Abstract
Hoeflea phototrophica Biebl et al. 2006 is a member of the family Phyllobacteriaceae in the order Rhizobiales, which is thus far only partially characterized at the genome level. This marine bacterium contains the photosynthesis reaction-center genes pufL and pufM and is of interest because it lives in close association with toxic dinoflagellates such as Prorocentrum lima. The 4,467,792 bp genome (permanent draft sequence) with its 4,296 protein-coding and 69 RNA genes is a part of the Marine Microbial Initiative.
Keywords: aerobic, rod-shaped, motile, photoheterotroph, Phenotype MicroArray, bacteriochlorophyll a, symbiosis, dinoflagellates, Prorocentrum lima, Phyllobacteriaceae
Introduction
Strain DFL-43T (= DSM 17068 = NCIMB 14078) is the type strain of Hoeflea phototrophica, a marine member of the Phyllobacteriaceae (Rhizobiales, Alphaproteobacteria) [1]. The genus, which was named in honor of the German microbiologist Manfred Höfle [2], contains four species, with H. marina as type species [2]; the name of a fifth member of the genus, 'Hoeflea siderophila', is until now only effectively published [3]. H. phototrophica DFL-43T and strain DFL-44 were found in the course of a screening program for marine bacteria containing the photosynthesis reaction-center genes pufL and pufM [4]. The species epithet 'phototrophica' refers to the likely ability of H. phototrophica strains to use light as an additional energy source [1]. Strain DFL-43T was isolated from single cells of a culture of the toxic dinoflagellate Prorocentrum lima maintained at the Biological Research Institute of Helgoland, Germany [1]. Here we present a summary classification and a set of features for H. phototrophica DFL-43T including so far undiscovered aspects of its phenotype, together with the description of the complete genomic sequencing and annotation.
This work is part of the Marine Microbial Initiative (MMI) which enabled the J. Craig Venter Institute (JCVI) to sequence the genomes of approximately 165 marine microbes with funding from the Gordon and Betty Moore Foundation. These microbes were contributed by collaborators worldwide, and represent an array of physiological diversity, including carbon fixers, photoautotrophs, photoheterotrophs, nitrifiers, and methanotrophs. The MMI was designed to complement other ongoing research at JCVI and elsewhere to characterize the microbial biodiversity of marine and terrestrial environments through metagenomic profiling of environmental samples.
Classification and features
16S rRNA analysis
A representative genomic 16S rRNA sequence of H. phototrophica DFL-43T was compared using NCBI BLAST [5,6] under default settings (e.g., considering only the high-scoring segment pairs (HSPs) from the best 250 hits) with the most recent release of the Greengenes database [7] and the relative frequencies of taxa and keywords (reduced to their stem [8]) were determined, weighted by BLAST scores. The most frequently occurring genera were Rhizobium (53.7%), Sinorhizobium (24.0%), Hoeflea (4.5%), Bartonella (4.5%) and Ahrensia (3.7%) (132 hits in total). Regarding the two hits to sequences from members of the species, both, the average identity within HSPs and the average coverage by HSPs were 100.0%. Regarding the single hit to sequences from other members of the genus, the average identity within HSPs was 98.2%, whereas the average coverage by HSPs was 100.0%. Among all other species, the one yielding the highest score was H. marina (AY598817), which corresponded to an identity of 98.2% and an HSP coverage of 100.0%. (Note that the Greengenes database uses the INSDC (= EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was AY922224 (Greengenes short name 'whalefall clone 131720'), which showed an identity of 98.1% and an HSP coverage of 97.5%. The most frequently occurring keywords within the labels of all environmental samples which yielded hits were 'bee' (3.1%), 'singl' (3.0%), 'abdomen, bumbl, distinct, honei, microbiota, simpl' (2.9%), 'microbi' (2.8%) and 'structur' (1.8%) (118 hits in total). Environmental samples which yielded hits of a higher score than the highest scoring species were not found, indicating that H. phototrophica is rarely found in environmental samples.
