Abstract
Bone development depends on environmental, nutritional and hormonal factors. Yet, an ordered and timed activation of genes and their associated molecular pathways are central for the growth and development of healthy bones. The correct expression of genes depends on both cis- and trans-regulatory elements. Of these, the elusive role of chromatin ultrastructure is just beginning to become appreciated. Changes in the higher-order structure of chromatin are affecting the expression of genes in response to intrinsic and environmental signals. Cohesin and condensin are members of the structural maintenance of chromosome (SMC) family of protein complexes, which mediate higher-order chromatin structure by tethering distinct regions of chromatin either inter- or intra-molecularly. In recent years, SMCs had been identified for their function in the regulation of gene expression and developmental processes, whereas malfunction of cohesin or condensin has an impact on human health. However, little is known about the specific roles of SMC complexes in bone development and their possible effect on bone health. Here, we review studies that suggest an intimate link between SMCs and bone development, as well as a plausible effect, direct or indirect, on the bone health. We describe genetic syndromes associated with SMCs with distinctive bone phenotypes and identify links between SMCs and bone-related molecular pathways. Future studies of the relationship between SMCs and bone development will reveal new understandings of both the cellular and molecular roles of SMC complexes and provide new insights into the growth and developmental processes in the bone.
Regulation of bone development
The human skeleton consists of >200 bones that serve as a mechanical scaffold, which supports and protects organs as well as anchors muscles. Early development of the skeleton in vertebrates depends on genes that are responsible for patterning cell distribution and proliferation into mesenchymal condensations at sites of future skeletal elements. Within these condensations, cells further differentiate into chondrocytes or osteoblasts and form cartilage and bone, respectively.1 During childhood and adolescence, bone undergoes longitudinal and radial growth through a mechanism called bone formation (osteogenesis).2 In most of the skeleton, osteogenesis involves differentiation of the mesenchymal tissue into bone and cartilage, which is later replaced by bone in a process called endochondral ossification. During this process, chondrocytes proliferate, undergo hypertrophy and die. Cartilage extracellular matrix is then invaded by blood vessels and becomes calcified; the scaffold is invaded by hematopoietic cells and osteoprogenitor cells (which differentiate into osteoblasts), the latter depositing bone on remnants of the cartilage matrix.3
Environmental, nutritional and hormonal factors are known to affect bone growth, development and repair. Yet, a large proportion of these processes are determined by genetics. The three major signaling cascades involved in osteoblast differentiation, bone formation and repair are WNT signaling, transforming growth factor-β/bone morphogenic protein, and fibroblast growth factor pathways.4 Genetic studies have uncovered hundreds of genes involved in these pathways and others that are related to embryonic cartilage development (chondrogenesis) and postnatal processes, such as osteoblasto- and osteoclastogenesis, endochondral bone formation and bone remodeling.4 The genetics of bone formation and modeling is complex and involves interactions between many genes and both cis- and trans-regulatory elements. Changes in the higher-order structure of chromatin are just beginning to become appreciated as a central player in the regulation of gene expression and developmental processes.5 The family of protein complexes called structural maintenance of chromosome (SMC) family facilitates higher-order chromatin organization and have key roles in chromosome stability and dynamics. Herein we present evidence for the involvement of SMC complexes in bone growth and development. In this review, we will discuss potential crossroads between SMC complexes and bone-related molecular pathways and propose possible mechanisms in which SMCs can affect these pathways.
