Key Points
DEPTOR is expressed in vascular endothelial cells and serves as an endogenous inhibitor of mTORC1, ERK1/2, and STAT1 activity.
DEPTOR is potent to regulate endothelial cell expression of chemokines and adhesion molecules, leukocyte-endothelial adhesion, and endothelial migratory responses.
Abstract
The maintenance of normal tissue homeostasis and the prevention of chronic inflammatory disease are dependent on the active process of inflammation resolution. In endothelial cells (ECs), proinflammation results from the activation of intracellular signaling responses and/or the inhibition of endogenous regulatory/pro-resolution signaling networks that, to date, are poorly defined. In this study, we find that DEP domain containing mTOR interacting protein (DEPTOR) is expressed in different microvascular ECs in vitro and in vivo, and using a small interfering RNA (siRNA) knockdown approach, we find that it regulates mammalian target of rapamycin complex 1 (mTORC1), extracellular signal-regulated kinase 1/2, and signal transducer and activator of transcription 1 activation in part through independent mechanisms. Moreover, using limited gene arrays, we observed that DEPTOR regulates EC activation including mRNA expression of the T-cell chemoattractant chemokines CXCL9, CXCL10, CXCL11, CX3CL1, CCL5, and CCL20 and the adhesion molecules intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 (P < .05). DEPTOR siRNA-transfected ECs also bound increased numbers of peripheral blood mononuclear cells (P < .005) and CD3+ T cells (P < .005) in adhesion assays in vitro and had increased migration and angiogenic responses in spheroid sprouting (P < .01) and wound healing (P < .01) assays. Collectively, these findings define DEPTOR as a critical upstream regulator of EC activation responses and suggest that it plays an important role in endogenous mechanisms of anti-inflammation and pro-resolution.
Introduction
The initiation of acute and chronic inflammation is characteristically associated with the activation of microvascular endothelial cells (ECs) responding to local proinflammatory cytokines released from infiltrating leukocytes and/or resident macrophages.1,2 The induced expression of adhesion molecules and chemokines by ECs initiates the recruitment of leukocytes into inflamed tissues, and the expression of cell surface molecules such as major histocompatibility complex molecules and costimulatory molecules by ECs serves to facilitate local lymphocyte activation responses in the course of a cell-mediated immune reaction.1,3 In addition, cell-mediated immune and delayed-type hypersensitivity reactions are associated with leukocyte-induced angiogenesis that fosters the development and progression of the chronic inflammatory microenvironment.1-3
Nevertheless, recent studies have also highlighted a paradigm where endogenous mechanisms of inflammation resolution function to maintain normal tissue homeostasis.4,5 Increasing data indicate that these mechanisms are critical for the prevention of chronic inflammatory diseases, in as much as the efficiency and/or the speed of pro-resolution determines the outcome of an inflammatory reaction.4 Pro-resolution is an active process involving the secretion of specific mediators, but is also dependent on the level of expression and/or the activity of intracellular regulatory adaptor molecules, kinases, and/or select inhibitory proteins.6-8
Mammalian target of rapamycin (mTOR)-mediated signaling responses have been reported to function in EC activation, including the induced expression of adhesion molecules and chemokines.9,10 In addition, mTOR activity facilitates EC proliferation and migration in vitro11 and is functional in tumor angiogenesis, wound healing angiogenesis, and leukocyte-induced angiogenesis that is characteristic of chronic inflammation in vivo.12-14 mTOR is an evolutionarily conserved serine/threonine kinase15 that forms 2 distinct multiprotein complexes, composed of either mTOR, raptor and mLST8 (called mTORC116) or mTOR, Rictor, Sin1, protor, and mLST8 (called mTORC217). mTORC1 controls cell growth in part by phosphorylating S6K1 and 4E-BP1,15,18 key regulators of protein synthesis. mTORC2 modulates cell survival and activation in response to growth factors by phosphorylating the Akt kinase19 or via activation of the protein kinase C pathway.20 Signaling in response to mTOR activation is tightly regulated by several negative feedback loops and by distinct proteins that serve to inhibit its activity. One such regulatory protein is DEP domain containing mTOR interacting protein (DEPTOR),21,22 which was recently reported to associate with both mTORC1 and mTORC2 and inhibit their activity in cancer cells,21,23,24 myocytes,25 and adipose tissue.26 Nevertheless, despite its potential importance as an upstream modulator/regulator of mTOR signaling, little is known about DEPTOR expression and function in normal cell types, and its effect(s) in EC-dependent mechanisms of inflammation is not known.