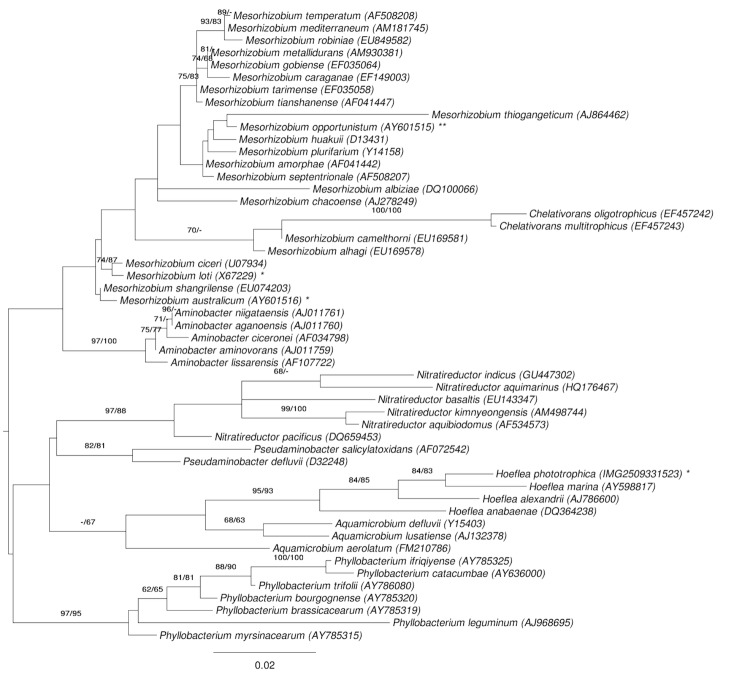
Figure 1 shows the phylogenetic neighborhood of H. phototrophica in a 16S rRNA based tree. The sequences of the two identical 16S rRNA gene copies in the genome differ by one nucleotide from the previously published 16S rRNA sequence (AJ582088)
Figure 1.
Phylogenetic tree highlighting the position of H. phototrophica relative to the type strains of the other species within the family Phyllobacteriaceae. The tree was inferred from 1,362 aligned characters [9,10] of the 16S rRNA gene sequence under the maximum likelihood (ML) criterion [11]. Rooting was done initially using the midpoint method [12] and then checked for its agreement with the current classification (Table 1). The branches are scaled in terms of the expected number of substitutions per site. Numbers adjacent to the branches are support values from 1,000 ML bootstrap replicates [13] (left) and from 1,000 maximum-parsimony bootstrap replicates [14] (right) if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [15] are labeled with one asterisk, those also listed as 'Complete and Published' (CP002279 for Mesorhizobium opportunistum) with two asterisks.
Table 1. Classification and general features of H. phototrophica DFL-43T according to the MIGS recommendations [16].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [17] | |
| Phylum Proteobacteria | TAS [18] | ||
| Class Alphaproteobacteria | TAS [19,20] | ||
| Order Rhizobiales | TAS [20,21] | ||
| Family Phyllobacteriaceae | TAS [20,22] | ||
| Genus Hoeflea | TAS [2] | ||
| Species Hoeflea phototrophica | TAS [1] | ||
| MIGS-7 | Subspecific genetic lineage (strain) | DFL-43T | TAS [1] |
| MIGS-12 | Reference for biomaterial | Biebl et al. 2006 | TAS [1] |
| Gram stain | Gram-negative | TAS [1] | |
| Cell shape | rod-shaped | TAS [1] | |
| Motility | motile | TAS [1] | |
| Sporulation | not reported | ||
| Temperature range | mesophile, 25-33°C | TAS [1] | |
| Optimum temperature | 31°C | TAS [1] | |
| Salinity | 0.5–7.0 % NaCl | TAS [1] | |
| MIGS-22 | Relationship to oxygen | aerobe | TAS [1] |
| Carbon source | acetate, malate | TAS [1] | |
| Energy metabolism | photoheterotroph | TAS [1] | |
| MIGS-6 | Habitat | marine | TAS [1] |
| MIGS-6.2 | pH | 6–9.0 | TAS [1] |
| MIGS-15 | Biotic relationship | host-associated | TAS [1] |
| MIGS-14 | Known pathogenicity | none | TAS [1] |
| MIGS-16 | Specific host | Prorocentrum lima ME130 | TAS [1] |
| MIGS-18 | Health status of Host | not reported | |
| Biosafety level | 1 | TAS [23] | |
| MIGS-19 | Trophic level | not reported | |
| MIGS-23.1 | Isolation | from a culture of Prorocentrum lima ME130 |
TAS [1] |
| MIGS-4 | Geographic location | not reported | |
| MIGS-5 | Time of sample collection | April 1, 2002 | TAS [1] |
| MIGS-4.1 | Latitude | 54.133 | TAS [1] |
| MIGS-4.2 | Longitude | 7.867 | TAS [1] |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). Evidence codes are from the Gene Ontology project [24].