SMC Complexes
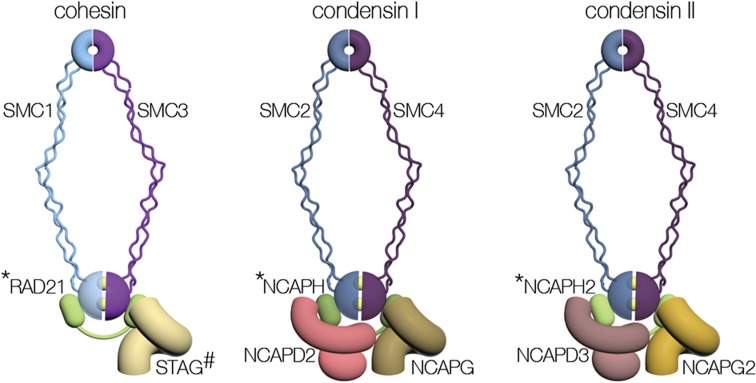
SMC complexes mediate higher-order chromatin structures by inter- or intra-molecularly tethering distinct regions of chromatin. SMCs are involved in the formation of chromatin loops, which can connect regulatory elements that are located at remote regions on the chromosome. These chromosome structures can either activate or suppress gene expression, define insulation regions and control the accessibility of transcription factors to the DNA. The SMC complexes cohesin and condensin that are evolutionary conserved in all eukaryotic cells feature a common molecular architecture but differ in their subunits (Figure 1 and Table 1).6,7 The heart of the SMC complex is a heterodimer composed of rod-like SMC proteins. The two ends of SMC proteins, called the hinge and the head, dimerize and form a ring structure. The dimerization of the SMC heads also creates two active ATPase domains. A kleisin superfamily protein is bound to the SMC heads and serves as the mediator of interaction with a set of the non-SMC regulatory subunits that vary between SMCs. Some of the mitotic subunits of cohesin are replaced by special subunits during meiosis (Table 1). SMC complexes are regulated by a large number of factors, generally referred to as auxiliary factors. Although the architecture shared by SMC complexes suggests a common biochemical mechanism, the molecular details of their activity remain elusive.6,7 SMCs are well known for their role in maintaining genome stability. However, this classical function of SMCs has been expanded in recent years to the regulation of gene expression.8 In addition, SMCs have been recognized as central players in developmental processes. Furthermore, studies have associated cohesin and condensin with several human developmental disorders and cancer.9,10,11
Figure 1.
Architecture of SMC protein complexes. The general architecture and subunit composition of cohesin, condensin I and condensin II are shown. The yellow balls between the SMC heads represent ATP molecules. * indicates the kleisin subunits STAG, NCAPH and NCAPH2. The cohesin subunit STAG is mark by # to indicate there are two paralogs called STAG1 and STAG2.
Table 1. Subunits of SMC complexes.
| Cohesina | Condensin I | Condensin II | |
|---|---|---|---|
| SMC protein | SMC1A/SMC1-βb | SMC2 | SMC2 |
| SMC protein | SMC3 | SMC4 | SMC4 |
| Kleisin | RAD21/REC8b/RAD21Lb | NCAPH | NCAPH2 |
| Non-SMC | STAG1/STAG2/STAG3b | NCAPG | NCAPG2 |
| Non-SMC | NCAPD2 | NCAPD3 |
Abbreviation: SMC, structural maintenance of chromosome.
aIsoforms are separated by a backslash.
bMeiotic isoform.
Cohesin
Cohesin is a four-subunit SMC complex that mediates sister chromatid cohesion, the process in which the newly replicated DNA strands, assembled into distinct chromatin fibers, are held together from the time of their formation until their separation during mitosis and meiosis (Figure 1). Cohesion maintains the fidelity of chromosome segregation by ensuring the bipolar attachment of the sister chromatids centromeres to the spindle.6,7 Cohesin was also identified as a key factor in non-mitotic processes. During the cellular response to double-strand breaks, cohesin induces cohesion between sister chromatids adjacent to the break in order to suppress nonhomologous recombination events.12 Most intriguingly, cohesin is also central to the regulation of gene expression.13 The complex is localized to a large number of regulatory elements on the chromosome and in many cases it is colocalized with the chromatin insulator CTCF (CCCTC-binding factor).14 Cohesin and CTCF mediate the formation of long-range chromatin interactions that are required for promoter choices and gene activation.14 It has been suggested that cohesin is recruited to gene promoters by interacting with the RNA polymerase II mediator complex and other transcriptional regulators, such as ATRX.15,16 Yet, the mechanism through which cohesin localizes to regulatory elements and affects gene expression is poorly understood. During development, sets of genes are activated or suppressed according to a precise program. Changes in the higher order of chromatin can affect gene expression levels directly or through the regulation of master transcription factors. Indeed, cohesin has been identified as a key factor in the development of the immune system and nerve cells.17,18 Several lines of evidence, discussed in the following sections, suggest that cohesin is also important for the proper development of bones.
