Abstract
The development of biomolecular imprinting over the last decade has raised promising perspectives in replacing natural antibodies with artificial antibodies. A significant number of reports have been dedicated to imprinting of organic and inorganic nanostructures, but very few were performed on nanomaterials with a transduction function. Herein we describe a relatively fast and efficient plasmonic hot spot-localized surface imprinting of gold nanorods using reversible template immobilization and siloxane co-polymerization. The technique enables a fine control of the imprinting process at the nanometer scale and provides a nanobiosensor with high selectivity and reusability. Proof of concept is established by the detection of neutrophil gelatinase-associated lipocalin (NGAL), a biomarker for acute kidney injury, using localized surface plasmon resonance spectroscopy. The work represents a valuable step towards plasmonic nanobiosensors with synthetic antibodies for label-free and cost-efficient diagnostic assays. We expect that this novel class of surface imprinted plasmonic nanomaterials will open up new possibilities in advancing biomedical applications of plasmonic nanostructures.
Keywords: Artificial antibodies, Gold nanorods, Localized surface plasmon resonance, Plasmonic biosensors, Macromolecular imprinting
1. Introduction
Biosensing platforms based on localized surface plasmon resonance (LSPR) or surface enhanced Raman scattering (SERS) hold enormous potential to provide highly sensitive, cost-effective, and point-of-care diagnostic tools.[1] However, similar to many other analytical methodologies such as enzyme-linked immunosorbent assays (ELISAs), plasmonic biosensors use natural antibodies. The use of natural antibodies in analytical methods is ubiquitous, with applications in disease diagnosis, toxicology testing, and biotechnology. Natural antibody production, either polyclonal antibodies achieved through animal immunization or monoclonal antibodies produced via cellular techniques, is expensive and time-consuming. Both the time and expense required for natural antibody production and their poor stability constitute a barrier to the rapid development and wide-spread application of plasmonic biosensors and clinical protocols for disease-specific screening. Macromolecular imprinting aims to overcome this problem by synthesizing specific artificial antibodies with high stability, reusability and cost-efficiency.[2, 3] Unlike other artificial antibodies such as aptamers that are made from biological molecules,[4-7] macromolecular imprinting typically involves the polymerization of an organic or organo-silicon monomer in the presence of a template (e. g., proteins). The resulting polymer presents a complementary conformation to the template and provides chemical interaction with its functional groups. A subsequent release of the template will then leave behind a polymeric recognition cavity with the desired shape and chemical functionality.
Despite the abundant literature dedicated to the imprinting of organic and inorganic nanostructures,[8-10] the implementation of molecular imprinting in plasmonic sensing has primarily been directed to planar gold surface[11-14] and/or mainly involved small molecule detection.[12, 13, 15-18] Although gold nanoparticles may enable LSPR spectroscopy and improve sensitivity, they have so far been used as a layer underneath or on top of a molecularly imprinted polymer (MIP) film.[13, 17] In these configurations, nanoparticles are not used as direct transduction elements but for enhancing Raman scattering from analyte molecules (SERS) or propagating SPR on planar gold surface. Other reported techniques involve embedding gold nanoparticles in a molecularly imprinted polymer or so-called Au-MIP nanocomposites,[13, 16, 19] which results in a random distribution of the nanoparticles and the molecular imprints. Biomacromolecular imprinting of noble-metal nanoparticles that takes full advantage of the unique structural and localized plasmonic properties of each individual nanoparticle continues to be a serious challenge.
Here, we demonstrate the first surface-imprinted LSPR nanosensor dedicated to protein detection and using sol-gel process in a localized fashion. Gold nanorods (AuNRs) were surface-imprinted by using siloxane co-polymerization to realize plasmonic nanostructures with built-in receptors. The fine control of the MIP film thickness by LSPR spectroscopy and the non-uniform distribution of the capping ligand around the nanorods were exploited to favor the molecular imprinting in areas that are most potent for plasmonic biosensing. Besides monitoring the imprinting process, LSPR spectroscopy is also used as a detection platform. The capture/release of the target biomolecule by the artificial antibodies induces a significant change in the refractive index of the layer surrounding the nanorods. The template recognition and binding is then detected as a shift in the LSPR wavelength that is dependent on the concentration of the analyte. The results were confirmed by Western blotting as an independent technique.
In this study, bovine serum albumin (BSA) bovine hemoglobin (Hb) and recombinant human neutrophil gelatinase-associated lipocalin (NGAL) are employed as target analytes. BSA and hemoglobin concentration measurement is among the most commonly performed blood tests, while NGAL is a novel promising specific urinary biomarker of acute kidney injury.[20, 21] Considering the high occurence of kideny-related diseases and clinical importance of their early diagnosis (see Supporting Information), there is a critical need for a low-cost, simple, stable and reliable NGAL assay.
2. Results and Discussion
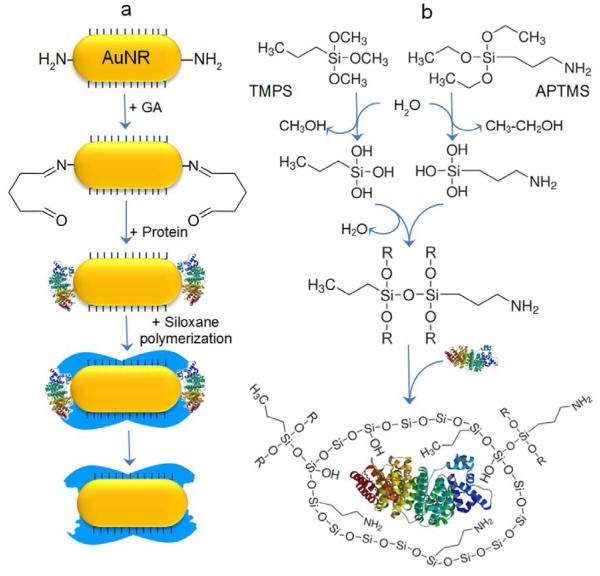
The imprinting process first involves the preparation of gold nanorods and their adsorption onto a glass substrate. Cetyltrimethyl-ammonium bromide (CTAB)-capped AuNRs with aspect ratio ranging from 2 to 3.5 are synthesized using a seed-mediated approach.[22, 23] The nanorods are then adsorbed onto poly(2-vinyl pyridine)-modified glass substrate (Supporting Information Figure S1). The immobilization of the protein templates is achieved by exposing the AuNRs to a mixture of p-aminothiophenol (p-ATP) and glutaraldehyde (GA). In aqueous solutions, p-ATP binds spontaneously to gold surface with its thiol group, while GA molecules form oligomers of variable size with a free aldehyde group at each end of the oligomer molecules. As a result, GA plays a role of a crosslinker between the amine groups of p-ATP molecules and the amine moieties on the side chains of the protein templates by forming unstable imine bonds in basic pH buffer solution (Figure 1a).[24] Following the immobilization of the template, the organo-siloxane monomers trimethoxypropylsilane (TMPS) and (3-aminopropyl)trimethoxysilane (APTMS), which are hydrolytically unstable, are co-polymerized onto the modified AuNR surface. While the Si-C bond and aminopropyl group cannot be cleaved, the ethoxy groups of APTMS and methoxy groups of TMPS undergo rapid hydrolysis to produce ethanol, methanol and trisilanols (Figure 1b). The subsequent condensation of the transient silanol groups yields an aminopropyl-functional amorphous polymer and entrapment of the protein templates. This sol-gel approach is known to be very versatile and flexible technique,[25-27] and has been used for molecularly imprinting in a variety of sensors.[28-33] The templates are finally removed by breaking the imine bonds of the cross-linker using a mixture of sodium dodecyl sulfate and oxalic acid.