Morphology and physiology
Cells of H. phototrophica are small rods of 0.3–0.5 μm in width and 0.7–2.0 μm length [1] (Figure 2) and motile by means of single, polar flagellum [1] (not visible in Figure 2). Depending on the availability of light, colonies are opaque to beige (grown in the dark) on marine agar 2216 [1]. The cultures are strictly aerobic and prefer microaerobic conditions. Good growth was detectable within a range of 25-33°C (1/5 limited growth rate below this value), concentration of sea salt from 0.5-7.0% and pH values from 6.0-9.0 [1]. Acetate and malate were accepted as carbon sources, whereas ethanol and methanol were not used for growth [1]. No hydrolysis of gelatin, starch, alginate or Tween 8 was observed [1].
Figure 2.
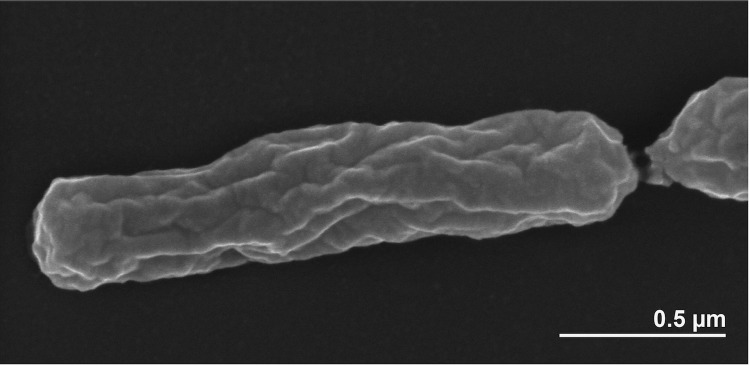
Scanning electron micrograph of H. phototrophica DFL-43T
The utilization of carbon compounds by H. phototrophica DFL-43T was also determined for this study using PM01 microplates in an OmniLog phenotyping device (BIOLOG Inc., Hayward, CA, USA). The microplates were inoculated at 28°C with a cell suspension at a cell density of 85% Turbidity and dye D. Further additives were artificial sea salts, vitamins, trace elements and NaHCO3. The exported measurement data were further analyzed with the opm package for R [25], using its functionality for statistically estimating parameters from the respiration curves such as the maximum height, and automatically translating these values into negative, ambiguous, and positive reactions. The strain was studied in two independent biological replicates, and reactions with a different behavior between the two repetitions were regarded as ambiguous and are not listed below.
H. phototrophica DFL-43T was positive for D,L-malic acid, D-cellobiose, D-fructose, D-galactonic acid-γ-lactone, D-galactose, D-galacturonic acid, D-gluconic acid, D-glucuronic acid, D-malic acid, D-mannitol, D-melibiose, D-sorbitol, D-trehalose, D-xylose, L-alanine, L-arabinose, L-glutamic acid, L-glutamine, L-lactic acid, L-lyxose, L-malic acid, L-proline, L-serine, acetic acid, adonitol, α-D-glucose, α-keto-glutaric acid, α-methyl-D-galactoside, β-methyl-D-glucoside, bromo-succinic acid, citric acid, ethanolamine, fumaric acid, m-inositol, maltose, maltotriose, mono-methyl succinate, propionic acid, pyruvic acid, succinic acid, sucrose and uridine. The strain was negative for 1,2-propanediol, 2'-deoxy-adenosine, D,L-α-glycerol-phosphate, D-alanine, D-aspartic acid, D-fructose-6-phosphate, D-glucosaminic acid, D-glucose-1-phosphate, D-glucose-6-phosphate, D-mannose, D-psicose, D-serine, D-threonine, L-alanyl-glycine, L-aspartic acid, L-fucose, L-galactonic acid-γ-lactone, L-rhamnose, L-threonine, N-acetyl-D-glucosamine, N-acetyl-β-D-mannosamine, acetoacetic acid, adenosine, α-D-lactose, α-hydroxy-butyric acid, α-hydroxy-glutaric acid-γ-lactone, α-keto-butyric acid, β-phenylethylamine, dulcitol, glycolic acid, glycyl-L-aspartic acid, glyoxylic acid, inosine, m-hydroxy-phenylacetic acid, m-tartaric acid, mucic acid, thymidine, tricarballylic acid, tween 40, tween 80 and tyramine.
