Abstract
WASF3 has been shown to be required for invasion and metastasis in different cancer cell types and knockdown of WASF3 leads to suppression of invasion/metastasis. Aberrant signaling through the interleukin 6/Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) axis in cancer cells has emerged as a major mechanism for cancer progression. In this study, we demonstrate that interleukin 6 induces both WASF3 expression and phosphoactivation in breast and prostate cancer cell lines through the JAK2/STAT3 pathway in two different ways. First, we show that STAT3 binds directly to the WASF3 promoter and increases transcription levels, which correlates with increased migration potential. Inactivation of STAT3 with short hairpin RNA, dominant negative constructs or S3I-201 leads to reduced WASF3 levels and reduced migration. Second, we have shown that JAK2, while activating STAT3, also interacts with and activates WASF3. Inhibition of JAK2 with short hairpin RNA or AG490 leads to loss of migration due to reduced WASF3 activation levels and prevention of its membrane localization. Together, these results define a novel signaling network whereby JAK2/STAT3 signaling creates a feed-forward loop to raise activated WASF3 levels that promote cancer cell motility.
Introduction
Metastasis is the primary cause of death in cancer patients and there is now convincing evidence that the acquisition of the metastatic phenotype is genetically controlled and apparently involves a wide variety of genes (1). On the one hand, a subclass of genes has been demonstrated to suppress metastasis while not affecting proliferation rate or tumorigenesis (2). On the other hand, individual genes have been shown to promote metastasis and if these genes are inactivated in any way, metastasis and invasion are suppressed (3–5). There have been many reports suggesting individual genetic events lead to suppression or promotion of metastasis, suggesting a complex and diverse series of pathways are potentially involved in this phenotype; although as we learn more about the function of some of these genes, it appears that these pathways may intersect and be coordinately regulated by a subset of master regulatory genes.
The Wiskott-Aldridge syndrome family (WASF) (6) of proteins carry motifs implicating them in actin cytoskeleton dynamics (7). Inactivation of WASF3 in breast and prostate cancer cells not only reduces motility and invasion in vitro but also metastasis in vivo (8,9). Cytoplasmic WASF proteins are largely retained in an inactive form (7) and are phosphoactivated in response to stimulation by growth factors such as platelet-derived growth factor (10). As a result, conformational changes expose motifs at the C-terminal end, which leads to recruitment of ARP2/3, promoting lamellipodia formation and increased cell motility and invasion (7).
We recently investigated how WASF3 regulates motility and invasion and have shown that knockdown of WASF3 leads to upregulation of the KISS1 metastasis suppressor gene (9). KISS1 normally suppresses activation of nuclear factor-kappaB (NF-κB) by promoting its interaction with inhibitor of NF-κB alpha. Downregulation of KISS1, a metastasis suppressor gene (11,12), releases the inhibition by inhibitor of NF-κB alpha allowing relocation of NF-κB to the nucleus in a WASF3-dependent manner (13), where one consequence is upregulation of Zinc Finger E box-binding homeobox 1 gene levels, which in turn represses the chromosome 1 miR-200 cluster, which normally suppress invasion and metastasis (14). Elevation of Zinc Finger E box-binding homeobox 1 gene also results in suppression of E-cadherin, loss of which is a critical step in the epithelial-to-mesenchyme transition associated with metastasis (14). WASF3 is present in a protein complex with the p85 subunit of phosphatidylinositol 3-kinase (8) and the Abelson murine leukemia viral oncogene homolog 1 kinase (15). Inactivation of either of these two kinases leads to reduced phosphorylation of WASF3, which is associated with loss of invasion. Expression of WASF3 has been reported to be elevated in advanced stage tumors (15) and overexpressing WASF3 in cancer cells leads to increased invasion potential (14). In a recent classification of breast cancer (16), the highly aggressive ‘claudin-low’ subgroup, which includes the triple negative tumors, showed increased expression of WASF3. How WASF3 is transcriptionally regulated in the cancer cell, however, is still unclear.
Aberrant expression of cytokines can profoundly affect tumor-cell processes including cell growth, survival, inflammation, migration and invasion (17,18). Although interleukin (IL)-6-induced Janus kinase (JAK) and signal transducer and activator of transcription 3 (STAT3) activation has been implicated in tumor-cell metastasis (19–22), the mechanism remains poorly understood. Our findings here provide new insights into the downstream molecular events of IL6/JAK2/STAT3 axis in breast and prostate cancer cell invasion and metastasis. We identified STAT3 as a direct regulator of WASF3 expression and show that IL6-induced JAK2/STAT3 activation leads to increased WASF3 expression. Moreover, we show that IL6-induced JAK2-WASF3 protein interaction leads to increased phosphoactivation of WASF3, which is independent of JAK2/STAT3 signal transduction. Thus, constitutive activation of the JAK2/STAT3 pathway in cancer cells appears to be one of the ways by which advanced stage cancers can upregulate WASF3, which promotes invasion and metastasis.