In these studies, we demonstrate that DEPTOR is expressed in normal human ECs where it functions as a cell intrinsic regulator of mTORC1, extracellular signal-regulated kinase (ERK)1/2, and signal transducer and activator of transcription (STAT)1 signaling and the inducible expression of adhesion molecules and chemokines. In addition, we show that it is potent to regulate leukocyte-EC adhesion and EC migration/angiogenesis. Our data define a paradigm where DEPTOR functions as a critical upstream regulator of EC activation responses and suggest that it plays an important role in endogenous mechanisms of inflammation resolution.
Materials and methods
For information on antibodies and reagents, please see supplemental Materials and methods on the Blood website.
Cell culture
Single donor human umbilical vein ECs (HUVECs) were purchased from Clonetics (Lonza, Allendale, NJ) and cultured in complete endothelial growth medium (EGM-2; Lonza), subcultured, and used between passages 3 and 6. For some experiments, we also used primary cultures of ECs isolated from umbilical cords and cultured as described.27 Human coronary artery ECs, human lung and dermal microvascular EC (HMVECs), and renal tubular epithelial cells were purchased from Lonza and cultured in the recommended growth medium; 786-0 cells were gifted to the laboratory (from Debabrata Mukhopadhyay, Mayo Clinic, Rochester, MN), and HEK293 cells were purchased from American Type Culture Collection (Manassas, VA).
Western blot analyses
Cells were lysed with radioimmunoprecipitation assay buffer (Boston Bioproducts, Boston, MA) containing protease and phosphatase inhibitors (Thermo Scientific, Rockford, IL). Proteins were separated on a sodium dodecyl sulfate-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). Membranes were blocked for 1 hour with Tris-buffered saline-0.1% Tween-20 containing 5% bovine serum albumin, and incubated with the primary antibody overnight at 4°C. Membranes were then washed and incubated with a species-specific secondary peroxidase-linked antibody for 1 hour at room temperature, and the protein of interest was detected by chemiluminescence (Thermo Scientific, Pierce).
Immunofluorescence microscopy
Human neonatal foreskins and discarded cardiac tissue from patients undergoing surgery were collected as approved by the Institutional Review Board at the Brigham and Women’s Hospital and Boston Children's Hospital. Informed consent was obtained in accordance with the Declaration of Helsinki. A description of the immunofluorescence technique used in these studies is provided in supplemental Materials and methods, as previously reported.28
Immunoprecipitation assays
HUVECs were lysed in 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate-containing lysis buffer lacking NaCl and supplemented with protease and phosphatase inhibitors (Thermo Scientific). Immunoprecipitations were performed using 500 μg of total protein and excess antibody with the Dynabeads Protein G immunoprecipitation kit, according to the manufacturer’s instructions (Life Technologies, Invitrogen, Grand Island, NY).
Small interfering RNA knockdown
Small interfering RNAs (siRNAs) for DEPTOR were purchased from Qiagen (Valencia, CA), and a negative control siRNA was purchased from Invitrogen. Transfection of HUVECs with siRNA (up to 50 nM) was performed using RNAimax lipofectamine (Life Technologies, Invitrogen), according to the manufacturer’s instructions. Following siRNA transfection, the efficiency of knockdown was assessed regularly by quantitative reverse transcriptase-polymerase chain reaction (PCR) and western blot analysis vs controls; our siRNAs were consistently found to be ∼90% efficient (supplemental Figure 1). All experiments were performed with 2 different siRNAs, as indicated.
Protein arrays
Protein arrays were performed using the Human Phospho-Kinase Array Kit (Proteome Profiler Array) purchased from R&D Systems (Minneapolis, MN), according to the manufacturer’s instructions.