Figure 1.
Concept and mechanism of plasmonic hot spot-localized imprinting of gold nanorods. a) Surface chemistry of the molecular imprinting process involving the preferential attachment of p-ATP to the ends of the AuNR, followed by glutaraldehyde (GA) interaction with the primary amine groups of p-ATP on one side and with the amine functions of the protein on the other side. Finally, siloxane monomers are polymerized in the presence of the protein templates. The release of the protein results in a polymeric recognition cavity (in blue). b) Co-polymerization reaction of the organo-siloxane monomers APTMS and TMPS. In aqueous environment, the monomers undergo hydrolysis then condensation to yield amorphous siloxane copolymer.
The use of APTMS and TMPS in aqueous media provides a polymer with amine (NH +3), hydroxyl (OH) and methyl (CH3) functional groups. This is of great importance as the concerted weak interactions, namely electrostatic, hydrogen bonding and hydrophobic interactions, are believed to be the most dominant form of interaction in template-receptor complexes.[34] The composition ratio of the siloxane co-polymerization has been adjusted to obtain the best trade-off between flexibility and mechanical strength.[35]
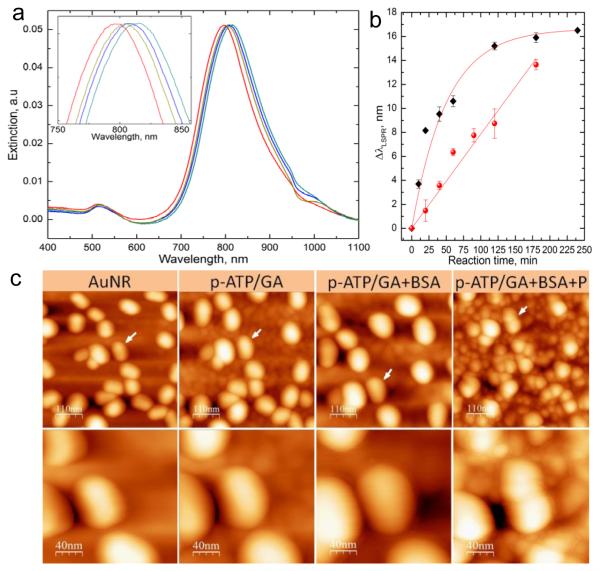
Effective application of molecular imprinting technology to plasmonic nanoparticles requires the consideration of two important parameters related to the transduction function properties. The first is the exponential decay of the electric field at the surface of the gold nanoparticles (Supporting Information Figure S2). Even though the decay length of the electric field in nanoparticles is around 20 nm, the sensitivity decreases dramatically with increasing distance from the surface and drops by more than 50% at a distance of 20 nm.[36] Only the close vicinity of the nanorod (< 10 nm) exhibits optimal refractive index sensitivity.[37] As a result, the thickness of the film separating the protein templates from the nanoparticles needs to be minimized. The functionalization of the AuNRs by the cross-linker, proteins or the siloxane polymer induces a shift in the LSPR wavelength (Figure 2a). Thus, we have used LSPR of AuNRs to monitor the growth of the crosslinker and siloxane copolymer layers at the nanoscale level. Figure 2b shows the increase in the LSPR wavelength shift as a function of the reaction time of both the mixture p-ATP/GA and the organo-siloxanes monomers. The growth of the crosslinker layer could be approximated with an exponential decay fit with a maximum shift of around 16 nm after 2 h of reaction. This limit does not reflect the limitation of the film growth but rather indicates the exponential decay in LSPR sensitivity with increasing distance from the nanorod surface. On the other hand, the growth of the siloxane co-polymer should theoretically follow the same exponential decay behavior. However, the reaction in this case is much slower and the initial stages of the copolymer growth could be approximated with a linear fit in the range studied here and exhibits a wavelength shift rate of Δλ/t ≈ 0.1 nm/min.
Figure 2.
Structural characterization of the protein imprinting procedure. a) Extinction spectra of the AuNR following each step. b) Plot depicting the shift in the LSPR wavelength as a function of the reaction time of p-ATP/GA (black squares) and siloxane co-polymerization (red circles). c) AFM images after treatment with the cross-linker (p-ATP/GA), immobilization of the protein (BSA) and co-polymerization of the siloxanes (P). The white arrows indicate the same nanorods after each step. Higher magnification AFM images are shown in the second row.
To evaluate the actual film thickness corresponding to the LSPR shift rate, we have performed AFM imaging of the same nanorods following each step of the imprinting process. As shown in Figure 2c, the size of the nanorods increases after each step and the thickness measurement indicates that a 1 nm shift in the LSPR wavelength corresponds to 1.2±0.3 nm organic layer thickness for both the cross-linker film and siloxane co-polymer. Based on these results, the optimal conditions for surface biomacromolecular imprinting were identified to be p-ATP/GA treatment for 10 min followed by 40 min polymerization of the siloxane monomers. These conditions lead to a ~3.5 nm thickness increase for each step, thus keeping the protein templates at less than 5 nm distance from the nanorods surface while resulting in a partial coverage of the protein templates with the siloxane copolymers.
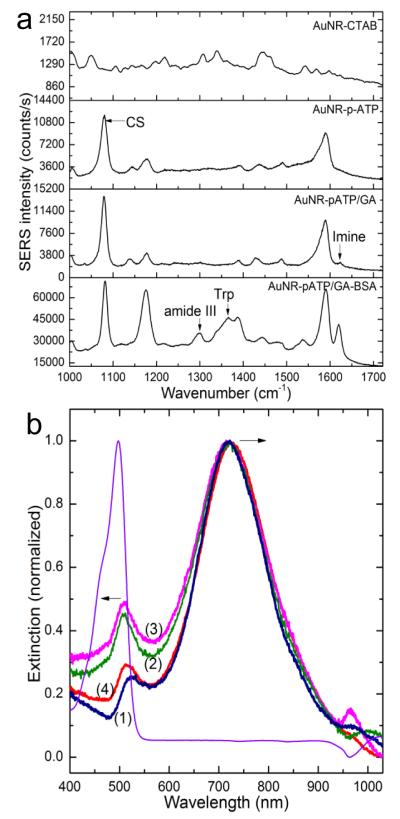
To further investigate the imprinting process, the chemical composition at the surface of plasmonic transducers after each step was analyzed by SERS spectroscopy. The functionalization of the AuNRs by p-ATP/GA and proteins results in the appearance of specific bands for each component as shown in Figure 3a. The functionalization of the AuNRs by p-ATP/GA results in the appearance of a specific band at 1083 cm−1 corresponding to CS stretching vibration with a contribution from C-N stretching of the amine group. The interaction of GA with p-ATP on one side and GA with the protein amine groups on the other side is evidenced by the appearance of the imine band at 1620 cm−1 after mixing GA with p-ATP and the significant increase in intensity of the same band after addition of the protein. The presence of the proteins is also confirmed by specific bands at 524 cm−1 and 724 cm−1 due to stretching of disulfide bridges (S-S) and C-S stretching of cysteine respectively. The bands at 833 cm−1, 1298 cm−1 and 1365 cm−1 are assigned to tyrosine, alpha helix of amide III and tryptophan, respectively. More details on the SERS bands are provided in Figure S3 and Table S1 in Supporting Information.
