Abstract
Although activation of tyrosine kinase pathways is a shared theme among myeloproliferative neoplasms, the pathogenetic basis of chronic neutrophilic leukemia (CNL) has remained elusive. Recently, we identified high-frequency oncogenic mutations in the granulocyte-colony stimulating factor receptor (CSF3R) in CNL and in some patients with atypical chronic myeloid leukemia. Inhibition of Janus kinase 2 or SRC kinase signaling downstream of mutated CSF3R is feasible and should be explored therapeutically. Herein, we discuss the potential impact of these findings for the classification and treatment of these disorders.
Introduction
In clinical practice, neutrophilia most commonly relates to leukemoid reactions due to chronic infections, inflammatory diseases, or various types of malignancies.1 Cytokine-driven neutrophilia accompanying plasma cell neoplasms is a well-described association.1,2 Chronic neutrophilic leukemia (CNL) is a rare myeloproliferative neoplasm, with only ∼200 patients reported to date, mostly culled from case reports and small case series.3 Atypical chronic myeloid leukemia (aCML) is an uncommon myelodysplastic/myeloproliferative (MDS/MPN) neoplasm, with a relative incidence estimated at 1 to 2 cases for every 100 patients with BCR-ABL1-positive chronic myeloid leukemia (CML).4,5
Histopathologic and clinical features of CNL and aCML
Absence of both BCR-ABL1 and rearrangement of PDGFRA, PDGFRB, or FGFR1 are minimal diagnostic requirements for CNL and aCML.3,4 The clinical and laboratory features of CNL include hepato/splenomegaly, persistent neutrophilic leukocytosis with minimal left shift often characterized by toxic granulation and Döhle bodies, and elevated leukocyte alkaline phosphatase and vitamin B12 levels.1,3,6,7 According to the 2008 World Health Organization (WHO) diagnostic criteria for CNL,3 the leukocytosis is ≥25 × 109/L; >80% of leukocytes are segmented neutrophils/band forms; and <10% are immature granulocytes. Granulocytic dysplasia is not present, and there is no monocytosis, eosinophilia, or basophilia. Examination of the bone marrow shows a myeloid hyperplasia with full maturation with <5% myeloblasts (<1% in the peripheral blood). Megakaryocytes are typically normal, but can include some small hypolobated megakaryocytes. Reticulin fibrosis is not significantly increased. Exclusionary criteria include no evidence of a reactive neutrophilia, other MPN, MDS, or overlap MDS/MPN disorder.3
Clinicopathologic characteristics of aCML include splenomegaly and a neutrophilic leukocytosis with left shift and prominent granulocytic dysplasia (eg, hypogranular and hypolobated neutrophils, abnormal chromatin clumping, pseudo–Pelger-Hüet neutrophils).4 The white blood count is ≥13 × 109/L, with immature granulocytes ≥10% of leukocytes and <20% blasts in the blood and marrow. Monocytosis and basophilia are not prominent (<2% peripheral blood basophils, <10% peripheral blood monocytes), and the leukocyte alkaline phosphatase level may be low, normal, or increased, therefore lacking diagnostic utility.4,8,9 A hypercellular bone marrow is observed with a myeloid hyperplasia and prominent granulocytic dysplasia, although multilineage dysplasia may be present.