RNA isolation and PCR arrays
Total mRNA was isolated from HUVECs 48 hours following control or DEPTOR siRNA transfection, using the RNeasy isolation kit (Qiagen, Valencia, CA). cDNA was generated using the qScript Supermix from Quanta Biosciences (Gaitherburg, MD), according to the manufacturer’s instructions and 2 commercial PCR arrays (“endothelial cell biology” and “chemokines and cytokines”; SABiosciences/Qiagen) were used to examine endothelial activation responses. Gene expression levels were evaluated using data analyzer template provided by the manufacturer, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, hypoxanthine phosphoribosyltransferase 1 (HPRT1), and ribosomal protein L13α were used as references.
Real-time PCR
Quantitative real-time PCR analyses were performed using the 7300 real-time PCR system and specific TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA). Relative expression of each gene vs the GAPDH control was calculated according to the 2−ΔΔCt method, as previously described.29
Enzyme-linked immunosorbent assay
The human CXCL10/IP-10 DuoSet enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems) was used for ELISA assays in culture supernatants, according to the manufacturer’s instructions.
Adhesion assays
Human peripheral blood mononuclear cells (PBMCs) were obtained from healthy volunteers, consented in accordance with Institutional Review Board approval at Boston Children’s Hospital. PBMCs (2.5 × 106/well) were cocultured with untreated or siRNA-transfected HUVECs in 6-well plates at 37°C for 60 minutes. Nonadherent PBMCs were removed by 3 phosphate-buffered saline washes, and adherent cells were evaluated by microscopy or were harvested using trypsin and processed for fluorescence-activated cell sorter (FACS) analysis (FACsCalibur; BD Biosciences, San Jose, CA) using anti-CD3, anti-CD45, and anti-CD31 fluorescent antibodies (BD Biosciences). Ten thousand cells per group were analyzed using FlowJo Software (TreeStar, Ashland, OR), and the adhesion index was calculated as the ratio of the percentage of PBMCs or CD3+ T cells within each experimental group compared with the untreated group.
For quantitative assays, CD3+ T cells were purified from PBMCs using the human Pan T cell Isolation Kit (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. Either PBMCs or CD3+ T cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE, 5 μM; Life Technologies) and were cocultured (5 × 105 per well) with confluent monolayers of untreated or siRNA-transfected HUVECs in 24-well cell culture plates at 37°C for 60 minutes. Nonadherent cells were removed by 3 phosphate-buffered saline washes, and adhesion was assessed by the measurement of fluorescence in each well using an automated plate reader (Victor; Perkin Elmer, Waltham, MA). The number of adherent cells in each well was calculated from a standard fluorescence intensity curve generated with increasing numbers of CFSE-labeled PBMCs or CD3+ T cells in control wells (supplemental Figure 2)
For each assay, experiments were performed with 2 siRNAs to DEPTOR, and results were pooled for final quantification. In addition, the adhesion of PBMCs or CD3+ T cells to tumor necrosis factor (TNF)α-treated HUVECs (100 U/mL for 6 hours) was evaluated in each experiment as a positive control.
Spheroid sprouting assays
HUVECs were transfected with control or DEPTOR siRNAs and cultured for 24 hours. Cells were then allowed to aggregate in hanging drops (500 cells per drop per spheroid), and after 24 hours, spheroids were embedded into a collagen type I matrix, as described.30 After another 24 hours, the spheroids were fixed for 15 minutes with 4% paraformaldehyde and were subsequently permeabilized for 5 minutes with 0.2% Triton X-100, and the F-actin cytoskeleton was stained with Alexa Fluor 488–conjugated phalloidin (Life Technologies, Invitrogen, Grand Island, NY). Samples were examined using a Perkin Elmer UltraVIEW Vox confocal microscope (Santa Clara, CA) using a Plan Apo 20× (0.75 NA) objective. Images were acquired using the Volocity Software (Improvision; Perkin Elmer), and the length of EC sprouts from 5 spheroids per condition was measured in 3 independent experiments, using ImageJ software.