Materials and methods
Cell culture and standard assays
All cell lines were purchased from American Type Culture Collection (Rockville, MD). Standard cell culture, transient transfections, luciferase reporter assays, western blotting, immunoprecipitation (IP), immunofluorescence, quantitative reverse transcription–polymerase chain reaction (QRT–PCR), cell migration assays and lentiviral transduction were carried out as described previously (9,14,23).
DNA constructs, antibodies and other reagents
The pGL-WASF3 promoter constructs (+494/−747, +494/−1101) were generated as described previously (24). The short hairpin RNAs (shRNAs) targeting JAK1 and JAK2 were kindly provided by Dr A.Levine (Memorial Sloan Kettering Cancer Center, NY) and Dr L.Staudt (Metabolism Branch of NCI, Bethesda, MD). pSIH1-puro-STAT3 shRNA was a gift of Dr FA.Sinicrope (Addgene, plasmid no. 26596) and EF.STAT3DN.Ubc.GFP was a gift of Dr L.M.Resar (Addgene, plasmid no. 24984). To construct the HA-WASF3 overexpression vector, the full-length human WASF3 was amplified from the template WASF3 cDNA clone BC050283 (Open Biosystems, Huntsville, AL) and was inserted into pCDH-CMV-MCS-EF1-puro lentiviral vector (System Biosciences, Mountain View, CA) as described previously (14). To stably knock down WASF3, pLKO.1 lentiviral vectors harboring shRNA-targeting WASF3 were obtained from Open Biosystems (Huntsville, AL). For western blot and IP assays, the following primary antibodies were used: WASF3, WASF2, JAK1, JAK2, p-JAK2 (Tyr1007/1008), STAT3, p-STAT3 (Tyr705), Rous Sarcoma viral oncogene (SRC), p-SRC (Tyr416), epidermal growth factor receptor (EGFR), p-EGFR (Tyr1068) (Cell Signaling Technology, Beverly, MA), HA, PY20 and glyceraldehyde 3-phosphate dehydrogenase (Sigma, St Louis, MO), GP130-blocking antibody BR-3 (Cell Sciences, Canton, MA). Recombinant Human IL6 was purchased from R&D Systems (Minneapolis, MN), the AG490 JAK inhibitor, EGFR inhibitor Gefitinib and STAT3 inhibitor S3I-201 were obtained from Selleckchem (Houston, TX), and the SRC inhibitor, Dasatinib, was purchased from ChemieTek (Indianapolis, IN).
Chromatin immunoprecipitation and oligonucleotide pull-down assays
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (14,24). IgG and the human RPL30 gene were used as negative controls (25). The primer set used to amplify the WASF3 promoter putative STAT3 binding sites was F 5′-ACACATCTGTAAAGAAAA-3′ and R 5′-TTTGCATGCCTTTATTCT-3′ (Supplementary Figure S1A, available at Carcinogenesis Online). The human RPL30 gene primers designed for ChIP assays were provided by Novus Biologicals (Littleton, CO). The oligonucleotide pull-down assay was performed as described previously (24). In this analysis, the wild-type 5′-biotinylated double-stranded oligonucleotides (5′-AGTGTTTCTTGAACTATTCATTAGAATTTAAAATTTTTTCCTAAAA GTTT-3′ and 5′-AAACTTTTAGGAAAAAATTTTAAATTCTAATGAA TAGTTCAAGAAACACT-3′) corresponding to the positions −930 to −882 of the WASF3 promoter harboring 3 STAT3 binding motifs were synthesized by Invitrogen. The control biotinylated oligonucleotides containing the mutated STAT3 binding site (5′-AGTGTA AGGGCATCTAAGCAGCAGTCTTTAAAATTTTGGAA GCTGAGTTT-3′ and 5′-AAACTCAGCTTCCAAAATTTTAAAGACTGCTG CTTAGATGCCCTTACACT-3′) in which the conserved nucleotides of the STAT3 consensus sequence were replaced.