Condensin
In eukaryotes, the canonical condensin I is conserved in almost all organisms, whereas the related condensin II complex is observed only in vertebrates.11 Both condensin I and II share core subunits and molecular architecture but differ in their kleisin and two non-SMC regulatory subunits (Figure 1 and Table 1).19 Condensins mediate the compaction of interphase chromatin before mitosis. Condensin II is localized on the chromatin throughout the cell cycle and functions in the early stages of chromatin condensation in prophase. In contrast, condensin I is cytoplasmic during interphase, and gains access to the chromatin only after nuclear membrane breakdown in pro-metaphase. Only then, condensin I and II collaborate to complete chromatin compaction.19 Condensin I and II distribute along the entire length of the chromosome in an alternated pattern. Depletion of condensin I or II subunits result in distinct swollen or curly chromosome morphology, respectively. Cloud-like chromosomes without observed sister chromatids cohesion were detected when both condensins were depleted.19 The spatial organization of condensin I and II, as well as the characteristic chromosome morphology obtained from their depletion suggest on a discrete function of each one of the complexes. The ratio between condensin I and II and the interplay with cohesin are essential for chromosome shaping.20 It is possible that after reformation of the nuclear envelope, a small fraction of condensin I remain in the nucleus, contributing to non-mitotic processes.21
The role of condensin in the regulation of gene expression is elusive. Condensin is involved in the expression regulation of the recombinant DNA and transfer RNA genes in budding and fission yeast and in the organization of polytene chromosomes in Drosophila.22,23,24 Condensin is involved in T-and B-cell development and maintenance of quiescence state by regulating the accessibility of transcription factors to chromatin.25,26 Many of our knowledge of the roles of condensin in the regulation of interphase chromosome come from studies of the dosage compensation complex (DCC) present in Caenorhabditis elegans. The DCC is a unique form of condensin I, which regulates X chromosome expression in C. elegans hermaphrodites. Unlike mammalian X inactivation, both X chromosomes in C. elegans hermaphrodite are actively transcribed but the expression of each X chromosome is reduced by 50%. In DCC, the SMC1 core subunit of condensin I is replaced by the related DPY27 subunit and the complex has a distinct set of non-SMC regulatory elements. However, it is unclear how this change affects the function of the DCC in comparison with condensin.27 Studies aiming to generalize the special function of the DCC to vertebrates and dissection of the full role of condensin in transcription, as well as the mechanisms of action are yet to be explored.
SMC-Related genetic disorders associated with bone abnormalities
The link between SMC complexes and bone has not been fully established, but an increasing number of reports suggest that the two are intimately associated. More specifically, syndromes caused by mutations in genes encoding SMC complex components are associated, in part, with skeletal abnormalities. In addition, SMC complexes have been identified as regulators of key factors in bone development. In-depth analysis of this intriguing interplay remains a future challenge for both bone and SMC fields.
Cohesinopathy is a group of human disorders associated with mutations in cohesin subunits or auxiliary factors, all of which express a bone-related phenotype. Cohesinopathy disorders are characterized by multisystematic growth retardation and intellectual disability. Robert's syndrome (RBS, OMIM number 268300) is a rare autosomal recessive genetic disorder caused by a mutation in the cohesin auxiliary factor ESCO2, an acetyltransferase that modifies the SMC3 core cohesin subunit. The skeletal aberrations in RBS patients include abnormal development of all four limbs. The bones of extremities are short, particularly the forearms and the tibia. RBS patients may also have abnormal or missing fingers and toes, and joint deformities mainly at the elbows and knees.28 A study in a ESCO2 knockout zebrafish model for RBS showed normal ossification; however, the shape of ossified structures was abnormal.29 Currently, the molecular pathway of cohesin in RBS is poorly understood.
Cornelia de Lange syndrome (CdLS, OMIM numbers 122470, 300590, 610759) is an additional cohesin-related genetic disorder with a prevalence of 1:10 000 live births. The disorder is classically manifested by prenatal and postnatal growth retardation and intellectual disability. CdLS patients often suffer from limb and digital anomalies, delayed skeletal maturation, abnormal thoracic configuration and flat acetabular angles in infancy. A mutation in one of five genes encoding a cohesin subunit or auxiliary factor is the cause of CdLS.30 The mutation in patients with CdLS results in transcriptional misregulation of genes, including MYC, a multilineage transcription factor involved in appendicular skeletal development and osteogenic differentiation.31,32,33,34 The combination of cohesin malfunction, misregulation of bone-related pathway and bone abnormalities implies on an unknown involvement of cohesin in bone growth and development.