In vitro migration assays
HUVECs were transfected with control or DEPTOR siRNAs, and after 48 hours, a linear wound was created in confluent cell monolayers by scratching with a pipette tip. After an additional 18 hours, cell migration into the wound was assessed by microscopy (10× objective) using a digital inverted microscope (AMG Evos XL Core; Fisher Scientific, Pittsburgh, PA). The degree of wound closure was measured as the percentage of the area covered by migrating cells in the initial wound in 9 wounds per test condition, using ImageJ software. Migration assays were performed using 2 siRNAs to DEPTOR, and results obtained with each siRNA were pooled for quantification and statistical analyses.
Statistical analyses
Statistical analyses were performed using the Student t test, and P < .05 was considered statistically significant.
Results
Expression of DEPTOR in vascular ECs
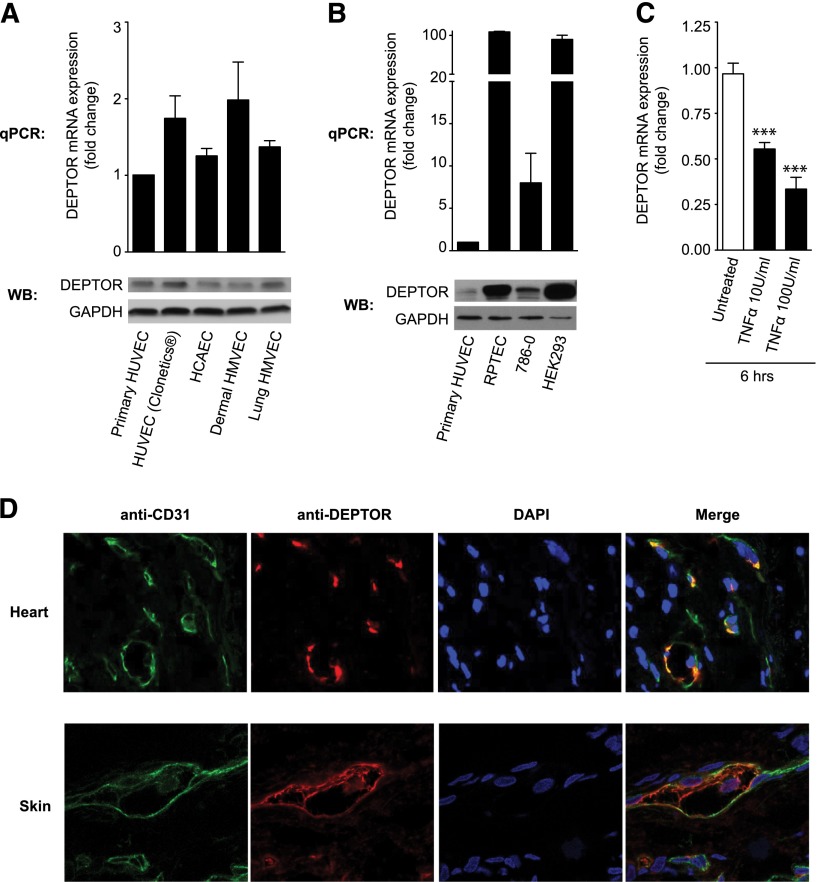
We initially evaluated the expression of DEPTOR at the mRNA and protein levels in HUVECs, human coronary artery ECs, and HMVECs. As illustrated in Figure 1A, using quantitative real-time PCR and western blot analysis, we found similar levels of expression of DEPTOR in each type of EC. Although DEPTOR mRNA expression was slightly higher in dermal HMVECs, compared with primary cultures of HUVECs, this difference was not statistically significant and was not translated into increased DEPTOR protein expression by western blot analysis (Figure 1A, lower). However, DEPTOR expression in HUVECs was low compared with other cell lines previously shown to express high levels of DEPTOR21 (Figure 1B). Furthermore, DEPTOR mRNA expression in the different ECs was quite variable, suggesting that DEPTOR expression may be regulated in these cells. Indeed, previous studies have shown that DEPTOR expression is rapidly down-regulated at the transcriptional and post-translational levels by mTOR itself, in response to growth signals.31-33 Consistent with this possibility, we found that stimulation of ECs with TNFα (10-100 U/mL) results in a rapid decrease in DEPTOR mRNA expression (Figure 1C).