Results
STAT3 upregulates WASF3 expression through direct binding to its promoter
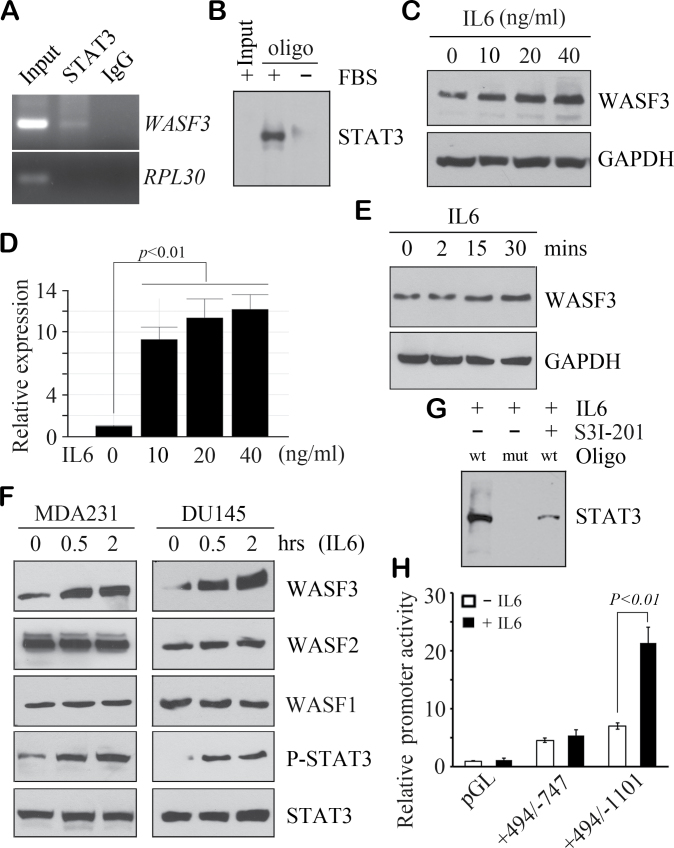
High-level WASF3 expression has been associated with aggressive breast tumors (15,16), although it is not known how its expression is regulated. Analysis of the DNA sequence within the ~1100 base pairs upstream of the initiation of transcription site, which we have shown to contain the minimal promoter (24), identified three potential STAT3 binding sites, with the canonical TTC(N)2–4GAA or TCC (N)2–6AA sequence (Supplementary Figure S1A, available at Carcinogenesis Online), located at upstream positions −894 to −886, −915 to −906 and −926 to −919. ChIP analysis from MDA-MB-231 (MDA231) cells demonstrated STAT3 binding to this region (Figure 1A) but not to the control RPL30 gene. To confirm this observation, we used biotinylated double-stranded oligonucleotides (Figure 1B), coupled to streptavidin agarose beads, to pull down proteins interacting with the STAT3 binding site sequence in MDA231 cells. Western blot analysis again demonstrated STAT3 binding to the WASF3 promoter sequences (Figure 1B). These results indicate that STAT3 binds directly to the corresponding sequences in the WASF3 promoter.
Fig. 1.
STAT3 upregulates WASF3 expression through direct binding to its promoter region. ChIP analysis in MDA231 cells shows the presence of the STAT3 sequences of the WASF3 promoter (A) in the input and STAT3 IP but not in the IgG control IP or ChIP with the RPL30 promoter, which is known not to be activated by STAT3. An oligonucleotide pull-down assay (B) confirmed that WASF3 is associated with a wild-type consensus STAT3 DNA binding site in the presence of fetal bovine serum but absent in starved cells. Western blot (C) and QRT–PCR analysis (D) show increased WASF3 protein and mRNA levels in MDA231 cells treated with different concentrations of IL6 for 30 min. In a time-course experiment (E), WASF3 levels were progressively increased after 15 min in MDA231 cells with IL6 treatment (20ng/ml). IL6 induction of WASF3 in MDA231 breast cancer cells and DU145 prostate cancer cells were remarkably induced after 30 min following activation of STAT3 (F). WASF1 and WASF2 levels were not affected by IL6 treatment as seen for WASF3 (F). When lysate from MDA231 cells was incubated with either the wild-type (wt) or the mutant (mut) STAT3 binding site, oligonucleotides strongly binding to the wt oligonucleotide was seen (G). No binding was seen for the mutant oligonucleotide, even in the presence of IL6. When cells were treated with the S3I-201 STAT3 inhibitor, binding to the wild-type oligonucleotide was reduced (G). When a luciferase reporter construct (+494/−1101), which included the three STAT3 binding sites, was introduced into MDA231 cells, luciferase activity was increased following treatment with IL6 for 30 min (H). No induction was seen in cells carrying either the luciferase empty vector (pGL) or the shorter (+494/−747) construct, which did not carry the STAT3 binding sites (H). The relative luciferase values are shown as the mean of three independent experiments (P < 0.01).
To determine whether there is a correlation between WASF3 protein levels and levels of activated STAT3, we analyzed the relative levels of these proteins in several breast cancer cell lines known to have varying WASF3 levels (Supplementary Figure S1B, available at Carcinogenesis Online). MDA231 and SKBR3 cells show high levels of WASF3 and activated STAT3. In contrast, T47D cells, which do not express WASF3, and MCF7 cells, which express low WASF3 levels, both showed very low levels of activated STAT3 (Supplementary Figure S1B, available at Carcinogenesis Online).