Chemotaxonomy
Phosphatidylglycerol, phosphatidylethanolamine and phosphatidylmonomethylethanolamine were the predominant polar lipids of the membrane. The most frequent cellular fatty acids in strain DFL-43T are the mono-unsaturated straight chain acids C18:1 ω7 (62.8%) and its methylated form C18:1 ω7 11Me (21%), followed by C16:0 (6.3%) and C19:1 (3.4%) [1]. The absorption spectrum of an acetone/methanol extract showed the presence of bacteriochlorophyll a and an additional carotenoid (possibly spheroidenone) in small amounts [1]. Further experiments indicated that the pigment production depends on the concentration of sea salts in the medium [1].
Genome sequencing and annotation
Genome project history
The genome was sequenced within the MMI supported by the Gordon and Betty Moore Foundation. Initial Sequencing was performed by the J. Craig Venter Institute, JCVI (Rockville, MD, USA), and a high-quality draft sequence was deposited at INSDC. The number of scaffolds and contigs was reduced and the assembly improved by a subsequent round of manual gap closure at HZI/DSMZ. A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High quality draft |
| MIGS-28 | Libraries used | Two genomic libraries: 40 kb fosmid library and 3 kb pUC18 plasmid library |
| MIGS-29 | Sequencing platform | ABI3730 |
| MIGS-31.2 | Sequencing coverage | 10.3 × Sanger |
| MIGS-30 | Assemblers | Consed 20.0 |
| MIGS-31.3 | Contig count | 5 |
| MIGS-32 | Gene calling method | Prodigal 2.0, Infernal 1.0.2 |
| INSDC ID | Final ID pending; previous version ABIA00000000 | |
| Genbank Date of Release | final version not yet available | |
| GOLD ID | Gi01415 | |
| NCBI project ID | 19311 | |
| Database: IMG | 2509276008 | |
| MIGS-13 | Source Material Identifier | DSM 17068 |
| Project relevance | Environmental, Marine Microbial Initiative |
Growth conditions and DNA extractions
Cells of strain DFL-43T were grown for two to three days on a LB & sea-salt agar plate, containing (l-1) 10 g tryptone, 5 g yeast extract, 10 g NaCl, 17 g sea salt (Sigma-Aldrich S9883) and 15 g agar. A single colony was used to inoculate LB & sea-salt liquid medium and the culture was incubated at 28°C on a shaking platform. The genomic DNA was isolated using the Qiagen Genomic 500 DNA Kit (Qiagen 10262) as indicated by the manufacturer. DNA quality and quantity were in accordance with the instructions of the genome sequencing center. DNA is available through the DNA Bank Network [26].
Genome sequencing and assembly
The genome was sequenced with the Sanger technology using a combination of two libraries. All general aspects of library construction and sequencing performed at the JCVI can be found on the JCVI website. Base calling of the sequences were performed with the phredPhrap script using default settings. The reads were assembled and assemblies analyzed using the phred/phrap/consed pipeline [27]. The last gaps were closed by adding new reads produced by recombinant PCR and PCR primer walks. In total 21 Sanger reads were required for gap closure and improvement of low quality regions. The final consensus sequence was built from 46,086 Sanger reads (10.3 × coverage).
Genome annotation
Gene prediction was carried out using GeneMark as part of the genome annotation pipeline in the Integrated Microbial Genomes Expert Review (IMG-ER) system [28]. To identify coding genes, Prodigal [29] was used, while ribosomal RNA genes within the genome were identified using RNAmmer [30]. Other non-coding genes were predicted using Infernal [31]. Manual functional annotation was performed within the IMG platform [28] and the Artemis Genome Browser [32].