Figure 3.
Characterization of protein immobilization and release. a) Surface enhanced Raman scattering spectra obtained from the AuNR at each step of the imprinting process. b) Normalized extinction spectra of fluorescein-conjugated BSA (f-BSA) at 498 nm (purple) and AuNR before (1) and after immobilization of the template f-BSA (2), followed by a treatment with oxalic acid (3) or a mixture of oxalic acid and SDS (4) to release the template. The black arrows indicate the shift in opposite direction of the longitudinal (720 nm) and transverse (523 nm) plasmon bands. The exposure of the AuNRs to BSA solution results in a red shift of the longitudinal band by 5 nm as expected, while the transverse band undergoes an apparent blue shift by 10 nm. The apparent blue-shift is due to the effect of fluorophore absorption band localized at slightly lower wavelength.
The confirmation of the protein template attachment is also achieved by using fluorescein-conjugated BSA (f-BSA). This experiment is also designed to ascertain that the shift in the resonance wavelength observed in LSPR spectroscopy is due to the protein templates. Fluorescein is a synthetic fluorophore with an absorption maximum at 497 nm and emission maximum at 521 nm in aqueous solution. The localization of the fluorophore absorption peak next to the transverse plasmonic band (at ~525 nm) makes it useful to detect the immobilization of the fluorescein-conjugated BSA on the nanorod surface by monitoring the fluorophore-induced changes of the extinction band (Figure 3b). The obtained results are in agreement with the previous results and confirms the effectiveness of the template immobilization procedure and its monitoring by LSPR. This experiment also reveals that the treatment with oxalic acid results in a release of 20 % ± 4% of the template (f-BSA), which is relatively close to the value reported in literature[35]. When a mixture of oxalic acid and Sodium dodecyl sulfate (SDS) is used, the template release improves to 58 ± 4% (Supporting Information, Figure S4). This efficiency is explained by the surfactant effect but also by the fact that the template is incubated in the siloxanes for only 2 h as opposed to 72 h in previous report.[35] The drawback of this technique is that the template release is accompanied with a material loss from the polysiloxanes, which could affect the number or specificity of the artificial antibodies.
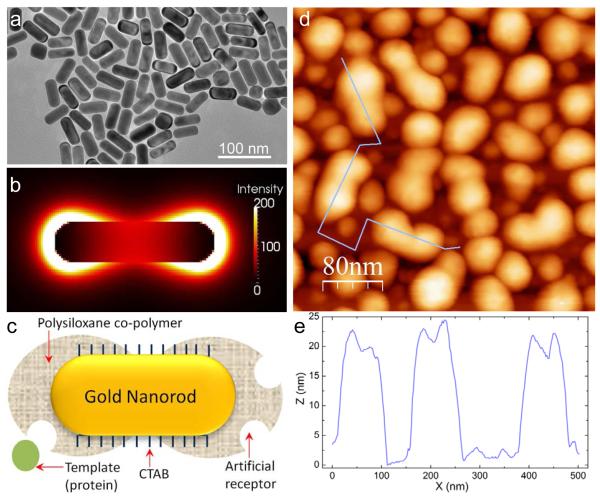
The second important consideration in molecular imprinting of gold nanoparticles is the localization of the plasmonic hot-spots. The distribution and concentration of the electric field around AuNPs mostly depend on their shape.[38] In the case of gold nanorods, the extinction spectrum exhibits a weak transverse (~515 nm) and much stronger and more sensitive longitudinal (~700 nm) plasmon resonances. The challenge is then to favor the molecular imprinting at the nanorod ends where the plasmonic hot-spots are localized (Figure 4a, b and c). The peanut-shaped structures observed in the AFM image represent imprinted nanorods at the optimum experimental conditions as discussed earlier (Figure 4d). The unique shape is explained by the distribution of the capping ligand, i.e. CTAB molecules on the nanorod surface. It is well-known that the nanorod ends are much less covered with CTAB molecules as compared to the sides, which enables their linear end-to-end assembly in some applications.[39] The same property leads here to a preferential adsorption of p-ATP/GA molecules at the nanorod ends and consequently to more available chemical anchors and faster growth of the siloxane polymer. The AFM profile shows that the siloxane polymer is 3-5 nm thicker than that on the nanorods sides, which suggests that there is no significant film growth on the side-wall surface of the nanorods. As a result, the molecular imprinting is mainly localized at the plasmonic hot-spots, which provides a maximum sensitivity in LSPR-based detection.
Figure 4.
Hot spot-localized imprinting of AuNRs. a) Representative TEM image of AuNR. b) Cross-sectional view of the electric field distribution around AuNR at the extinction maximum of the longitudinal band (724 nm in Fig. 2e). The image is obtained by finite-difference time-domain (FDTD) modeling. c) Scheme showing the preferential growth of the siloxane co-polymer at the nanorods ends where the capping ligand (CTAB) is sparse. Figures b and c show the spatial matching of the imprinted area with the localization of the plasmonic hot-spots. d) AFM image (scan size 400 nm × 400 nm) depicting the peanut-shaped MIP-AuNRs, corresponding to the scheme in Fig. c. The violet line indicates the localization the height profile represented in Fig. e. We can see that MIP-AuNR thickness id 3-5 nm higher at the ends than in the center of the nanorods.