The disease course of CNL is variable, but acceleration is typically characterized by refractory neutrophilia, worsening organomegaly, and blastic transformation. In a literature review of 40 patients meeting WHO criteria for true CNL, the median survival was 23.5 months (range, 1-106 months).7 Median time to acute myeloid leukemia (AML) transformation was 21 months (range, 3-94 months). The most frequent causes of death were intracranial hemorrhage, progressive disease/blastic transformation, and regimen-related toxicity from induction chemotherapy or transplantation.6,7
The largest series of WHO-defined aCML consists of 55 cases from an Italian cohort.5 The overall median survival was 25 months compared with survivals ranging from 14 to 30 months gleaned from 3 smaller studies.8-10 In the Italian report, transformation to acute leukemia occurred in 22 patients (40%), with a median time from diagnosis of 18 months.5 A multivariate analysis revealed that shorter survival was associated with older age (>65 years), female gender, leukocyte count >50 × 109/L, and the presence of immature circulating precursors.5
No standard of care exists for CNL or aCML. Therapy has primarily consisted of hydroxyurea or other oral chemotherapeutics, as well as interferon-α.1,5-11 These agents can elicit improvement in blood counts but exhibit no proven disease-modifying benefit. Although splenic irradiation and splenectomy may provide transient palliation of symptomatic splenomegaly, the latter has been associated with anecdotal worsening of neutrophilic leukocytosis in CNL. The limited experience with induction-type chemotherapy for blastic transformation is generally poor with death related to resistant disease or regimen-related toxicities. Allogeneic transplantation may result in favorable long-term outcomes in selected patients, particularly when undertaken in the chronic phase of disease.1,5-7,9
Cytogenetic/molecular features
Although clonality has been demonstrated in CNL,12,13 the majority of patients exhibit normal cytogenetics.1,6,7 The frequency of chromosomal changes in aCML is more variable, ranging from 20% to 88% in 4 series.5,8-10 In both diseases, trisomy 8 and del (20q) are the most common nonspecific chromosomal abnormalities observed at diagnosis or at the time of progressive disease. The e19/a2 type BCR/ABL mRNA transcript (p230) that was initially reported as the molecular basis for some cases of CNL is instead now considered related to an uncommon neutrophilic variant of CML.14 Notwithstanding case reports of JAK2 V617F positivity in selected cases of CNL and aCML,11,15,16 no other recurrent genetic mutations had been identified in these diseases until the recent discoveries of mutant SETBP1 and CSF3R.15,17 In addition to these newly defined, high-frequency mutations in SETBP1 and CSF3R, other recurrent mutations in a variety of genes including NRAS, IDH2, and CBL have recently been described in aCML at lower frequencies.17
New genetic findings in CNL and aCML
CSF3R mutations
We recently reported that ∼50% to 60% of patients with CNL or aCML harbor mutations in the receptor for colony-stimulating factor 3 (CSF3R; GCSFR).15 Among a total of 29 patients, 8 of 9 CNL (89%) and 8 of 20 aCML (40%) cases exhibited CSF3R mutations. These mutations fall into 2 classes: nonsense or frame-shift mutations that lead to premature truncation of the cytoplasmic tail of the receptor (truncation mutations) and point mutations in the extracellular domain of CSF3R (membrane proximal mutations). The most common CSF3R mutation in CNL/aCML is the membrane proximal mutation: T618I. CSF3R is known to signal downstream through both Janus kinase (JAK)18 and SRC tyrosine kinase pathways,19 and the 2 classes of CSF3R mutations that we observed exhibit different downstream signaling and kinase inhibitor sensitivities. Although CSF3R truncation mutations may exhibit sensitivity to JAK kinase inhibition in the context of high concentrations of the CSF3R ligand, downstream signaling operates predominantly through SRC kinases and exhibits drug sensitivity to SRC kinase inhibitors such as dasatinib. In contrast, CSF3R membrane proximal mutations strongly activate the JAK/signal transducer and activator of transcription pathway and are sensitive to JAK kinase inhibitors such as ruxolitinib. A CNL patient with the CSF3R T618I membrane proximal mutation was treated with ruxolitinib and exhibited a marked, durable decrease in white blood cell and absolute neutrophil counts and resolution of thrombocytopenia (initially reported as a 5-month response, now still responding after 11 months). This case, although anecdotal, provides a strong rationale for the investigation of tyrosine kinase inhibitors (TKIs) in CNL and aCML patients. Further investigation in clinical trial settings will be required to determine whether JAK kinase inhibition in the setting of CSF3R mutation will lead to greater decreases in mutation allele burden compared with the responses observed in the setting of JAK2V617F. This will also be informative regarding the extent to which CSF3R mutation is a primary driver of disease.
Under normal circumstances, the CSF3R ligand, granulocyte-colony-stimulating factor (G-CSF), promotes growth and survival of myeloid precursor cells, ultimately leading to differentiation of these myeloid precursors into neutrophils. In keeping with the prominent role of G-CSF in the production of neutrophils, deletion of CSF3R leads to neutropenia in mouse models.20 In addition to regulating normal neutrophil homeostasis, G-CSF levels rapidly increase during infection, resulting in elevated levels of neutrophils as a component of the immune response.21 The normal role of CSF3R in promoting neutrophil production is biologically consistent with our observation of CSF3R activating mutations in hematologic malignancies characterized by high levels of neutrophils.