Genome properties
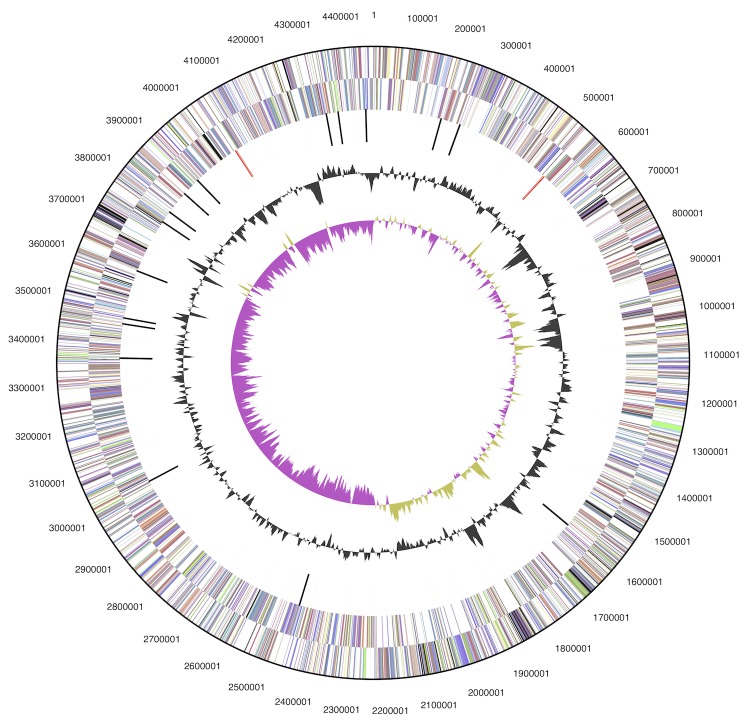
The draft genome consists of one circular scaffold with a total length of 4,467,822 bp containing five large contigs with a total length of 4,467,792 bp and a G+C content of 59.8%. Contig lengths vary from 133,683 bp to 2,215,172 bp (Figure 3); genome statistics are provided in Table 3. Of the 4,296 genes predicted, 4,227 were protein-coding genes, and 69 RNAs; pseudogenes were not identified. The majority of the protein-coding genes (83.1%) were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Figure 3.
Graphical map of the chromosome. From outside to the centerp: Genes on forward strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,467,832 | 100.00 |
| DNA coding region (bp) | 4,006,040 | 89.66 |
| DNA G+C content (bp) | 2,671,973 | 59.81 |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 4,296 | 100.00 |
| RNA genes | 69 | 1.61 |
| rRNA operons | 2 | |
| tRNA genes | 47 | 1.09 |
| Protein-coding genes | 4,227 | 98.39 |
| Pseudo genes | 0 | |
| Genes with function prediction | 3,574 | 83.19 |
| Genes in paralog clusters | 1,423 | 33.12 |
| Genes assigned to COGs | 3,525 | 82.05 |
| Genes assigned Pfam domains | 3,580 | 83.33 |
| Genes with signal peptides | 927 | 21.58 |
| Genes with transmembrane helices | 994 | 24.57 |
| CRISPR repeats | 0 |
Table 4. Number of genes associated with the general COG functional categories.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 178 | 4.58 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.00 | RNA processing and modification |
| K | 274 | 7.05 | Transcription |
| L | 162 | 4.17 | Replication, recombination and repair |
| B | 2 | 0.05 | Chromatin structure and dynamics |
| D | 27 | 0.69 | Cell cycle control, cell division, chromosome partitioning |
| Y | - | - | Nuclear structure |
| V | 39 | 1.00 | Defense mechanisms |
| T | 175 | 4.50 | Signal transduction mechanisms |
| M | 205 | 5.27 | Cell wall/membrane/envelope biogenesis |
| N | 60 | 1.54 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | - | - | Extracellular structures |
| U | 66 | 1.70 | Intracellular trafficking, secretion, and vesicular transport |
| O | 135 | 3.47 | Posttranslational modification, protein turnover, chaperones |
| C | 226 | 5.81 | Energy production and conversion |
| G | 325 | 8.36 | Carbohydrate transport and metabolism |
| E | 405 | 10.41 | Amino acid transport and metabolism |
| F | 80 | 2.06 | Nucleotide transport and metabolism |
| H | 157 | 4.04 | Coenzyme transport and metabolism |
| I | 188 | 4.83 | Lipid transport and metabolism |
| P | 178 | 4.58 | Inorganic ion transport and metabolism |
| Q | 130 | 3.34 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 524 | 13.47 | General function prediction only |
| S | 353 | 9.08 | Function unknown |
| - | 773 | 18.00 | Not in COGs |
Acknowledgements
Sequencing, assembly, annotation and data analysis for the first draft version were supported by the Gordon and Betty Moore Foundation Marine Microbiology Initiative, as part of its Marine Microbial Sequencing Project (http://www.moore.org/marinemicro). Support for the subsequent gap closure and analysis via the German Research Foundation (DFG) SFB/TRR 51 is gratefully acknowledged. We also thank the European Commission which supported phenotyping via the Microme project 222886 within the Framework 7 program.