STAT3 is required for IL6-induced WASF3 expression in breast and prostate cancer cells
Phosphoactivation of STAT3 leads to oligomerization (26), which is required for its function as a transcription factor. IL6 has been shown to activate STAT3 signaling in many different cell types, and treatment of WASF3 expressing MDA231 cells with IL6 shows upregulation of WASF3 in a concentration-dependent manner using western blotting (Figure 1C) and QRT–PCR (Figure 1D). WASF3 induction by IL6 is also time dependent, with high levels of induction after only 15 min (Figure 1E). WASF3 levels were also dramatically increased by IL6 treatment in other breast cancer cell lines, SKBR3, MCF7 and MDA453 (Supplementary Figure S1C, available at Carcinogenesis Online). Starved MDA231 cells express low levels of WASF3 and low levels of activated STAT3 (Figure 1F). Treatment of these starved cells with IL6 induces STAT3 activation after 30 min, with a concomitant increase in WASF3 levels. We have shown previously that knockdown of WASF3 in DU145 prostate cancer cells also prevents invasion and metastasis (27), and treatment of these starved cells with IL6 also resulted in increased WASF3 levels and STAT3 activation (Figure 1F) but not WASF1 and WASF2 levels (Figure 1F), providing further evidence for the lack of involvement of other WASF family members in invasion and metastasis. Using the oligonucleotide pull-down assay described above, STAT3 was shown to bind to the wild-type sequence but not an oligonucleotide with mismatched sequences in the STAT3 binding sites, even in the presence of IL6 (Figure 1G). When these cells were treated with the S3I-201 inhibitor of STAT3 activation, binding to the wild-type oligonucleotide is greatly reduced (Figure 1G). Using a WASF3 promoter reporter assay (Figure 1H), IL6 strongly increases activity with the +449/−1101 construct that contains the STAT3 binding sites but does not enhance luciferase activity in the +449/−747 construct lacking the STAT3 binding sites. Together, these results demonstrate that activated STAT3 is a potent regulator of WASF3.
To investigate the relationship between STAT3 activation and WASF3 expression further, we used shRNA knockdown of STAT3 in MDA231, which reduces WASF3 protein levels and expression levels (Figure 2A). Expressing a dominant negative (DN) construct (Y705F) that inactivates STAT3 in both MDA231 and DU145 cells leads to a concomitant reduction in WASF3 levels, which was the same as that seen in shSTAT3 knockdown cells (Figure 2B). Luciferase assays, using the +474/−1101 promoter construct in MDA231 cells, showed that inhibition of STAT3 using either DN constructs or specific shRNA leads to significant downregulation of WASF3 promoter activation (Figure 2C). Furthermore, shRNA knockdown of STAT3 renders MDA231 cells relatively insensitive to IL6 induction of WASF3 (Figure 2D), and IL6 cannot induce luciferase activity in the +474/−1101 WASF3 promoter in these cells (Figure 2E). Using transwell migration assays, IL6 treatment, which normally leads to increased migration (Figure 2F), does not affect this phenotype in MDA231 cells in which either WASF3 or STAT3 has been knocked down by shRNAs (Figure 2F), suggesting the function of WASF3 may be involved in STAT3-mediated cancer cell migration.
Fig. 2.
Phosphorylation of STAT3 regulates WASF3 expression. Western blot and QRT–PCR analysis (A) shows decreased WASF3 mRNA and protein levels in stable STAT3 knockdown MDA231 cells (shSTAT3) compared with cells treated with a control shRNA designed against green fluorescent protein (shGFP). In MDA231 and DU145 cells, either inhibition of STAT3 transcripts by shRNA (shSTAT3) or inactivation of STAT3 by expressing a STAT3 DN construct significantly suppresses WASF3 protein levels (B). Increased STAT3 levels in the STAT3 DN cells reflect the expression of the exogenous gene. Luciferase assays show that the WASF3 promoter (+494/−1101) activity is decreased after either knockdown of STAT3 or DN inactivation of STAT3 in MDA231 (C). In stable STAT3 knockdown MDA231 cells, no change in WASF3 protein levels (D) or WASF3 promoter activity (E) was observed following IL6 treatment compared with cells expressing the control shRNA. The relative luciferase values in (C) and (E) are shown as the mean of three independent experiments (P < 0.01). Knockdown of WASF3 or STAT3 in MDA231 cells (F) reduces migration potential compared with control cells (shGFP). Data are shown as mean ± SD from three independent experiments (P < 0.01).
IL6-induced expression of WASF3 is dependent on GP130/JAK/STAT3 signal transduction
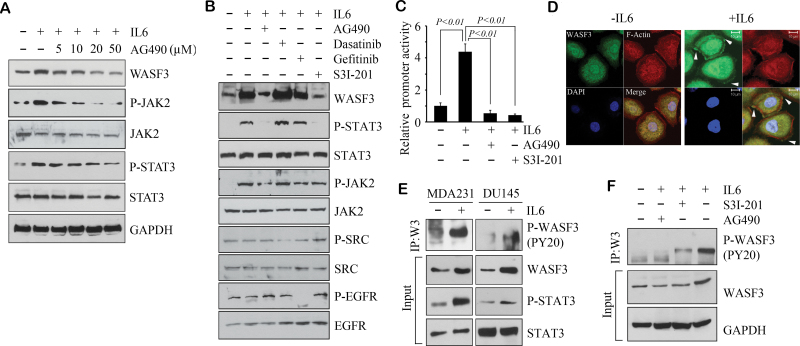
Activation of STAT3 is achieved predominantly through the JAK1/2 kinases, which suggests that JAK1/2 may be responsible for regulation of WASF3. As seen in Figure 3A, application of IL6 to the cells in which JAK1/2 activity was suppressed by pan-JAK inhibitor AG490 led to reduced activation of STAT3 and a concomitant reduction in WASF3 levels. To determine the role of other kinases in mediating the IL6/JAK/STAT3/WASF3 axis, we pretreated with different kinase inhibitors before IL6 stimulation. Inhibition of STAT3 activation with S3I-201 or AG490 in the presence of IL6 leads to a loss of phospho-STAT3, which leads to reduced WASF3 levels (Figure 3B). However, inhibition of SRC or EGFR activation in IL6-treated cells with Dasatinib or Gefitinib could not block IL6-induced JAK2/STAT3 phosphorylation and WASF3 protein levels (Figure 3B) Using the luciferase promoter assay (Figure 3C), MDA231 cells treated with IL6 show increased levels of promoter activation, but this effect is significantly reduced when the cells are treated concomitantly with AG490 or S3I-201. IL6 activation of STAT3 is largely achieved through its preferred GP130 receptor (26), although other receptors are also used. When GP130 was blocked in MDA231 cells using the BR-3 monoclonal antibody, no effect on WASF3 protein levels was detected following IL6 treatment (Supplementary Figure S2A, available at Carcinogenesis Online). Thus, IL6-induced expression of WASF3 is probably dependent on GP130/JAK/STAT3 signal transduction.