Mutations in CSF3R have previously been identified in patients with severe congenital neutropenia (SCN).22-24 These SCN patients harbor germ-line mutations in neutrophil elastase or other genes necessary for production of neutrophils. To avoid infections associated with neutropenia, a large percentage of these patients are treated chronically with G-CSF to improve neutrophil production. Approximately one-third of SCN patients eventually acquire truncation mutations in CSF3R (analogous to the truncation mutations we have observed in de novo CNL and aCML), which are associated with transformation to myelodysplasia or AML.22,23 Additionally, similar to our observations in CNL/aCML, SCN patients can acquire compound CSF3R mutations. A recent paper described a case of a SCN patient who acquired a CSF3R truncation mutation followed by a T618I point mutation (reported as T595I using the traditional CSF3R numbering system that does not include the signal peptide).24
CSF3R truncation mutations result in a loss of a portion of the cytoplasmic domain of the receptor, suggesting possible molecular mechanisms for receptor activation. Truncated CSF3R may lack the di-leucine internalization motif, resulting in an increase in cell surface expression of the receptor.25 These truncated receptors may also lack the binding site for suppressor of cytokine signaling 3, which reduces trafficking of CSF3R to the lysosome. The functional consequence of these truncations is altered and increased receptor activity conferring ligand hypersensitivity. In contrast, the membrane proximal mutations exhibit receptor activation in the complete absence of ligand,15,24 suggesting distinct mechanisms of activation of these classes of CSF3R mutations. These distinct mechanisms may contribute to the differential downstream signaling and drug sensitivity of truncation vs membrane proximal mutations. In fact, 4 of 5 patients with truncation mutations also had the T618I point mutation, indicating that acquisition of both classes of mutations may be advantageous for tumor pathogenesis.
SETBP1 mutations
Recurrent mutations in SETBP1 were recently identified in 25% of aCML patients.17 Set binding protein (SETBP1) interacts with SET, a negative regulator of the tumor suppressor protein phosphatase 2A (PP2A).26 SETBP1 protects SET from protease cleavage, thus increasing the amount of SET available to repress the activity of PP2A.27 In AML, SETBP1 overexpression is significantly associated with reduced survival, indicating that SETBP1 may be relevant to leukemia oncogenesis.27 SETBP1 is mutated at lower frequencies in unclassified MDS/MPN (10%) and CMML (4%), but no SETBP1 mutations are found in AML, acute lymphoblastic leukemia, chronic lymphocytic leukemia, or solid tumors.17 SETBP1 mutations in aCML were associated with higher white blood cell counts at diagnosis and poorer survival.17 Similarly, overall survival was significantly worse for CMML patients with SETBP1 mutations.28,29
In aCML, the majority of SETBP1 mutations are located in a 14 amino acid stretch that is also mutated in Schinzel-Giedion syndrome, a rare genetic disease characterized by congenital malformations, mental retardation, and a high prevalence of epithelial tumors.30 When phosphorylated, this mutational hotspot is bound by the E3 ubiquitin ligase subunit β-TrCP1, leading to ubiquitination of SETBP1 and subsequent degradation.17 The SETBP1 mutants disrupt this consensus β-TrCP motif, leading to increased SETBP1 and SET expression, which lowers PP2A activity and increases proliferation of cells.17 This increased cellular proliferation may be relevant to the pathobiology of aCML.
CSF3R and SETBP1 mutational overlap
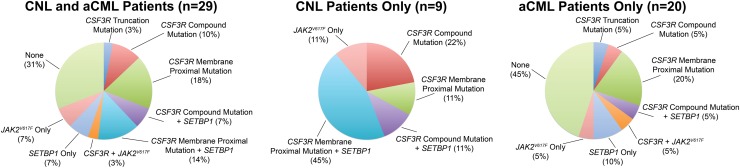
CSF3R and SETBP1 mutations are not mutually exclusive in CNL/aCML. In follow-up sequencing of an expanded cohort of 29 patients from the New England Journal of Medicine report,15 21% have exhibited both CSF3R and SETBP1 mutations, with 31% of samples having CSF3R mutations only and 7% with SETBP1 mutations only (Figure 1). These combinatorial mutations suggest a requirement for different therapeutic approaches tailored to the molecular profile of individual patients. Although these recent studies have increased our understanding of the genetic drivers of CNL and aCML, this knowledge is lacking in ∼30% of patients. Functional genomic studies will permit a more comprehensive understanding of the molecular complexity of these diseases.
Figure 1.