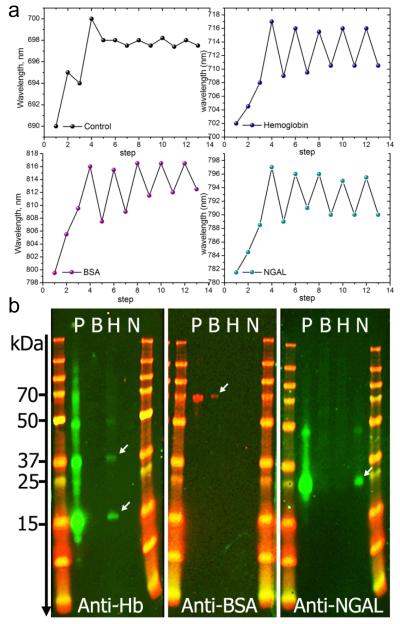
To demonstrate the template rebinding, reproducibility and reusability of the imprinted nanosensors, three different nanorod-coated glass substrates were imprinted with BSA, hemoglobin or NGAL proteins. A fourth sample that underwent the same imprinting procedure but without using protein templates was used as a control. As shown by the extinction spectra (Supporting Information Figure S5) and Figure 5a, the accumulated shift due to the imprinting process is ~16 nm. This is true for all the samples used except the control that only shows ~10 nm shift. This is expected as no protein is used in this sample. Instead of a red-shift, a blue-shift is observed for the control sample at step 2 (Figure 5a), which is likely due to a loss of weakly adsorbed material (CTAB, GA) after 2 h of incubation in PBS buffer. Likewise, the first release of the template in the different samples suggests that the protein template removal is accompanied by the loss of weakly polymerized siloxane, leading to a blue shift of ~8 nm rather than the expected ~5 nm for protein removal. The following cycles of protein capture and release show a better stability of the imprinted AuNR surface and demonstrate excellent reproducibility for all the proteins used. The shifts induced by the capture/release cycles are around 5 nm for the molecularly imprinted supports, while they are 10 times smaller for the control, demonstrating a very good efficiency of the artificial antibodies. The small shifts observed for the control are mostly caused by non-specific adsorption of the protein. The same results were obtained with two other proteins, i.e immunoglobulin G (IgG) and allophycocyanin (Supporting Information Figure S6)
Figure 5.
Reproducibility and selectivity of the MIP-AuNR nanosensors. a) Shift of the LSPR wavelength following the different steps of the imprinting process. Each measurement point represents the shift obtained at the end of each step indicated with numbers. Steps 1 to 4 correspnond to AuNR, AuNR-pATP/GA, AuNR-pATP/GA-protein and AuNR-pATP/GA-protein-siloxane copolymer, respectively. At step 3, the control was exposed to PBS buffer solution without proteins. Steps 5 to 13 correspond to 4 cycles of protein capture and release, resulting in a red-shift or blue-shift, respectively. The same procedure is applied to hemoglobin, BSA and NGAL biomarker. The control was not treated with proteins. b) Western blotting of the elute solutions obtained from molecularly imprinted nanorod substrate prepared with 3 different protein templates: Bovine Hemoglobin (Hb), Bovine Serum Albumin (BSA) and recombinant human NGAL biomarkers. Each panel contains two molecular weight marker columns flanking 4 migration lanes: P is the protein mixture (containing Hb, BSA and NGAL) applied to all prepared MIP-AuNR substrates. Lines B, H and N contain the elute solutions obtained from the MIP-AuNR substrates imprinted separately with BSA, Hb and NGAL respectively. The treatment with the antibody Anti-Hb, reveals 2 clearly identified bands in the H lane indicating the presence of denaturated Hb with a single sub-unit (~ 17 kDa) or two sub-units (~ 37 kDa). A weak band at ~ 50 kDa indicates the presence of a 3 sub-units Hb. The second panel treated with anti-BSA reveals a weak band at ~70 kDa demonstrating the presence of BSA. The anti-NGAL treatment of the last paned shows a clear band at ~ 25 kDa, corresponding to NGAL molecular weight. A weak band is also observed at ~ 45 kDa indicating the presence of NGAL concatemers due to a small amount of recombinant NGAL resulting from plasmid concatemerization.
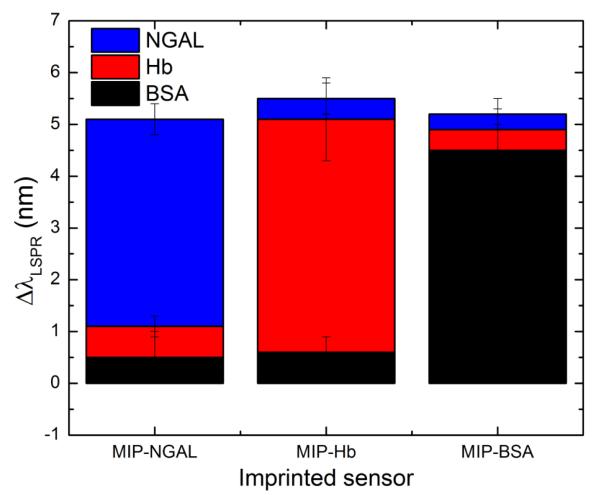
The reproducibility of the detection immediately raises the next important question in molecular imprinting which is the specific recognition capability of the imprinted cavities. To investigate this aspect, competitive binding experiment was performed. The prepared substrates were challenged with a mixture of the three proteins. After thorough rinsing, the proteins were released and the elute solutions were separately analyzed with Western blotting (Figure 5b). Despite the exposure of the MIP-AuNR substrates to a mixture of three different proteins, only one protein was recognized in each panel, corresponding to the template used for imprinting. This is in agreement with the LSPR measurements performed with the imprinted sensors on different protein mixtures (Supporting Information Figure S7). It is interesting to note that the LSPR shift of the extinction spectra is also accompanied by an increase in the full width at half maximum (FWHM) (Supporting Information Figure S8). Both Western blotting and LSPR results clearly demonstrate the high specificity and selectivity of the imprinted nanorods. Cross-binding experiment was also realized to assess the contribution of non-specific protein adsorption (Figure 6). The three imprinted sensors were separately and successively exposed to each protein and the results show that non-specific binding induces a shift of less than 1 nm in all cases, which is an indication of a relatively low level of adsorption.
Figure 6.
Cross-binding experiment for the evaluation of non-specific adsorption. The sensors imprinted with three different proteins (MIP-NGAL, MIP-Hb and MIP-BSA) were separately and successively exposed to NGAL, Hb and BSA proteins and the induced LSPR wavelength shift was monitored.
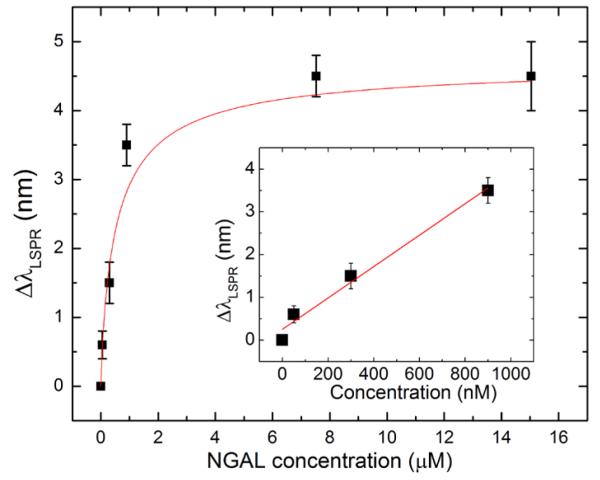
To complete the analytical investigation of the molecularly imprinted nanorods, different concentrations of NGAL were used to evaluate the detection sensitivity. Figure 7 clearly shows that a plateau is reached at concentrations higher than 16 μM (400 μg/mL). However, a linear relationship could be applied to concentrations lower than 1 μM. This dynamic range offers a LSPR sensitivity of 0.25 nm/nM and a detection limit of 13 nM or 0.32 μg/mL of NGAL (the wavelength precision of the optical detection system is ± 0.1 nm). This performance surpasses that afforded by the molecular imprinting of other traditional techniques such as quartz crystal microbalance and liquid chromatography,[40] and is very suitable for the detection of many proteins at clinical concentrations including NGAL biomarkers after kidney injury.[21] However, the challenge remains to approach the performance of natural antibodies in a comparable detection platform.[41] The sensitivity could be significantly improved in the future by achieving molecular imprinting of strongly coupled plasmonic nanostructures, which offer significantly higher sensitivity.[42]
Figure 7.