Rare chromosomal aberration may reveal new information regarding the correlation between a genotype and a phenotype. A 17-Mbp chromosomal deletion in chromosome 9 that includes the SMC2 condensin subunit locus has been reported.35 The patient examined at the age of 1 year showed abnormalities in cranium, fingers, forehead and cranial sutures. However, because of the large size of the deletion, further studies are required to determine a direct link between SMC2 haploinsufficency in this patient and the skeletal abnormalities. Additional research will also be necessary to fully understand the effect of SMC2 on skeletal development.
Possible involvement of SMCs in molecular pathways of bone development
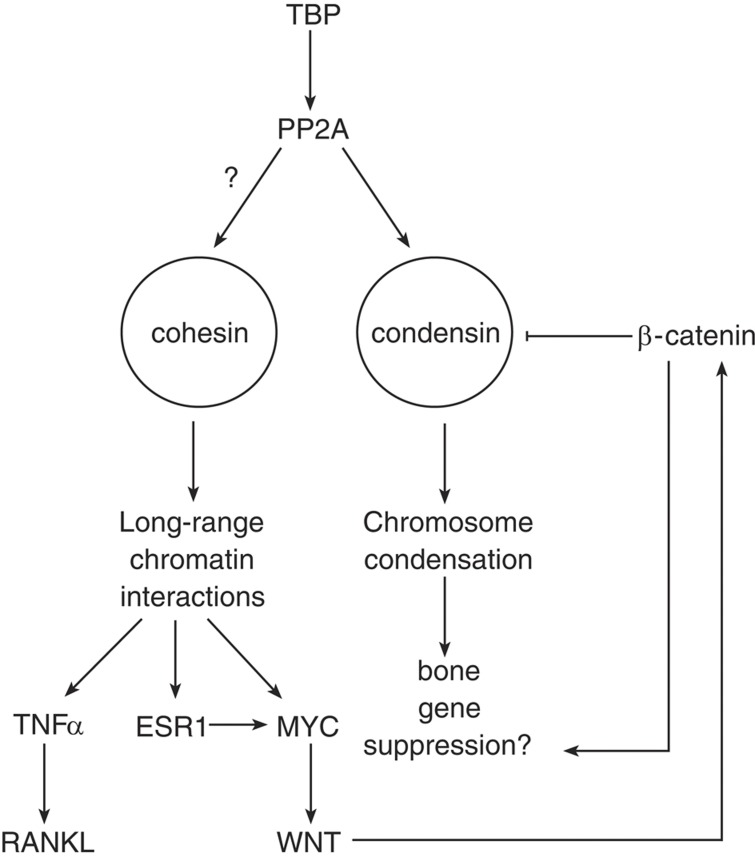
Several links between cohesin and molecular pathways involved in bone development have been identified, and they are summarized in Figure 2 and Table 2. The expression of MYC, which is misregulated in CdLS, is directly and positively regulated by cohesin.36 MYC is a central component of the mitogenic signals of WNT, a central pathway in the bone and cartilage differentiation.37 Two unanswered questions remain: is misregulation of MYC a primary cause for bone abnormalities in cohesinopathy? and do mutations in cohesin disrupt the WNT pathway? The answers to these questions would define a new pathway in bone development but remain a future challenge.
Figure 2.
Possible pathways linking cohesin, condensin and bone development. See text for details. The ‘?' symbol next to the arrow connecting protein phosphatase 2A (PP2A) and cohesin indicates a link between PP2A and cohesin. However, this interaction has been characterized only on mitotic chromosomes. Similar interaction in the context of cohesin role in transcription has not been described.
Table 2. Suggested bone genetic-related mechanisms involving SMC complexes.
| Gene | Gene function | Role in bone development | Relationship with SMC complexes | Reference |
|---|---|---|---|---|
| MYC | Transcription factor, central regulator of cell growth, proliferation, and apoptosis | Involved in appendicular skeletal development and osteogenic differentiation | Expression is regulated by cohesin | 31,34,36,39 |
| Wnt | Wnt genes encode secreted glycoproteins, which mediate intercellular signaling | Has a central role in bone and cartilage differentiation | Inhibits cell proliferation by suppressing the expression of the condensin subunit SMC2 | 4,33,37,49 |
| ESR1 | Receptor for estrogen | Changes in bone mineral density; A suggested role in osteoporosis | Cohesin binds chromatin and regulates gene expression in response to estrogen | 40,41,42,43 |
| TNF-α | An inflammatory cytokine involved in immunity and inflammation | Promotes osteolysis by activation of osteoblasts | The cohesin subunit STAG2 is as a transcriptional coactivator of TNF-α; | 44,45 |
| NCAPG | Condensin complex subunit 3 | Associated with height | Non-SMC subunit of condensin I | 47,48 |
Abbreviations: SMC, structural maintenance of chromosome; TNF-α, tumor necrosis factor-α.