Mutations in CSF3R and SETBP1 are common in CNL and aCML. Percentages of CSF3R, SETBP1, and JAK2 V617F mutations in 29 patients with CNL or aCML are shown. CSF3R mutations arise in 2 classes, nonsense or frameshift mutations that truncate the cytoplasmic tail (truncation mutations) and point mutations in the extracellular domain (membrane proximal mutation), and some cases exhibit both classes of mutations on the same allele (compound mutations). These mutations can occur in isolation or in combination with other mutant genes, with 21% of patients having both CSF3R and SETBP1 mutations. One patient exhibited mutations in both CSF3R (G683R) and JAK2 (V617F); however, the clonality of this double mutation could not be established due to limited material, presenting the possibility of polyclonal populations of tumor cells with distinct mutational profiles. The frequencies of each class of CSF3R mutation alone or in combination with SETBP1 or JAK2 are shown for a combined cohort of CNL and aCML (n = 29), the CNL cases only (n = 9), and the aCML cases only (n = 20).
Summary
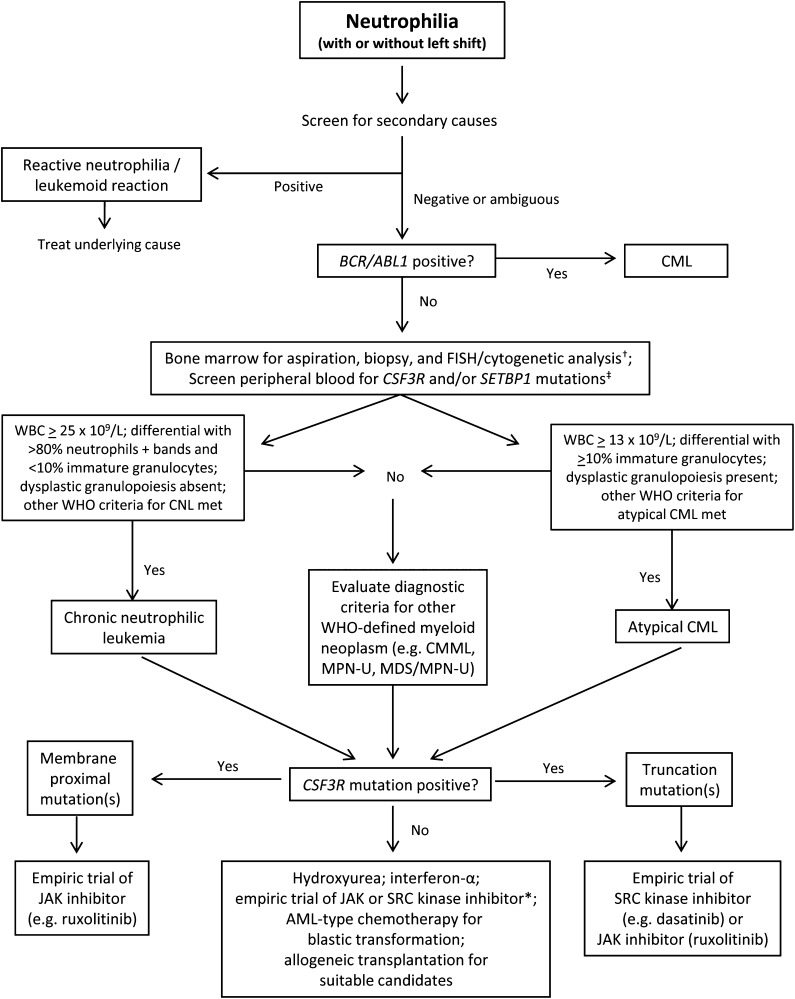
The discovery of high-frequency CSF3R mutations in CNL, and to a lesser extent in aCML, identifies a new major diagnostic criterion for these diseases and suggests a close relationship between these neutrophilic leukemias. These findings may also lend more specificity to the WHO diagnostic criteria for CNL and aCML, which are variably applied in routine clinical practice. In patients for whom the cause of neutrophilia is not easily discernible, the incorporation of CSF3R mutation testing can be a useful point-of-care diagnostic to evaluate for the presence of a clonal myeloid disorder, as well as the potential for genetically informed therapy (Figure 2).
Figure 2.
Provisional diagnostic algorithm for neutrophilia and genetically informed treatment options. †FISH probes for myeloid-associated cytogenetic abnormalities can be used to complement standard karyotype analysis to establish the presence of a clonal myeloid disorder. Cytogenetic/FISH or molecular evaluation of rearranged PDGFRA/B, and FGFR1 should also be considered if eosinophilia is present. ‡Testing for JAK2 V617F, infrequently identified in CNL or atypical CML, should also be considered. The list of relevant mutations, including molecular abnormalities in complementary signaling pathways to CSF3R and SETBP1, may expand over time. *For patients who are CSF3R-mutation negative, use of a JAK inhibitor (particularly if JAK2 V617F positive) or SRC kinase inhibitor could be considered since mutations in the same or related signaling pathways may be present.