Fig. 3.
JAK regulates WASF3 at both the transcription and phosphorylation levels. IL6 normally induces WASF3 expression in MDA231 cells, but co-treatments with AG490 (a pan-JAK inhibitor) show a dose-dependent reduction of both WASF3 levels and activated STAT3 levels (A). Inhibition of JAK1/2 with AG490 or STAT3 with S3I-201 leads to reduced STAT3 activation and reduced WASF3 levels. In contrast, treatment with Dasatinib (SRC inhibitor) or Gefitinib (EGFR inhibitor) does not significantly affect activated STAT3 or WASF3 levels (B). In these experiments, MDA231 cells were pretreated with specific inhibitors for 4 h before IL6 stimulation. In luciferase activity promoter assays for WASF3 (C), IL6 clearly increases luciferase activity in MDA231 cells but coincident treatment with AG490 or the S31-201 STAT3 inhibitor suppresses promoter activity. The relative luciferase values are shown as the mean of three independent experiments (P < 0.01). Confocal microscopy analysis of DU145 cells shows that treatment with IL6 leads to relocation of WASF3 to the membrane (D). Activation of JAK/STAT3 signaling in MDA231 and DU145 cells by IL6 (E) leads to increased activated WASF3 levels, as well as overall WASF3 levels and phosphoactivated STAT3 levels. When MDA231 cells, stimulated with IL6 are pretreated with a JAK inhibitor (AG490), levels of activated WASF3 are suppressed. In contrast, inhibition of STAT3 with S3I-201 has only a mild effect on WASF3 activation levels (F).
JAK2 is required for IL6-induced WASF3 expression and activation in breast and prostate cancer cells
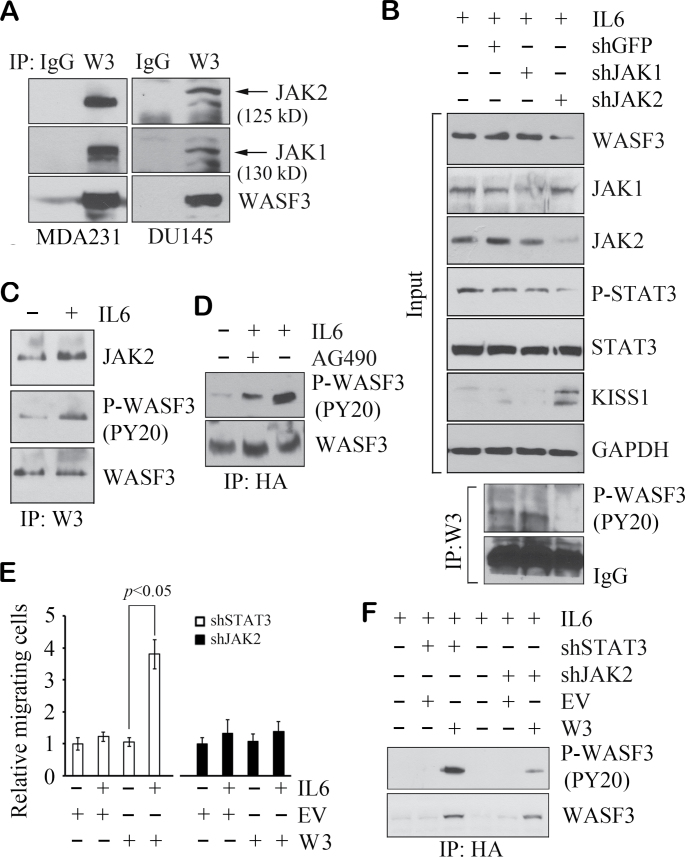
Functional activation of WASF3 is believed to lead to activation of cytoskeleton reorganization promoting invasion and metastasis (7). Our previous studies suggested that phosphoactivation of WASF3 is required for migration and invasion of breast cancer cells (15). Confocal microscopy (Figure 3D) of starved DU145 cells shows a flattened morphology allowing easy visualization of protein movement to the membrane. In starved DU145 cells (Figure 3D), WASF3 is found in the cytoplasm but when treated with IL6, a subfraction of WASF3 moves to plasma membrane protrusions at the leading edge of the cell, which demonstrates a functional consequence of IL6 stimulation of WASF3 activation. Consistently, IP analysis shows increased activated WASF3 levels, as well as WASF3 protein levels and activated STAT3 levels following IL6 treatment (Figure 3E). To determine whether activation of WASF3 is affected by manipulation of the JAK/STAT3 pathway, we pretreated MDA231 cells with either AG490 or S3I-201 before IL6 stimulation. When the cells were pretreated with AG490, WASF3 was not activated by IL6 treatment (Figure 3F). S3I-201 treatment, however, shows reduced WASF3 activation, presumably due to reduced total WASF3 levels (Figure 3F). Confocal microscopy (Supplementary Figure S2B, available at Carcinogenesis Online) shows a dramatic relocation of WASF3 to the membrane following addition of IL6, but this relocation to the membrane was prevented when JAK function is inactivated with AG490. Inactivation of STAT3 with S3I-201 suppresses WASF3 transcription, although it does not affect relocation of WASF3 to the membrane in the presence of IL6 (Supplementary Figure S2B, available at Carcinogenesis Online). These studies further demonstrate the central importance of JAKs in the functional activation of WASF3. On reanalysis of our mass spectroscopy data for WASF3-interacting proteins (23), we found both JAK1 and JAK2 were present in the WASF3 immunocomplex in PC3 cells (Supplementary Figure S3, available at Carcinogenesis Online). This interaction was confirmed in western blot analysis of an IP of WASF3 from DU145 cells and also in MDA231 cells (Figure 4A), where both JAK1 and JAK2 were present in the immunocomplex.
Fig. 4.