The estrogen-signaling pathway is also tightly involved in skeletal development, bone maturation and closure of epiphyseal plates in growing bones in both females and males.38 The activation of MYC in response to changes of estradiol levels is regulated by cohesin.39 ESR1, which encodes the estrogen receptor, is associated with changes in the bone mineral density and has a major role in osteoporosis and bone's osteogenic response.40,41,42 Cohesin is enriched in promoters of estrogen-responsive genes, pointing out a possible collaboration between it and ESR1. Furthermore, depletion of the cohesin subunit SMC3 significantly impaired the estrogen-regulated transcriptome.43 Taken together, these findings suggest that cohesin modifies the higher-order structure of chromatin in estrogen pathway-related genes and serves toward establishment of the correct pattern of gene expression. Although most of these effects of estrogen were observed in oncogenic models,39 similar processes may affect normal bone growth and development and are awaiting to be characterized.
The cohesin subunit STAG2 has been identified as a transcriptional coactivator of the tumor necrosis factor-α (TNFα), an inflammatory cytokine involved in immunity and inflammation.44 TNF-α promotes osteolysis, or pathological bone resorption, by activation of osteoblasts and tissue stromal cells to express receptor activator of nuclear factor-κB ligand (RANKL), which induces osteoclast differentiation and activity function.45 In addition, TNF-α can act directly on osteoclast precursors, often via parallel signaling pathways and in synergy with RANKL, to promote osteoclastogenesis. Remarkably, while TNF-α stimulated waves of transcription of target genes in human umbilical vein endothelial cells, cohesin bound at the boundaries of the target genes stalls or slows down RNA polymerase, suggesting that cohesin antagonizes the TNF-α-dependent activation of genes.46 To this end, it is unclear how does the stalling of RNA polymerase by cohesin reflect on the developmental program of the cell.
To make the picture even more tantalizing, a genome-wide analysis identified an association between the height of German warmblood horses and the non-SMC condensin I complex subunit NCAPG47 Such correlation was also found in cattle, suggesting an evolutionarily conserved effect of condensin I on the growth of bones.48 A recent study revealed a new link between the WNT pathway and condensin. The β-catenin/TCF4 transcription factor binds to the SMC2 promoter, suppressing the gene expression and inhibiting cell proliferation.49
The different studies and their findings clearly demonstrate a relationship between SMCs and key factors in bone-related pathways. Therefore, the next challenge is to dissect the molecular mechanisms, which are affecting bone development. The first mechanism involving direct regulation of differentiation genes such as MYC and ESR1, is described above. In such mechanism, SMCs mediate changes in the chromatin structure that in turn control the expression levels of the master regulators themselves. In the second possible mechanism, inspired by the functional cooperation of STAG2 and TNF-α, SMCs collaborate with master regulators of transcription and modulate the chromatin structure of target genes, allowing their activation or repression.
An alternative and exciting mechanism in which SMCs can affect bone development is mitotic bookmarking. At the end of each cell cycle, chromosomes are condensed in order to allow their proper segregation to the two daughter cells. Bookmarking of genes, mediated by binding of specific factors to the promoter before the onset of mitosis or by epigenetic signals, allows cells to maintain a certain differentiation pathway by reactivating the same set of genes when chromosomes decondense and re-enter the cell cycle. DNA methylation and histone modifications are involved in bookmarking.50 It is possible that cohesin is a key factor in mitotic bookmarking. Evidence comes from the altered DNA methylation pattern of chromosomes derived from CdLS patients. It was suggested that cohesin marks CpG islands for methylation. Furthermore, cohesin preferably binds hypomethylated regions and in turn alters the higher-order structure of the chromatin. Cohesin-mediated structure may protect regions of the chromosomes from condensation and allow their rapid activation after mitosis.