Figure 1.
DEPTOR expression in human vascular ECs. (A) DEPTOR expression was analyzed at the mRNA level by quantitative PCR (qPCR) and at the protein level by western blot analysis in primary cultures of HUVECs, commercially available HUVECs, human coronary artery ECs, and dermal and lung human microvascular ECs (HMVECs). Illustrated is the mean fold change in mRNA expression ± standard error of the mean (SEM) from 3 independent experiments. (B) qPCR and western blot analysis of DEPTOR expression in HUVECs compared with renal proximal tubule epithelial cells (RPTECs), renal cancer 786-0 cells, and human embryonic kidney HEK293 cells. Representative of 3 experiments. (C) HUVECs were treated with TNFα (10 or 100 U/mL) for 6 hours, and DEPTOR mRNA expression was subsequently analyzed by qPCR. The bar graph represents the mean fold change in DEPTOR mRNA expression (±SEM) in 5 independent experiments (***P < .001). (D) Human cardiac tissue (atrium) and skin were evaluated by double immunofluorescence for the expression of DEPTOR or endothelial-specific CD31, as indicated. Illustrated are representative confocal images where DEPTOR (red) was found to colocalize with ECs (green) in each tissue. The yellow staining in the merged image is representative of colocalization.
We also evaluated the pattern of expression of DEPTOR by immunofluorescence in situ in human heart (Figure 1D, upper) and human skin (Figure 1D, lower) and found a prominent expression pattern within vascular ECs. However, expression was focal, and levels of expression varied among ECs, again suggesting that DEPTOR is regulated in vivo.
Function of DEPTOR in the regulation of mTOR signaling in ECs
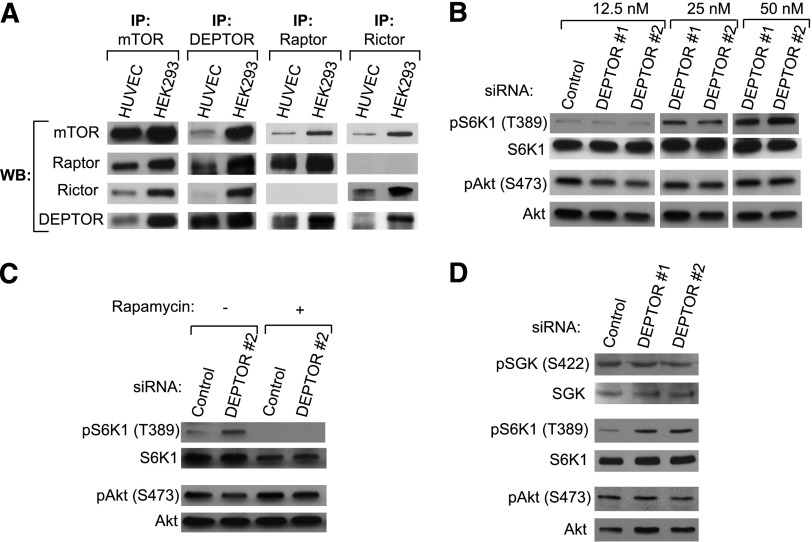
DEPTOR has consistently been found to associate with both mTORC1 and mTORC2 and to inhibit their activity in different types of cancer cells.21,34 We initially evaluated whether it also interacts with mTOR complexes in EC (Figure 2). Using immunoprecipitation assays, we observed that DEPTOR forms a complex with mTOR and raptor (mTORC1) in ECs but minimally associates with Rictor (mTORC2; Figure 2A). We transfected HUVECs with increasing concentrations of 2 siRNAs to DEPTOR, or a control siRNA, and after 48 hours, we analyzed levels of pS6K1 (T389) and pAkt (S473) by western blot. As anticipated,21,25,34 we found that knockdown of DEPTOR in ECs led to a marked increase in the phosphorylation of S6K1 (Figure 2B), but surprisingly, knockdown had little effect on the level of expression of pAkt (S473) (Figure 2B; supplemental Figure 3A). Similar results were obtained following DEPTOR siRNA transfection of coronary artery, dermal, and lung ECs (supplemental Figure 3B).
