL. R-Body-producing bacteria. Microbiol Rev 1989; 53:25-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrallhammer M. The killer trait of Paramecium and its causative agents. Palaeodiversity 2010; 3(Supplement):79-88 [Google Scholar]
- 29.Beier CL, Horn M, Michel R, Schweikert M, Görtz HD, Wagner M. The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales ( Alphaproteobacteria) and to Francisella tularensis ( Gammaproteobacteria). Appl Environ Microbiol 2002; 68:6043-6050 10.1128/AEM.68.12.6043-6050.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernández-Romero D, Lucas-Elío P, López-Serrano D, Solano F, Sanchez-Amat A. Marinomonas mediterranea is a lysogenic bacterium that synthesizes R-bodies. Microbiology 2003; 149:2679-2686 10.1099/mic.0.26524-0 [DOI] [PubMed] [Google Scholar]
- 31.Phrap and Phred for Windows. MacOS, Linux, and Unix. www.phrap.com
- 32.Markowitz VM, Mavromatis K, Ivanova NN, Chen IA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 33.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagesen K, Hallin PF, Rødland E, Stærfeldt HH, Rognes T, Ussery DW. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009; 25:1335-1337 10.1093/bioinformatics/btp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics 2000; 16:944-945 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 37.Heruth DP, Pond FR, Dilts JA, Quackenbush RL. Characterization of genetic determinants for R body synthesis and assembly in Caedibacter taeniospiralis 47 and 116. J Bacteriol 1994; 176:3559-3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai M, Wang L, Cai H, Li Y, Tang YQ, Wu XL. Rubrimonas shengliensis sp. nov. and Polymorphum gilvum gen. nov., sp. nov., novel members of Alphaproteobacteria from crude oil contaminated saline soil. Syst Appl Microbiol 2011; 34:321-327 10.1016/j.syapm.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 39.Pradella S, Allgaier M, Hoch C, Päuker O, Stackebrandt E, Wagner-Döbler I. Genome Organization and localization of the pufLM genes of the photosynthesis reaction center in phylogenetically diverse marine Alphaproteobacteria. Appl Environ Microbiol 2004; 70:3360-3369 10.1128/AEM.70.6.3360-3369.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto UM, Pappas KM, Winans SC. The ABCs of plasmid replication and segregation. Nat Rev Microbiol 2012; 10:755-765 10.1038/nrmicro2882 [DOI] [PubMed] [Google Scholar]
- 41.Petersen J. Phylogeny and compatibility: plasmid classification in the genomics era. Arch Microbiol 2011; 193:313-321 [DOI] [PubMed] [Google Scholar]
- 42.Petersen J, Brinkmann H, Pradella S. Diversity and evolution of repABC type plasmids in Rhodobacterales. Environ Microbiol 2009; 11:2627-2638 10.1111/j.1462-2920.2009.01987.x [DOI] [PubMed] [Google Scholar]
- 43.Zielenkiewicz U, Ceglowski P. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim Pol 2001; 48:1003-1023 [PubMed] [Google Scholar]
- 44.Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol 1996; 50:753-789 10.1146/annurev.micro.50.1.753 [DOI] [PubMed] [Google Scholar]
- 45.Barkay T, Miller SM, Summers AO. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 2003; 27:355-384 10.1016/S0168-6445(03)00046-9 [DOI] [PubMed] [Google Scholar]
- 46.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol 2003; 1:137-149 10.1038/nrmicro753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giraud MF, Naismith JH. The rhamnose pathway. Curr Opin Struct Biol 2000; 10:687-696 10.1016/S0959-440X(00)00145-7 [DOI] [PubMed] [Google Scholar]
- 48.Laus MC, Logman TJ, Lamers GE, Van Brussel AA, Carlson RW, Kijne JW. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol 2006; 59:1704-1713 10.1111/j.1365-2958.2006.05057.x [DOI] [PubMed] [Google Scholar]