JAK2 binds to, and specifically activates, WASF3. IP analysis demonstrates the presence of endogenous JAK1 and JAK2 in WASF3 immunoprecipitates from MDA231 and DU145 cell lysates (A). In MDA231 cells, WASF3 is remarkably suppressed by knockdown of JAK2 but not by loss of JAK1 (B, above). This is reflected in phosphoactivation of WASF3 (B, below). Reduced WASF3 levels correlate with increased KISS1 levels. In (C), IP analysis shows treatment with IL6 for 2 min leads to increased activation of WASF3 and the interaction between JAK2 and WASF3. Starved MCF7 cells forced to express an HA-tagged WASF3 protein show low levels of activated WASF3, but when stimulated with IL6, a dramatic increase in activated WASF3 levels is seen (D). Inactivation of JAK2 in these IL6-treated cells with AG490 leads to reduced levels of activated WASF3 (D). In the MCF7 cells expressing exogenous WASF3 (W3), when STAT3 is knocked down by shRNA, IL6 treatment leads to a significant increase in cell migration (E) compared with untreated cells and cells in which transfected the empty vector (EV). When JAK2 is knocked down in these cells, even in the presence of exogenous WASF3, no difference in migration was detected (E). Data are shown as mean ± SD from three independent experiments (P < 0.05). Analysis of activated WASF3 in these cells shows robust levels in the absence of STAT3 but greatly reduced levels in the absence of JAK2 (F).
To target specific members of the JAK protein family, we used shRNA approaches to knock down either JAK1 or JAK2 in MDA231 cells. Specific knockdown of JAK1 does not lead to reduced WASF3 protein levels (Figure 4B). In contrast, knockdown of JAK2 (Figure 4B) led to a significant reduction in WASF3 levels, which is presumably due to loss of STAT3 activation (see above). Reduction in WASF3 levels is correlated with increased KISS1 levels following knockdown of JAK2. This is consistent with the inverse relationship between expression of these two genes as we demonstrated previously (9). This effect was also reflected in phosphoactivation levels of WASF3, which were only affected by JAK2 knockdown (Figure 4B), and was maintained using different JAK2 shRNAs (Supplementary Figure S4, available at Carcinogenesis Online), mitigating any off-target effects. We further determined the relationship between protein–protein interaction levels and phosphorylation. In the presence of IL6, increased JAK2 protein and higher phosphorylation levels of WASF3 were found in the WASF3 immunocomplex compared with cells that were not treated with IL6 (Figure 4C), suggesting JAK2 activates WASF3 through direct interaction.
JAK2 plays a critical role in WASF3-mediated cancer cell migration
Non-invasive MCF7 cells express relative low levels of WASF3 (14) but do not show significant levels of activated WASF3 even in the presence of serum. We created MCF7 cells stably expressing an HA-tagged WASF3 protein, which in the presence of IL6 show increased levels of phosphoactivation of WASF3 (Figure 4D). Treatment with AG490 reduced activated WASF3 levels implicating JAK1/2 in the activation process (Figure 4D). To determine whether JAK2/STAT3 signaling affects WASF3-dependent cell migration in MCF7 cells forced to express WASF3, we used standard transwell migration assays (Figure 4E). In STAT3-knockdown MCF7 cells, IL6 treatment did not result in altered cell migration but in STAT3-silenced cells, exogenous expression of WASF3 led to a dramatic increase in migration (Figure 4E). This would be expected because, even though loss of STAT3 suppresses endogenous WASF3 transcription, JAK2 can still activate exogenous WASF3 driven by a constitutive promoter, which leads to increased cell migration. When JAK2 is knocked down, however, even in the presence of IL6, cell migration potential is not affected whether WASF3 is expressed or not (Figure 4E), which indicates that JAK2-mediated phosphorylation of WASF3 is critical for MCF7 cell migration. This was confirmed in Figure 4F where the MCF7 cells expressing exogenous WASF3 show increased activated WASF3 levels in the presence of IL6 even though STAT3 expression is inhibited, but when JAK2 is knocked down, activated WASF3 levels are dramatically reduced.