The basis for phenotypic differences between CSF3R mutation-positive CNL and aCML may relate to multiple factors. These likely include patient genetic background, characteristics of the distinct CSF3R mutation (eg, hematopoietic cell of origin, allele burden, activation of distinct signaling cascades), and the accompanying clonal architecture of these neoplasms (eg, mutations of SETBP1, JAK2 V617F, or other genes). CNL may be the archetype of an MPN whose pathogenesis is predominantly characterized by mutations of CSF3R, whereas aCML may be more genetically heterogeneous.
Given the poor prognosis of these disorders, the potential applicability of JAK or SRC kinase inhibitors is an important bench-to-bedside implication of the discovery of activating CSF3R mutations. These drugs are expected to be more effective in the chronic phase of disease before increasing genetic complexity associated with disease transformation supervenes, and therapeutic inhibition of CSF3R-related signaling may be less relevant. Future experience with these agents will inform whether complete hematologic remissions and in-depth molecular responses are realized, similar to experience with TKIs in CML and PDGFRA/B-rearranged myeloid neoplasms associated with eosinophilia. Ultimately, comparison of TKI-treated patients to historical controls will help determine whether these therapies impact disease endpoints such as leukemia-free progression and overall survival.
Acknowledgments
We apologize to investigators whose work was not included and referenced in this review due to space restrictions.
J.G. is supported by the Charles and Ann Johnson Foundation. J.E.M. is supported in part by the Training Program in Molecular Hematology 5T32HL007781-20. J.W.T. is supported by grants from The Leukemia and Lymphoma Society, the V Foundation for Cancer Research, the Gabrielle’s Angel Foundation for Cancer Research, the William Lawrence and Blanche Hughes Fund, and the National Cancer Institute (4 R00CA151457-03).
Authorship
Contribution: J.G., J.E.M., T.I.G., and J.W.T. contributed to the writing of the manuscript.
Conflict-of-interest disclosure: J.G. and J.W.T. receive funding for administration of clinical trials from Incyte, manufacturer of ruxolitinib, and J.G. additionally serves on an Incyte advisory board. The remaining authors declare no competing financial interests.
Correspondence: Jason Gotlib, Department of Medicine (Hematology), Stanford Cancer Institute, 875 Blake Wilbur Dr, Room 2324, Stanford, CA 94305-5821; e-mail: jason.gotlib@stanford.edu.
References
- 1.Reilly JT. Chronic neutrophilic leukaemia: a distinct clinical entity? Br J Haematol. 2002;116(1):10–18. doi: 10.1046/j.1365-2141.2002.03234.x. [DOI] [PubMed] [Google Scholar]
- 2.Kohmura K, Miyakawa Y, Kameyama K, Kizaki M, Ikeda Y. Granulocyte colony stimulating factor-producing multiple myeloma associated with neutrophilia. Leuk Lymphoma. 2004;45(7):1475–1479. doi: 10.1080/10428190310001645870. [DOI] [PubMed] [Google Scholar]
- 3.Bain BJ, Brunning RD, Vardiman JW, Thiele J. Chronic neutrophilic leukaemia. In: Swerdlow SH, Campo E, Lee Harris N, et al., editors. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. pp. 38–39. [Google Scholar]
- 4.Vardiman JW, Bennett JM, Bain BJ, Brunning RD, Thiele J. Atypical chronic myeloid leukaemia, BCR-ABL1 negative. In: Swerdlow SH, Campo E, Lee Harris N, et al., editors. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. pp. 80–81. [Google Scholar]
- 5.Breccia M, Biondo F, Latagliata R, Carmosino I, Mandelli F, Alimena G. Identification of risk factors in atypical chronic myeloid leukemia. Haematologica. 2006;91(11):1566–1568. [PubMed] [Google Scholar]
- 6.Böhm J, Schaefer HE. Chronic neutrophilic leukaemia: 14 new cases of an uncommon myeloproliferative disease. J Clin Pathol. 2002;55(11):862–864. doi: 10.1136/jcp.55.11.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott MA, Hanson CA, Dewald GW, Smoley SA, Lasho TL, Tefferi A. WHO-defined chronic neutrophilic leukemia: a long-term analysis of 12 cases and a critical review of the literature. Leukemia. 2005;19(2):313–317. doi: 10.1038/sj.leu.2403562. [DOI] [PubMed] [Google Scholar]
- 8.Kurzrock R, Bueso-Ramos CE, Kantarjian H, et al. BCR rearrangement-negative chronic myelogenous leukemia revisited. J Clin Oncol. 2001;19(11):2915–2926. doi: 10.1200/JCO.2001.19.11.2915. [DOI] [PubMed] [Google Scholar]