Interestingly, condensin may indicate another mechanism of mitotic bookmarking in differentiated bone cells. The expression of lineage-specific genes is mediated by maintaining the TATA-binding protein (TBP) in their promoters in condensed chromosomes.51,52 TBP has been shown to antagonize condensin binding through the dephosphorylation of its NCAPG subunit by recruiting protein phosphatase 2A (PP2A) in HeLa cells, inactivating condensin at these regions, and create a mitotic bookmark.52
Future perspective
An increasing number of observations and circumstantial evidence suggest a link between the SMC complexes, bone growth and development. Up to this point, two basic facts have been established experimentally. First, cohesin and condensin regulate the expression of key factors in bone development (Figure 2). Second, human disorders associated with mutations in genes encoding these complexes show a clear bone development phenotype. We are currently looking up in the genome-wide association study results of bone phenotype of children in order to find genetic association between SMC-encoding genes and bone development,53 (MCZ, DEK and IO; unpublished).
The next fundamental enigmas that need to be explored are the specific effects of cohesin in bone development and the mechanism of its regulation. Studies showed that the expression of a gene is correlated to the amount of cohesin bound to its promoter. An intensive genomic effort is now required in order to gain new insight into the process of osteoblast differentiation. Chromatin immunoprecipitation-sequence platform (or similar technology) can be used to analyze the cohesin occupancy in promoters by experiments in order to identify the specific genes that are being regulated by cohesin in bone cells. Chromosome conformation capture is a useful tool to study long-range interactions in chromatin, as had been shown in the ENCODE project.14 A future goal is to use this technique to define the specific interactions between cis-regulatory modules and promoters in osteoblasts. Other new technologies such as ChIA-PET will allow combining the knowledge regarding the chromosomal localization of the SMC complexes and the higher-order structure of the chromosome in osteoblasts. These data will allow integrating the biology of the bone with the nuclear architecture of the nucleus.
On the other hand, experts in the SMC field need to explore how and when SMCs are recruited to promoters in osteoblasts and, specifically, how they gain their specificity. This is likely to involve interaction with tissue-specific transcription factors and chromatin remodelers. Identifying such bone-related factors and characterizing the temporal and spatial relationship between SMCs and bone pathways is a necessary step for understanding the functions of SMCs in the bone. The ongoing effort to dissect the biochemical details of cohesin and condensin activities will also elucidate their bone-related functions. An interesting future goal is to test whether environmental signals affect bones through the modulation of SMCs activity. These intriguing questions are to be explored. Finally, this multidisciplinary effort will provide new diagnostic techniques and therapeutic targets for osteoporosis and other bone health-related diseases.
Acknowledgments
We thank Dr Avi Matityahu for critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
References
- Galotto M, Campanile G, Robino G, Cancedda FD, Bianco P, Cancedda R. Hypertrophic chondrocytes undergo further differentiation to osteoblast-like cells and participate in the initial bone formation in developing chick embryo. J Bone Miner Res 1994;9:1239–1249. [DOI] [PubMed] [Google Scholar]
- Clarke B. Normal bone anatomy and physiology. Clin J Am Soc Nephrol 2008;3(Suppl 3): S131–S139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol 2008;40:46–62. [DOI] [PubMed] [Google Scholar]
- Macsai CE, Foster BK, Xian CJ. Roles of Wnt signalling in bone growth, remodelling, skeletal disorders and fracture repair. J Cell Physiol 2008;215:578–587. [DOI] [PubMed] [Google Scholar]
- Vogelmann J, Valeri A, Guillou E, Cuvier O, Nollmann M. Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus 2011;2:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 2008;24:105–129. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem 2005;74:595–648. [DOI] [PubMed] [Google Scholar]
- Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol 2013;25:327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Print CG, Monnich M. Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies. Front Genet 2012;3:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, Musio A. The dark side of cohesin: the carcinogenic point of view. Mutat Res 2011;728:81–87. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 2012;26:1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 2007;317:245–248. [DOI] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 2005;132:4743–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature 2012;489:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010;467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, Eubanks JH, Mann MR et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell 2010;18:191–202. [DOI] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol 2009;182:44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM et al. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development 2007;134:3191–3201. [DOI] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 2003;115:109–121. [DOI] [PubMed] [Google Scholar]