Discussion
Constitutive activation of STAT3 has been reported in a wide variety of human cancers (28), in many cases associated with advanced stage tumors. In experimental systems, increased STAT3 activation correlates with increased invasion and metastasis (29,30). IL6 signaling, through activation of JAK2, is a significant means of activating STAT3, allowing dimerization and relocation to the nucleus (26), where it is suggested that specific STAT3 target genes that are related to enhancement of metastasis are activated. We have now shown that one of these genes is WASF3, which is central to the JAK2/STAT3 regulation of invasion and metastasis. Previously, we demonstrated that inactivation of WASF3 in breast and prostate cancer cells that have functional JAK2/STAT3 signaling potential leads to loss of invasion and metastasis (8,9). Furthermore, cells that have not undergone the epithelial-to-mesenchyme transition, such as MCF7 cells, express very low levels of WASF3 (14). Activation of JAK/STAT signaling in these cells by IL6 leads to increased WASF3 expression and invasion. Overexpressing WASF3 in MCF7 cells leads to increased sensitivity to JAK2-induced cell motility. In the reciprocal experiment, Gu et al. (31) demonstrated that WASF3 was one of the genes downregulated as a result of inactivation of STAT3 in prostate cancer cells. In this same series of experiments, WASF3 expression was not affected by knockdown of STAT5a/b, which showed a preferential association with cell growth and viability, compared with loss of STAT3 that specifically affected metastasis. These observations are consistent with our analysis of the WASF3 promoter region that did not contain consensus STAT5 binding motifs (data not shown). Thus, it appears that WASF3 provides the conduit through which extracellular cytokine signals involving JAK2/STAT3, and promoting invasion and metastasis, are processed. WASF3 is present throughout the cytoplasm of starved cells in its inactive, non-phosphorylated form. Stimulation with IL6 leads to WASF3 activation within 2 min, which is associated with relocation to the membrane to engage with JAK2. This clearly defines the critical response to cytokine, and possibly other growth factor, stimulation. Upregulation of WASF3 transcription by STAT3 occurs later than JAK2-induced WASF3 activation (~15 min), which increases WASF3 levels, which we have also shown is also related to invasion potential (9,14). These observations collectively suggest that JAK2 is a critical factor affecting WASF3 function in IL6 signaling, which interacts with and activates WASF3 almost immediately and subsequently upregulates WASF3 expression following JAK2 activation of STAT3.
Cytokines are produced by cells such as leukocytes and macrophages, which have important influence in the role of the microenvironment and metastasis (17,18). Cancer cells, however, also demonstrate autocrine action of IL6 signaling as a result of increased NF-κB expression, which regulates IL6 expression (32). We have previously shown that WASF3 expression leads indirectly to the activation of NF-κB, through suppression of KISS1 function (14). Increased NF-κB activity, therefore, increases IL6 production, which in turn can activate WASF3 through JAK2 and increased expression through STAT3. This autocrine loop, therefore, provides stimulation of the metastatic phenotype, which is enhanced by other related effects of WASF3 expression such as upregulation of ZEB1, which suppresses E-cadherin expression and downregulates the miR-200 family (14). Thus, activation of WASF3 has central influence over many of the mechanisms that promote invasion and metastasis.
WASF3 is a member of a three-gene family, which shares the structural motifs that facilitate actin cytoskeleton reorganization (7) and both WASF1 and WASF2 have been implicated in cell movement but not specifically in invasion and metastasis. We have reported previously that the other WASF family members cannot compensate for loss of WASF3 in its functional control of invasion (10). Knockdown of WASF1 or WASF2 does not affect KISS1 expression and does not have consequences on NF-κB signaling (9). Here we show that STAT3 does not affect expression of WASF1 or WASF2 and IL6 does not induce their expression either. Analysis of the ~1000bp of the initiation of transcription site in WASF1 and WASF2 failed to identify consensus STAT3 binding sites, providing one possible explanation why these two genes have not been implicated in invasion in response to cytokine stimulation. Thus, it appears that regulation of WASF3 and its influence on invasion and metastasis are specific to this family member in cell system studies so far. WASF3, therefore, presents a potential target to suppress metastasis.
Supplementary material
Supplementary Figures S1–S4 can be found at http://carcin. oxfordjournals.org/
Funding
National Institutes of Health (CA120510). J.K.C. is a Georgia Cancer Coalition Scholar.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- ChIP
chromatin immunoprecipitation
- DN
dominant negative
- IL
interleukin
- IP
immunoprecipitation
- JAK
Janus kinase
- NF-κB
nuclear factor-kappaB
- QRT–PCR
quantitative reverse transcription–polymerase chain reaction
- shRNA
short hairpin RNA
- SRC
Rous Sarcoma viral oncogene
- STAT3
signal transducer and activator of transcription 3
- WASF
Wiskott-Aldridge syndrome family.