References
- 1.Biebl H, Tindall BJ, Pukall R, Lünsdorf H, Allgaier M, Wagner-Döbler I. Hoeflea phototrophica sp. nov., a novel marine aerobic alphaproteobacterium that forms bacteriochlorophyll a. Int J Syst Evol Microbiol 2006; 56:821-826 10.1099/ijs.0.63958-0 [DOI] [PubMed] [Google Scholar]
- 2.Peix A, Rivas R, Trujillo ME, Vancanneyt M, Velazquez E, Willems A. Reclassification of Agrobacterium ferrugineum LMG 128 as Hoeflea marina gen. nov., sp. nov. Int J Syst Evol Microbiol 2005; 55:1163-1166 10.1099/ijs.0.63291-0 [DOI] [PubMed] [Google Scholar]
- 3.Sorokina AI, Chernousova EI, Dubinina GA. Hoeflea siderophila sp. nov., new neutrophilic iron-oxidizing bacteria. Mikrobiologiia 2012; 81:64-71 [PubMed] [Google Scholar]
- 4.Pradella S, Allgaier M, Hoch C, Päuker O, Stackebrandt E, Wagner-Döbler I. Genome organization and localization of the pufLM genes of the photosynthesis reaction center in phylogenetically diverse marine Alphaproteobacteria. Appl Environ Microbiol 2004; 70:3360-3369 10.1128/AEM.70.6.3360-3369.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korf I, Yandell M, Bedell J. BLAST, O'Reilly, Sebastopol, 2003. [Google Scholar]
- 6.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410 [DOI] [PubMed] [Google Scholar]
- 7.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-5072 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter MF. An algorithm for suffix stripping. Program: electronic library and information systems 1980; 14:130-137.
- 9.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 10.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 11.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 12.Hess PN, De Moraes Russo CA. An empirical test of the midpoint rooting method. Biol J Linn Soc Lond 2007; 92:669-674 10.1111/j.1095-8312.2007.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? Lect Notes Comput Sci 2009; 5541:184-200 10.1007/978-3-642-02008-7_13 [DOI] [Google Scholar]
- 14.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), Version 4.0 b10. Sinauer Associates, Sunderland, 2002. [Google Scholar]
- 15.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2012; 40:D571-D579 10.1093/nar/gkr1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: DJ Brenner, NR Krieg, JT Staley, GM Garrity (eds), Bergey's Manual of Systematic Bacteriology, second edition, vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria), Springer, New York, 2005, p. 1. [Google Scholar]
- 19.Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 1. [Google Scholar]
- 20.Validation List No 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 21.Kuykendall LD. Order VI. Rhizobiales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 324. [Google Scholar]
- 22.Mergaert J, Swings J. Family IV. Phyllobacteriaceae fam. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 393. [Google Scholar]
- 23.BAuA. 2012-update, Classification of Bacteria and Archaea in risk groups. http://www.baua.de TRBA 466, p. 104.
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaas LAI, Sikorski J, Michael V, Göker M, Klenk HP. Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 2012; 7:e34846 10.1371/journal.pone.0034846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW. The DNA Bank Network the start from a German initiative. [ 10.1089/bio.2010.0029] Biopreserv Biobank 2011; 9:51-55. [DOI] [PubMed]
- 27.Phrap and Phred for Windows, MacOS, Linux, and Unix.www.phrap.com
- 28.Markowitz VM, Mavromatis K, Ivanova NN, Chen IA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 29.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagesen K, Hallin PF, Rødland E, Stærfeldt HH, Rognes T, Ussery DW. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009; 25:1335-1337 10.1093/bioinformatics/btp157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics 2000; 16:944-945 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]