- Lam WW, Peterson EA, Yeung M, Lavoie BD. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev 2006;20:2973–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Ogushi S, Saitou M, Hirano T. Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol Biol Cell 2011;22:3465–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T. RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells 2007;12:759–771. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 2008;22:2204–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl TA, Sweeney SJ, Knepler PJ, Bosco G. Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet 2008;4:e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Gatzka M, Thomas PG, Ihle JN. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J 2011;30:263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Papasani M, Hao Y, Calamito M, Wei F, Quinn WJ III et al. YY1 controls Igkappa repertoire and B-cell development, and localizes with condensin on the Igkappa locus. EMBO J 2013;32:1168–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR et al. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 2009;23:602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Berg DJ, Francke U. Robert's syndrome: a review of 100 cases and a new rating system for severity. Am J Med Genet 1993;47:1104–1123. [DOI] [PubMed] [Google Scholar]
- Monnich M, Kuriger Z, Print CG, Horsfield JA. A zebrafish model of Robert's syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PLoS One 2011;6:e20051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet 2009;76:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZQ, Shung CY, Ota S, Akiyama H, Keene DR, Hurlin PJ. Sequential and coordinated actions of c-Myc and N-Myc control appendicular skeletal development. PLoS One 2011;6:e18795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiehl JB. Complex ankle fracture dislocations with syndesmotic diastasis. Orthop Rev 1990;19:499–507. [PubMed] [Google Scholar]
- Sathi GA, Tsujigiwa H, Ito S, Siar CH, Katase N, Tamamura R et al. Osteogenic genes related to the canonic WNT pathway are down-regulated in ameloblastoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2012;114:771–777. [DOI] [PubMed] [Google Scholar]
- Battaglino R, Kim D, Fu J, Vaage B, Fu XY, Stashenko P. c-myc is required for osteoclast differentiation. J Bone Miner Res 2002;17:763–773. [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet 2009;84:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ et al. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol 2010;344:637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao DY, Jonason JH, Zhang Y, Hsu W, Chen D, Hilton MJ et al. Cartilage-specific beta-catenin signaling regulates chondrocyte maturation, generation of ossification centers, and perichondrial bone formation during skeletal development. J Bone Miner Res 2012;27:1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren A, Lundmark P, Axelsson T, Lind L, Syvanen AC. Association of the estrogen receptor 1 (ESR1) gene with body height in adult males from two Swedish population cohorts. PLoS One 2008;3:e1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan MV, Eccles MR, Horsfield JA. Cohesin is required for activation of MYC by estradiol. PLoS One 2012;7:e49160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart F, Marini F, Bianchi G, Minisola S, Luisetto G, Pirazzoli A et al. Age-specific effects of estrogen receptors' polymorphisms on the bone traits in healthy fertile women: the BONTURNO study. Reprod Biol Endocrinol 2009;7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Ralston SH, Bennett ST, Brandi ML, Grinberg D, Karassa FB et al. Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. JAMA 2004;292:2105–2114. [DOI] [PubMed] [Google Scholar]
- Lee K, Jessop H, Suswillo R, Zaman G, Lanyon L. Endocrinology: bone adaptation requires oestrogen receptor-alpha. Nature 2003;424:389. [DOI] [PubMed] [Google Scholar]
- Rhodes JM, McEwan M, Horsfield JA. Gene regulation by cohesin in cancer: is the ring an unexpected party to proliferation? Mol Cancer Res 2011;9:1587–1607. [DOI] [PubMed] [Google Scholar]
- Lara-Pezzi E, Pezzi N, Prieto I, Barthelemy I, Carreiro C, Martinez A et al. Evidence of a transcriptional co-activator function of cohesin STAG/SA/Scc3. J Biol Chem 2004;279:6553–6559. [DOI] [PubMed] [Google Scholar]
- Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB. TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med 2012;209:319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K et al. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci USA 2009;106:18357–18361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetens J, Widmann P, Kuhn C, Thaller G. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim Genet 2013;44:161–171. [DOI] [PubMed] [Google Scholar]
- Lindholm-Perry AK, Sexten AK, Kuehn LA, Smith TP, King DA, Shackelford SD et al. Association, effects and validation of polymorphisms within the NCAPG—LCORL locus located on BTA6 with feed intake, gain, meat and carcass traits in beef cattle. BMC Genet 2011;12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Suarez-Lopez L, Castano J, Messent A, Abasolo I, Fernandez Y et al. Human SMC2 protein, a core subunit of human condensin complex, is a novel transcriptional target of the WNT signaling pathway and a new therapeutic target. J Biol Chem 2012;287:43472–43481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino M, van Wijnen AJ, Stein JL, Lian JB et al. Bookmarking the genome: maintenance of epigenetic information. J Biol Chem 2011;286:18355–18361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle 2009;8:818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol 2008;10:1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gomez C, Kemp JP, Estrada K, Eriksson J, Liu J, Reppe S et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet 2012;8:e1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]