Sensitivity of the MIP-AuNR nanosensor expressed by the shift in the LSPR wavelength in nm as a function of NGAL concentration in μM. The data are fitted with an exponential decay function. The linear fit (in the inset) could be applied for protein concentrations lower than 1 μM, providing a sensitivity of 0.6 nm/μM (R2=0.98).
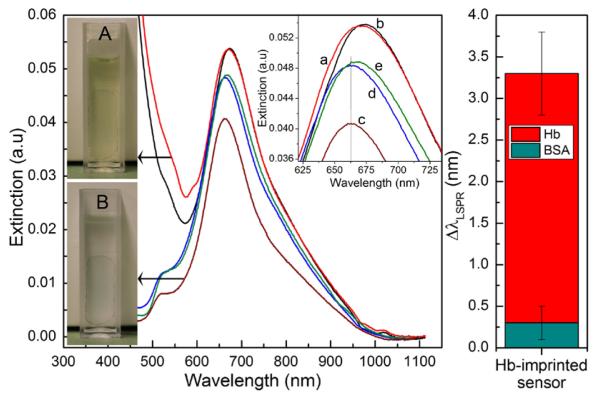
To further demonstrate the implementation of the imprinted AuNR sensors in real-world and complex samples, we have performed hemoglobin detection in a normal urine sample (Figure 8). The presence of hemoglobin in urine; i.e., hemoglobinurea is a pathological condition, which can lead to acute tubular necrosis. A Hb-imprinted sensor was first immersed in a urine solution to check the effect of this complex sample containing a variety of organic and inorganic components on the extinction spectrum. As shown in Figure 8, the longitudinal peak at λ≈ 662 nm used for sensing remains intact, while the transverse peak is overlapped by the huge absorbance of urine at λ < 500 nm. This absorbance is caused by urine pigments including urobilin which is the final product of heme breakdown. To decrease the effect of this absorbance/fluorescence on the plasmon resonance peaks, the urine sample was diluted 5 times before use. The AuNR sensor was then incubated in a urine sample containing 30 μg/mL of either hemoglobin or BSA. After washing with distilled water, the extinction spectra of the sensors were obtained in water. The results show that a very significant LSPR shift is observed for hemoglobin (Δλ=3.5 nm ± 0.5 nm), while the change caused by BSA remains insignificant (Δλ < 0.5 nm). This is a clear indication of the successful specific capture of hemoglobin by the imprinted nanorods and a demonstration of the feasibility of antibody-free plasmonic nanosensing for real-world applications.
Figure 8.
Application of AuNR-imprinted sensors for the detection of hemoglobin in a real urine sample. (A) Image of the sensor substrate during incubation in urine sample. (a) and (b) represent respectively the extinction spectra of the sensor immersed in cuvette (A) before and immediately after addition of 30 μg/mL hemoglobin. After incubation in urine sample without or with the presence of target proteins, the sensor is washed and immersed in water (cuvette B) for LSPR analysis. The graphs (c-e) represent respectively the extinction spectra of the sensor substrate after incubation in normal urine (c) or after incubation in urine sample containing either BSA (d) or hemoglobin (e). The diagram on the right depicts the shift in the maximum wavelength of the Hb-imprinted nanorods after exposition to urine containing either Hemoglobin or BSA.
3. Conclusions
In conclusion, we have demonstrated an efficient technique for biomacromolecular imprinting of gold nanorods using reversible cross-linking and organo-siloxane co-polymerization. The anisotropic distribution of the nanorods capping ligand was successfully exploited to specifically localize the protein imprinting at the plasmonic hot-spots. The technique provides polymer artificial antibodies with a high selectivity. The localized plasmonic activity of the nanorods enables not only the step-by-step monitoring of the imprinting process, which provides a new practical tool for molecular imprinting research, but also the direct detection of the protein capture and release at physiologically relevant concentrations. Molecular imprinting of nanorods to achieve synthetic antibodies on their surface opens up a novel class of plasmonic nanostructures with built-in biorecognition capability. The technical and conceptual development reported here are likely to open up new possibilities in biomedical applications of plasmonic nanostructures particularly in label-free and antibody-free diagnostic tools.
4. Experimental Section
Nanoparticle synthesis and deposition
AuNRs are synthesized using a seed-mediated approach.[22, 23] Seed solution was prepared by adding 1 ml of an ice-cold solution of 10 mM sodium borohydride to 10 ml of 0.1 M cetyltrimethylammonium bromide (CTAB) and 2.5 × 10−4 M HAuCl4 (aq.) solution under magnetic stirring at room temperature. The color of the solution changed from yellow to brown. Growth solution was prepared by mixing 95 ml of 0.1 M CTAB, 1 ml of 10 mM silver nitrate, 5 ml of 10 mM HAuCl4, and 0.55 ml of 0.1 M ascorbic acid in the same order. The solution was homogenized by gentle stirring. To the resulting colorless solution, 0.12 ml of freshly prepared seed solution was added and set aside in the dark for 14 h. The solution turned from colorless to violet brown with most of the color change happening in the first hour. Prior to use, AuNR solution is centrifuged twice at 13,000 rpm for 10 min to remove excess CTAB and then redispersed in nanopure water. The synthesized nanorods have an average size 20 nm x 60 nm.
Nanorod-coated glass and silica substrates were prepared by first exposing the piranha-cleaned glass surface to 4% poly(2-vinylpyridine) (P2VP) solution in ethanol for 1 h, followed by rinsing with ethanol. The modified glass substrates were then exposed to nanorod solution overnight. Finally the substrates were thoroughly rinsed with water to remove the weakly adsorbed nanorods and then dried with nitrogen flux.
Macromoleular imprinting procedure
The molecularly imprinted AuNRs were prepared in three steps: first, the nanorod-coated glass substrate were immersed in a freshly prepared 2 mL phosphate buffer saline solution (PBS, pH8.3) containing 20 μL of glutaraldehyde (25%) and 20 μL of pATP (4 mM in ethanol). The immersion time determines the thickness of the crosslinker film (p-ATP/GA). An immersion time of 10 min provides an LSPR shift of 2-3 nm, corresponding to ~3 nm thickness. The substrates are then rinsed gently with buffer and kept in the same buffer solution. Protein immobilization was performed by exposing the functionalized substrates to 1 mg/mL bovine serum albumin (BSA; pdb ID: 1E7I, Ip=4.7, MW=66.5 kDa), bovine hemoglobin (Hb; pdb ID: 2HHB, Ip= 6.8, MW=64.5 kDa) or 0.5 mg/mL recombinant human neutrophil gelatinase-associated lipocalin (NGAL; pdb ID: 3TZS, Ip= 8.9, MW=25 kDa) all prepared in PBS solution (pH 8.3). The exposure was performed under gentle shaking for 30 min followed by 2h incubation at 4°C. After rinsing with the same buffer solution, the protein-coated substrates were immersed in 3 mL phosphate buffer (pH 7.5) to which 5 μL TMPS and 5 μL APTMS were freshly added. The immersion time determines the thickness of the polysiloxane co-polymer. An immersion time of 40 min results in a polymer thickness of ~5 nm. The samples were then gently rinsed with PBS solution (pH 7.5) and stored in the same buffer overnight at 4°C. The last step of the molecular imprinting process is the template release. The captured protein templates inside the siloxane co-polymer is extracted by exposing the imprinted substrates to a mixture of 2% SDS and 2 mM oxalic acid for 1 h.