Figure 2.
DEPTOR associates with mTORC1 and inhibits its activity in ECs, but it minimally interacts with mTORC2. (A) DEPTOR, mTOR, Raptor, and Rictor were immunoprecipitated (IP) from HUVECs and HEK293 cells, and western blot (WB) analysis was subsequently performed with anti-DEPTOR, -mTOR, -Raptor, and -Rictor. As illustrated, DEPTOR associates with mTOR and raptor, but minimally associates with Rictor in HUVECs. (B) HUVECs were transfected with a control or 2 DEPTOR siRNAs (#1 and #2), and the expression of pS6K1 (T389), S6K1, pAkt (S473), and Akt was evaluated by western blot analysis. (C) Control or DEPTOR siRNA (50 nM)-transfected ECs were cultured for 48 hours and treated with rapamycin (10 ng/mL) for the last 1 hour of culture. Cell lysates were analyzed by western blot for the expression of pS6K1 (T389), S6K1, pAkt (S473), and Akt. (D) The expression of pSGK (S422), SGK, pS6K1 (T389), S6K1, pAkt (S473), and Akt was analyzed by western blot in HUVECs transfected with a control or 2 DEPTOR siRNAs (50 nM). All data presented are representative of ≥3 independent experiments.
We postulated that increased mTORC1 activity and augmented phosphorylation of S6K1 in DEPTOR siRNA-transfected ECs may result in an inhibition of mTORC2 activity via well-established negative feedback loops35,36 and may be responsible for the apparent absence of function of DEPTOR on the regulation of mTORC2 activity. Therefore, to further test whether DEPTOR directly regulates mTORC2, we transfected HUVECs with DEPTOR siRNA, and we treated the cells with rapamycin (10 ng/mL for 1 hour) to target mTORC1. Although rapamycin inhibited the phosphorylation of S6K1, DEPTOR knockdown again failed to increase the phosphorylation of Akt (S473) (Figure 2C). We also analyzed the levels of pSGK (S422), another target of mTORC2, in HUVECs transfected with control or 2 DEPTOR siRNAs. As illustrated in Figure 2D, knockdown of DEPTOR in HUVECs similarly had no effect on the level of phosphorylation/activation of SGK. Thus, DEPTOR both associates with and regulates mTORC1, but it has no effect on mTORC2 activity in ECs.
Function of DEPTOR in the regulation of ERK1/2 and STAT1 activity in ECs
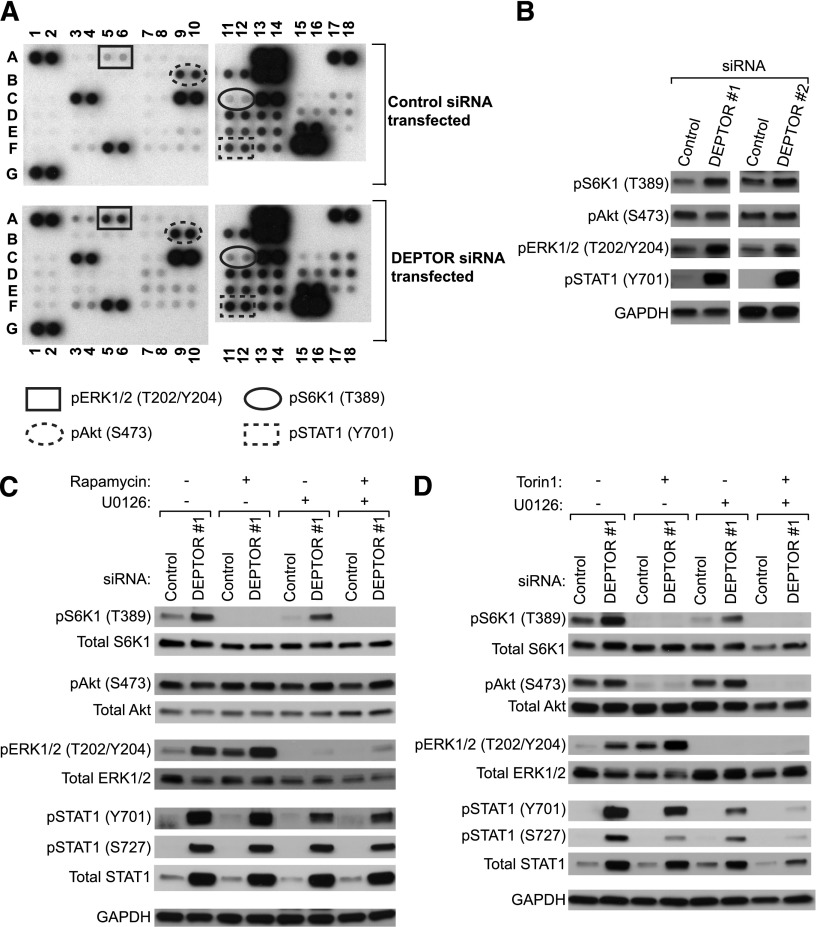