- 9.Martiat P, Michaux JL, Rodhain J. Philadelphia-negative (Ph-) chronic myeloid leukemia (CML): comparison with Ph+ CML and chronic myelomonocytic leukemia. The Groupe Français de Cytogénétique Hématologique. Blood. 1991;78(1):205–211. [PubMed] [Google Scholar]
- 10.Hernández JM, del Cañizo MC, Cuneo A, et al. Clinical, hematological and cytogenetic characteristics of atypical chronic myeloid leukemia. Ann Oncol. 2000;11(4):441–444. doi: 10.1023/a:1008393002748. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Pan J, Guo J. Presence of the JAK2 V617F Mutation in a Patient with Chronic Neutrophilic Leukemia and Effective Response to Interferon Alfa-2b. Acta Haematol. 2013;130(1):44–46. doi: 10.1159/000345851. [DOI] [PubMed] [Google Scholar]
- 12.Böhm J, Kock S, Schaefer HE, Fisch P. Evidence of clonality in chronic neutrophilic leukaemia. J Clin Pathol. 2003;56(4):292–295. doi: 10.1136/jcp.56.4.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong YL, Cheng G. Clonal nature of chronic neutrophilic leukemia. Blood. 1993;82(3):1035–1036. [PubMed] [Google Scholar]
- 14.Pane F, Frigeri F, Sindona M, et al. Neutrophilic-chronic myeloid leukemia: a distinct disease with a specific molecular marker (BCR/ABL with C3/A2 junction). Blood. 1996;88(7):2410–2414. [PubMed] [Google Scholar]
- 15.Maxson JE, Gotlib J, Pollyea DA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368(19):1781–1790. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steensma DP, Dewald GW, Lasho TL, et al. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005;106(4):1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piazza R, Valletta S, Winkelmann N, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45(1):18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson SE, Oates AC, Harpur AG, Ziemiecki A, Wilks AF, Layton JE. Tyrosine kinase JAK1 is associated with the granulocyte-colony-stimulating factor receptor and both become tyrosine-phosphorylated after receptor activation. Proc Natl Acad Sci USA. 1994;91(8):2985–2988. doi: 10.1073/pnas.91.8.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corey SJ, Burkhardt AL, Bolen JB, Geahlen RL, Tkatch LS, Tweardy DJ. Granulocyte colony-stimulating factor receptor signaling involves the formation of a three-component complex with Lyn and Syk protein-tyrosine kinases. Proc Natl Acad Sci USA. 1994;91(11):4683–4687. doi: 10.1073/pnas.91.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 1996;5(5):491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami M, Tsutsumi H, Kumakawa T, et al. Levels of serum granulocyte colony-stimulating factor in patients with infections. Blood. 1990;76(10):1962–1964. [PubMed] [Google Scholar]
- 22.Dong F, Brynes RK, Tidow N, Welte K, Löwenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333(8):487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 23.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;109(1):93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 24.Beekman R, Valkhof MG, Sanders MA, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119(22):5071–5077. doi: 10.1182/blood-2012-01-406116. [DOI] [PubMed] [Google Scholar]
- 25.Ward AC, van Aesch YM, Schelen AM, Touw IP. Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood. 1999;93(2):447–458. [PubMed] [Google Scholar]
- 26.Minakuchi M, Kakazu N, Gorrin-Rivas MJ, Abe T, Copeland TD, Ueda K, Adachi Y. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001;268(5):1340–1351. doi: 10.1046/j.1432-1327.2001.02000.x. [DOI] [PubMed] [Google Scholar]
- 27.Cristóbal I, Blanco FJ, Garcia-Orti L, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115(3):615–625. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 28.Damm F, Itzykson R, Kosmider O, et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013;27(6):1401–1403. doi: 10.1038/leu.2013.35. [DOI] [PubMed] [Google Scholar]
- 29.Laborde RR, Patnaik MM, Lasho TL, et al. SETBP1 mutations in 415 patients with primary myelofibrosis or chronic myelomonocytic leukemia: independent prognostic impact in CMML [published online ahead of print April 5, 2013]. Leukemia. doi: 10.1038/leu.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoischen A, van Bon BW, Gilissen C, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42(6):483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]