Characterization techniques
The synthesis, deposition and functionalization of AuNRs were monitored through their UV-visible extinction spectra collected using Shimadzu 1800 spectrophotometer. TEM images were obtained using a field emission transmission electron microscope (JEM-2100F, JEOL) operating at an accelerating voltage of 200 kV. AFM images were acquired using a Dimension 3000 AFM system (Bruker) in tapping mode using a silicon nitride cantilever. The change in shape and thickness of the modified nanorods during the imprinting process was monitored by imaging the same nanorods after each step of the process. This is made possible by performing successive scans starting with a large 50 μm × 50 μm scan size and ending with a 200 nm × 200 nm scan size (Supporting Information Figure S9).
The chemical composition of the imprinted gold nanorods was studied by comparing their surface enhanced Raman spectra collected using a Renishaw inVia confocal Raman spectrometer mounted on a Leica microscope with 50X objective (NA =0.90) in the range of 100–3200 cm−1. A diode laser of 785 nm wavelength (0.5 mW) is used for sample excitation.
Electromagnetic modelling
The modeling of the electromagnetic field distribution around plasmonic nanorods was performed using three-dimensional finite-difference time-domain (FDTD) technique with commercially available software (EM Explorer). FDTD simulations exploit the time and position dependence of Maxwell’s equations to model electromagnetic waves in rectangular 3D cells of finite volume called Yee cells[43]. A single AuNR of 20 nm x 80 nm size is modeled in a simulation domain of 300 nm × 300 nm × 200 nm. First, a wavelength scanning mode (300 nm-1100 nm) was performed to obtain the extinction profile of the nanorod using p-polarized incident plane wave for illumination. Perfectly matched layer (PML) absorbing boundary conditions were applied in all directions. Then, a higher resolution simulation (Yee cell size of 2 nm) was run at the extinction maximum wavelength (λ = 724 nm) to obtain the electromagnetic field distribution. The complex refractive index of gold at this frequency was set to n = 0.18+i 4.96.[44]
Competitive and cross-binding experiments
The competitive binding experiment was achieved by immersing each one of the three imprinted sensors (MIP-NGAL, MIP-BSA and MIP-Hb) in a mixture of proteins containing 0.5 mg/mL BSA, 0.5 mg/mL Hb and 0.5 mg/mL NGAL and incubated for 30 min at room temperture. The sensors were then washed with PBS buffer in three different bath for 5 min each and under gentle shaking. Each sensor substrate is finally and separately exposed to a mixture of SDS (2%) and oxalic acid (2 mM) for 30 min under gentle shaking, and the eluate is analysed by Western-blotting. Th cross-binding experiment was realized by exposing succcessively each imprinted sensor in 0.5 mg/mL NGAL, BSA and finally Hb for 30 min each. After each treatment, the shift in the maximum resonance wavelength was monitored with the UV-visible spectrometer (Figure 6).
Western blotting protocol
Samples of the initial applied mixture of the 3 proteins and each specific eluate (30 μL) were mixed with 10 μl of 4x SDS sample buffer (LI-COR Biosciences, Lincoln, NE) containing mercaptoethanol and heated at 90C for 10 min. The sample representing the applied protein mixture was further diluted 10-fold with 1x SDS sample buffer. Five μL of each sample was applied to a 17-well 4-12% acrylamide BIS-TRIS gel (Invitrogen) and electrophoresed for 32 min at 200V, and the proteins transferred to nitrocellulose by means of an i-BLOT (Invitrogen, a division of Life Technologies, Grand Island, NY). The membrane was blocked in LI-COR blocking buffer, cut into segments, and incubated with antibodies specific to mouse albumin (abcam, Cambridge, MA) rabbit anti-mouse antibody that detects human serum albumin and that of other species with less affinity 1/10,000, human hemoglobin (R&D Systems, Minneapolis, MN) goat antibody 0.2 μg/ml, or human NGAL (R&D Systems, Minneapolis, MN) goat antibody 0.2 μg/ml; each diluted in LI-COR blocking buffer containing 0.1% Tween-20 (Sigma Chemical Company, St. Louis, MO) overnight. Membranes were washed 3 times in phosphate buffered saline (PBS) containing 0.1% Tween-20 and each membrane incubated with donkey anti-rabbit IgG IRDye 680 or donkey anti-goat IgG IRDye 800 (LI-COR Biosciences, Lincoln, NE) as appropriate for 1 h. The membranes were washed 3 times as above, one time with PBS, and visualized by means of an Odyssey Infrared Imager (LI-COR Biosciences, Lincoln, NE).
Supplementary Material
Figure S1: Extinction spectra of gold nanorods in (a) solution and on (b) glass substrates. For comparison purposes, the blue line (c) represents spectrum b with a different scale. The transfer of the nanorods from solution to a glass substrates results in a decrease of the extinction intensity by 10 to 50 times and a red-shift by ~30 nm. This shift is caused by the significant increase in refractive index in one side of the nanorods from n=1.33 (water) to n=1.5 (glass substrate).
Figure S2. Profile and distribution of the electric field intensity around the gold nanorods. The intensity at the nanorods ends is one order of magnitude higher than that at the center of the nanorods. The intensity, however, decreases exponentially by increasing distance from the nanorods surface. The x-axis is in nm.
Figure S3. Surface enhanced Raman scattering of molecularly imprinted AuNRs after BSA protein capture. The assignments of the different bands are indicated in Table S1.
Figure S4. Template release using oxalic acid and SDS. Fitting of the extinction spectra provides the integrated surface area (A) of the peak due to the template fluorescein conjugated BSA at λ≈ 500 nm. (1) AuNR before template immobilization (A= 4.28), (2) AuNR after template immobilization (A= 9.39), (3) template release using oxalic acid (A= 8.33), template release using a mixture of oxalic acid and SDS (A= 6.4).
Figure S5. Extinction spectra of Hemoglobin (a, b), NGAL (c, d) and BSA (e, f) capture/release cycles with molecularly imprinted AuNRs. The spectra represent AuNRs (black line) before and after successive addition of glutaraldehyde/p-ATP (pink line), proteins (green line) and siloxane copolymers (blue line with a star). The red and blue lines represent different cycles of protein capture (blue) and release (red). These different steps and cycles correspond to the data plotted in Figure 5a.