References
- 1. Nasr Z., et al. (2012). Tumor progression and metastasis: role of translational deregulation. Anticancer Res., 32, 3077–3084 [PubMed] [Google Scholar]
- 2. Steeg P.S., et al. (2003). Metastasis suppressor genes: basic biology and potential clinical use. Clin. Breast Cancer, 4, 51–62 [DOI] [PubMed] [Google Scholar]
- 3. Adorno M., et al. (2009). A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell, 137, 87–98 [DOI] [PubMed] [Google Scholar]
- 4. Pollock C.B., et al. (2005). Oncogenic K-RAS is required to maintain changes in cytoskeletal organization, adhesion, and motility in colon cancer cells. Cancer Res., 65, 1244–1250 [DOI] [PubMed] [Google Scholar]
- 5. Sossey-Alaoui K., et al. (2005). WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp. Cell Res., 308, 135–145 [DOI] [PubMed] [Google Scholar]
- 6. Sossey-Alaoui K., et al. (2002). WAVE3, an actin-polymerization gene, is truncated and inactivated as a result of a constitutional t(1;13)(q21;q12) chromosome translocation in a patient with ganglioneuroblastoma. Oncogene, 21, 5967–5974 [DOI] [PubMed] [Google Scholar]
- 7. Takenawa T., et al. (2007). The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol., 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 8. Sossey-Alaoui K., et al. (2007). Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am. J. Pathol., 170, 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teng Y., et al. (2011). Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. Int. J. Cancer., 129, 2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sossey-Alaoui K., et al. (2005). WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J. Biol. Chem., 280, 21748–21755 [DOI] [PubMed] [Google Scholar]
- 11. Lee J.H., et al. (1997). Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res., 57, 2384–2387 [PubMed] [Google Scholar]
- 12. Jiang Y., et al. (2005). KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha. Clin. Exp. Metastasis., 22, 369–376 [DOI] [PubMed] [Google Scholar]
- 13. Cho S.G., et al. (2009). KiSS1 suppresses TNFalpha-induced breast cancer cell invasion via an inhibition of RhoA-mediated NF-kappaB activation. J. Cell. Biochem., 107, 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teng Y., et al. (2013). WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene.,10.1038/onc.2012.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sossey-Alaoui K., et al. (2007). c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J. Biol. Chem., 282, 26257–26265 [DOI] [PubMed] [Google Scholar]
- 16. Prat A., et al. (2010). Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res., 12, R68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baldridge M.T., et al. (2011). Inflammatory signals regulate hematopoietic stem cells. Trends Immunol., 32, 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elsawa S.F., et al. (2011). Comprehensive analysis of tumor microenvironment cytokines in Waldenstrom macroglobulinemia identifies CCL5 as a novel modulator of IL-6 activity. Blood., 118, 5540–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sullivan N.J., et al. (2009). Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene., 28, 2940–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walter M., et al. (2009). Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene, 28, 2745–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Queen M.M., et al. (2005). Breast cancer cells stimulate neutrophils to produce oncostatin M: potential implications for tumor progression. Cancer Res., 65, 8896–8904 [DOI] [PubMed] [Google Scholar]
- 22. Ara T., et al. (2010). Interleukin-6 in bone metastasis and cancer progression. Eur. J. Cancer., 46, 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teng Y., et al. (2012). HSP90 and HSP70 proteins are essential for stabilization and activation of WASF3 metastasis-promoting protein. J. Biol. Chem., 287, 10051–10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghoshal P., et al. (2012). Hypoxia-induced upregulation of the WASF3 metastasis associated gene leads to enhanced invasion in breast cancer cells. Int. J. Cancer., 129, 2825–2835 [Google Scholar]
- 25. Li P., et al. (2010). Stat3 activates the receptor tyrosine kinase like orphan receptor-1 gene in chronic lymphocytic leukemia cells. PLoS One, 5, e11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu H., et al. (2004). The STATs of cancer–new molecular targets come of age. Nat. Rev. Cancer, 4, 97–105 [DOI] [PubMed] [Google Scholar]
- 27. Teng Y., et al. (2010). Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br. J. Cancer., 103, 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buettner R., et al. (2002). Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res., 8, 945–954 [PubMed] [Google Scholar]
- 29. Xiong H., et al. (2012). Constitutive activation of STAT3 is predictive of poor prognosis in human gastric cancer. J. Mol. Med. (Berl)., 90, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 30. Abdulghani J., et al. (2008). Stat3 promotes metastatic progression of prostate cancer. Am. J. Pathol., 172, 1717–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gu L., et al. (2010). Transcription factor Stat3 stimulates metastatic behavior of human prostate cancer cells in vivo, whereas Stat5b has a preferential role in the promotion of prostate cancer cell viability and tumor growth. Am. J. Pathol., 176, 1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu H., et al. (2009). STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer., 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.