Figure S6. Extinction spectra of IgG (a) and allophycocyanin (b) capture/release cycles with molecularly imprinted AuNRs. The LSPR shifts are depicted in the insets. NR: imprinted nanorods, C: template capture, R: template release.
Figure S7. Localized surface plasmon resonance analysis of different protein mixtures using three different imprinted sensors with a specificity for (a) hemoglobin, (b) BSA and (c) NGAL proteins. The black line labelld C represents the extinction spectrum of the sensor after expositition to buffer. The green and red spectra are respectively obtained after exposition of the imprinted sensor to mixtures of two or three different proteins. A shift of 4-5 nm is observed only when the sensor is exposed to a mixture containing the protein used for imprinting (red line). The other spectra exhibit less than 1 nm LSPR shift.
Figure S8. Change of the full width at half maximum (FWHM) of the extinction spectra before and after protein capture with the imprinted sensors. The data are obtained from the analysis of the spectra depicted in Figure S7. In all cases, the increase in FWHM (Δ FWHM) is lower than 10 %. This change seems to increase with increasing protein size but more investigation with a larger number of proteins is needed to understand the different contributions to the increase in FWHM.
Figure S9: Atomic Force Microscopy (AFM) images depicting the multiple scan size used to localize and monitor thickness change of the MIP-AuNRs. The scan sizes are (a) 50 um x 50 um, (b) 5 um x 5 um, (c) 2 um x 2 um, (d) 760nm x 760 nm, (e) 368 nm x 368 nm.
Table S1. Selected SERS bands of CTAB, p-ATP and Proteins with their vibrational
Acknowledgments
This work was supported by Office of Congressionally Directed Medical Research Programs (DoD-Army) (W81XWH-11-1-0439) and St. Louis Institute of Nanomedicine and the National Institutes of Health R01CA141521 to JJM.
Footnotes
Supporting Information Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Abdennour Abbas, Department of Mechanical Engineering and Materials Science. Washington University in St. Louis, St Louis, MO, 63130 (USA).
Dr. Limei Tian, Department of Mechanical Engineering and Materials Science. Washington University in St. Louis, St Louis, MO, 63130 (USA).
Prof. Jeremiah J. Morrissey, Department of Anesthesiology, Division of Clinical and Translational Research, Washington University in St. Louis, St Louis, MO, 63110, (USA)
Prof. Evan D. Kharasch, Department of Anesthesiology, Division of Clinical and Translational Research, Washington University in St. Louis, St Louis, MO, 63110, (USA)
Prof. Srikanth Singamaneni, Department of Mechanical Engineering and Materials Science. Washington University in St. Louis, St Louis, MO, 63130 (USA)..
References
- [1].Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Nat. Mater. 2008;7:442. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- [2].Ye L, Mosbach K. Chem. Mater. 2008;20:859. [Google Scholar]
- [3].Hayden O, Lieberzeit PA, Blaas D, Dickert FL. Adv. Funct. Mater. 2006;16:1269. [Google Scholar]
- [4].Guo L, Kim D-H. Chem. Commun. 2011;47:7125. doi: 10.1039/c1cc11865j. [DOI] [PubMed] [Google Scholar]
- [5].Song S, Wang L, Li J, Fan C, Zhao J. TrAC, Trends Anal. Chem. 2008;27:108. [Google Scholar]
- [6].Kim NH, Lee SJ, Moskovits M. Adv. Mater. 2011;23:4152. doi: 10.1002/adma.201101847. [DOI] [PubMed] [Google Scholar]
- [7].Fabris L, Schierhorn M, Moskovits M, Bazan GC. Small. 2010;6:1550. doi: 10.1002/smll.201000262. [DOI] [PubMed] [Google Scholar]
- [8].Hu X, Li G, Li M, Huang J, Li Y, Gao Y, Zhang Y. Adv. Funct. Mater. 2008;18:575. [Google Scholar]
- [9].Carter SR, Rimmer S. Adv. Funct. Mater. 2004;14:553. [Google Scholar]
- [10].Cai D, Ren L, Zhao H, Xu C, Zhang L, Yu Y, Wang H, Lan Y, Roberts MF, Chuang JH, Naughton MJ, Ren Z, Chiles TC. Nature nanotech. 2010;5:597. doi: 10.1038/nnano.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pernites R, Ponnapati R, Felipe MJ, Advincula R. Biosens. Bioelectron. 2011;26:2766. doi: 10.1016/j.bios.2010.10.027. [DOI] [PubMed] [Google Scholar]
- [12].Frasconi M, Tel-Vered R, Riskin M, Willner I. Anal. Chem. 2010;82:2512. doi: 10.1021/ac902944k. [DOI] [PubMed] [Google Scholar]
- [13].Matsui J, Akamatsu K, Hara N, Miyoshi D, Nawafune H, Tamaki K, Sugimoto N. Anal. Chem. 2005;77:4282. doi: 10.1021/ac050227i. [DOI] [PubMed] [Google Scholar]
- [14].Lautner G, Kaev J, Reut J, Öpik A, Rappich J, Syritski V, Gyurcsányi RE. Adv. Funct. Mater. 2011;21:591. [Google Scholar]
- [15].Bompart M, De Wilde Y, Haupt K. Adv. Mater. 2010;22:2343. doi: 10.1002/adma.200904442. [DOI] [PubMed] [Google Scholar]
- [16].Tokareva I, Tokarev I, Minko S, Hutter E, Fendler JH. Chem. Commun. 2006:3343. doi: 10.1039/b604841b. [DOI] [PubMed] [Google Scholar]
- [17].Bompart M, Haupt K, Ayela C. In: Molecular Imprinting. Haupt K, editor. Vol. 325. Springer; Berlin / Heidelberg: 2012. p. 83. [Google Scholar]
- [18].Liao Y-J, Shiang Y-C, Huang C-C, Chang H-T. Langmuir. 2012;28:8944. doi: 10.1021/la204651t. [DOI] [PubMed] [Google Scholar]
- [19].Riskin M, Tel-Vered R, Willner I. Adv. Mater. 2010;22:1387. doi: 10.1002/adma.200903007. [DOI] [PubMed] [Google Scholar]
- [20].Mishra J, Dent C, Tarabishi R, Mitsnefes MM, Ma Q, Kelly C, Ruff SM, Zahedi K, Shao M, Bean J, Mori K, Barasch J, Devarajan P. The Lancet. 365:1231. doi: 10.1016/S0140-6736(05)74811-X. [DOI] [PubMed] [Google Scholar]
- [21].Parikh CR, Jani A, Mishra J, Ma Q, Kelly C, Barasch J, Edelstein CL, Devarajan P. Am. J. Transplant. 2006;6:1639. doi: 10.1111/j.1600-6143.2006.01352.x. [DOI] [PubMed] [Google Scholar]
- [22].Lee K-S, El-Sayed MA. J. Phys. Chem. B. 2005;109:20331. doi: 10.1021/jp054385p. [DOI] [PubMed] [Google Scholar]
- [23].Gole A, Murphy CJ. Langmuir. 2007;24:266. doi: 10.1021/la7026303. [DOI] [PubMed] [Google Scholar]
- [24].Abbas A, Vercaigne-Marko D, Supiot P, Bocquet B, Vivien C, Guillochon D. Colloids Surf. B. Biointerfaces. 2009;73:315. doi: 10.1016/j.colsurfb.2009.06.007. [DOI] [PubMed] [Google Scholar]
- [25].Whitcombe MJ, Chianella I, Larcombe L, Piletsky SA, Noble J, Porter R, Horgan A. Chem. Soc. Rev. 2011;40:1547. doi: 10.1039/c0cs00049c. [DOI] [PubMed] [Google Scholar]
- [26].Díaz-García ME, Laínño RB. Microchimica Acta. 2005;149:19. [Google Scholar]
- [27].Wulff G. Angew. Chem. Int. Ed. 1995;34:1812. [Google Scholar]
- [28].Holthoff EL, Bright FV. Acc. Chem. Res. 2007;40:756. doi: 10.1021/ar700087t. [DOI] [PubMed] [Google Scholar]
- [29].Dickert FL, Hayden G. Anal. Chem. 2002;74:1302. doi: 10.1021/ac010642k. [DOI] [PubMed] [Google Scholar]
- [30].Zhang Z, Liao H, Li H, Nie L, Yao S. Anal. Biochem. 2005;336:108. doi: 10.1016/j.ab.2004.09.027. [DOI] [PubMed] [Google Scholar]
- [31].Carlson CA, Lloyd JA, Dean SL, Walker NR, Edmiston PL. Anal. Chem. 2006;78:3537. doi: 10.1021/ac051375b. [DOI] [PubMed] [Google Scholar]
- [32].Gao R, Kong X, Wang X, He X, Chen L, Zhang Y. J. Mater. Chem. 2011;21:17863. [Google Scholar]
- [33].Lin Z, Yang F, He X, Zhao X, Zhang Y. J. Chromatogr. 2009;1216:8612. doi: 10.1016/j.chroma.2009.10.025. [DOI] [PubMed] [Google Scholar]
- [34].Chen L, Xu S, Li J. Chem. Soc. Rev. 2011;40:2922. doi: 10.1039/c0cs00084a. [DOI] [PubMed] [Google Scholar]
- [35].Shiomi T, Matsui M, Mizukami F, Sakaguchi K. Biomaterials. 2005;26:5564. doi: 10.1016/j.biomaterials.2005.02.007. [DOI] [PubMed] [Google Scholar]
- [36].Nusz GJ, Curry AC, Marinakos SM, Wax A, Chilkoti A. ACS Nano. 2009;3:795. doi: 10.1021/nn8006465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mayer KM, Hafner JH. Chem. Rev. 2011;111:3828. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- [38].Nehl CL, Hafner JH. J. Mater. Chem. 2008;18:2415. [Google Scholar]
- [39].Abbas A, Tian L, Kattumenu R, Halim A, Singamaneni S. Chem. Commun. 2012;48:1677. doi: 10.1039/c2cc16356j. [DOI] [PubMed] [Google Scholar]
- [40].Zhou X, Li W, He X, Chen L, Zhang Y. Separ. Purif. Rev. 2007;36:257. [Google Scholar]
- [41].Mayer KM, Lee S, Liao H, Rostro BC, Fuentes A, Scully PT, Nehl CL, Hafner JH. ACS Nano. 2008;2:687. doi: 10.1021/nn7003734. [DOI] [PubMed] [Google Scholar]
- [42].Halas NJ, Lal S, Chang W-S, Link S, Nordlander P. Chem. Rev. 2011;111:3913. doi: 10.1021/cr200061k. [DOI] [PubMed] [Google Scholar]
- [43].Yee KS. IEEE Trans. Antennas Propag. 14:302. [Google Scholar]
- [44].Palik ED, editor. Handbook of Optical Constantsof Solids. Academic, Boston; MA: 1985. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Extinction spectra of gold nanorods in (a) solution and on (b) glass substrates. For comparison purposes, the blue line (c) represents spectrum b with a different scale. The transfer of the nanorods from solution to a glass substrates results in a decrease of the extinction intensity by 10 to 50 times and a red-shift by ~30 nm. This shift is caused by the significant increase in refractive index in one side of the nanorods from n=1.33 (water) to n=1.5 (glass substrate).
Figure S2. Profile and distribution of the electric field intensity around the gold nanorods. The intensity at the nanorods ends is one order of magnitude higher than that at the center of the nanorods. The intensity, however, decreases exponentially by increasing distance from the nanorods surface. The x-axis is in nm.
Figure S3. Surface enhanced Raman scattering of molecularly imprinted AuNRs after BSA protein capture. The assignments of the different bands are indicated in Table S1.
Figure S4. Template release using oxalic acid and SDS. Fitting of the extinction spectra provides the integrated surface area (A) of the peak due to the template fluorescein conjugated BSA at λ≈ 500 nm. (1) AuNR before template immobilization (A= 4.28), (2) AuNR after template immobilization (A= 9.39), (3) template release using oxalic acid (A= 8.33), template release using a mixture of oxalic acid and SDS (A= 6.4).
Figure S5. Extinction spectra of Hemoglobin (a, b), NGAL (c, d) and BSA (e, f) capture/release cycles with molecularly imprinted AuNRs. The spectra represent AuNRs (black line) before and after successive addition of glutaraldehyde/p-ATP (pink line), proteins (green line) and siloxane copolymers (blue line with a star). The red and blue lines represent different cycles of protein capture (blue) and release (red). These different steps and cycles correspond to the data plotted in Figure 5a.
Figure S6. Extinction spectra of IgG (a) and allophycocyanin (b) capture/release cycles with molecularly imprinted AuNRs. The LSPR shifts are depicted in the insets. NR: imprinted nanorods, C: template capture, R: template release.
Figure S7. Localized surface plasmon resonance analysis of different protein mixtures using three different imprinted sensors with a specificity for (a) hemoglobin, (b) BSA and (c) NGAL proteins. The black line labelld C represents the extinction spectrum of the sensor after expositition to buffer. The green and red spectra are respectively obtained after exposition of the imprinted sensor to mixtures of two or three different proteins. A shift of 4-5 nm is observed only when the sensor is exposed to a mixture containing the protein used for imprinting (red line). The other spectra exhibit less than 1 nm LSPR shift.
Figure S8. Change of the full width at half maximum (FWHM) of the extinction spectra before and after protein capture with the imprinted sensors. The data are obtained from the analysis of the spectra depicted in Figure S7. In all cases, the increase in FWHM (Δ FWHM) is lower than 10 %. This change seems to increase with increasing protein size but more investigation with a larger number of proteins is needed to understand the different contributions to the increase in FWHM.
Figure S9: Atomic Force Microscopy (AFM) images depicting the multiple scan size used to localize and monitor thickness change of the MIP-AuNRs. The scan sizes are (a) 50 um x 50 um, (b) 5 um x 5 um, (c) 2 um x 2 um, (d) 760nm x 760 nm, (e) 368 nm x 368 nm.
Table S1. Selected SERS bands of CTAB, p-ATP and Proteins with their vibrational