To study whether DEPTOR regulates additional signaling pathways in ECs, we next performed a protein kinase array and thus compared the relative levels of phosphorylation of multiple kinases and/or their protein substrates in control siRNA- and DEPTOR siRNA-transfected HUVECs. As illustrated in Figure 3A and supplemental Table 1, the array confirmed a regulatory effect of DEPTOR on mTORC1 activity, but there were notable additional effects on the phosphorylation/activation of ERK1/2 and STAT1. Other phosphokinases and adaptor molecules were also increased in DEPTOR siRNA-transfected EC (vs controls), including p38α, Paxillin, PLCγ-1, and FAK, but each to a lesser extent than ERK1/2. The marked regulatory effect of DEPTOR on pERK1/2 (T202/Y204) and pSTAT1 (Y701) expression was confirmed by western blot analysis (Figure 3B).
Figure 3.
DEPTOR inhibits ERK1/2 and STAT1 activity in ECs. HUVECs were transfected with control or DEPTOR siRNAs (#1 and #2), and after 48 hours, a phosphokinase protein array was performed on cell lysates to analyze the relative expression of 46 individual phosphokinase proteins. (A) A representative blot (of n = 2) showing the levels of phosphorylation of individual kinases and their protein substrates in control and 2 DEPTOR siRNA-transfected ECs (#1 and #2). Transfection with either siRNA resulted in similar findings by phosphokinase array. (B) Expression of pS6K1 (T389), pAkt (S473), pERK1/2 (T202/Y204), pSTAT1 (Y701), and GAPDH was examined in control or DEPTOR siRNAs-transfected HUVECs by western blot analysis. Representative results of >3 independent experiments are shown. (C-D) HUVECs were transfected with control or DEPTOR siRNAs, cultured for 48 hours, and treated with (C) rapamycin (10 ng/mL) or (D) Torin1 (1 μM) and/or U0126 (10 μM) for the last 18 hours of cell culture. Cells lysates were analyzed by western blot analysis for the expression of pS6K1 (T389), total S6K1, pAkt (S473), total Akt, pERK1/2 (T202/Y204), total ERK1/2, pSTAT1 (Y201 and S727), total STAT1, and GAPDH, as illustrated. Representative blots from 3 independent experiments are shown.
To determine whether DEPTOR regulates these signaling pathways through interrelated mechanisms, we transfected HUVECs with control or DEPTOR siRNAs and treated the cells with U0126 (10 μM for 18 hours) and/or with rapamycin (10 ng/mL for 18 hours) or Torin1 (1 μM for 18 hours) to inhibit ERK1/2 or mTOR activity, respectively. As illustrated in Figure 3C-D, we found that DEPTOR knockdown resulted in a marked increase in the phosphorylation of S6K1, in the absence or presence of pharmacological ERK1/2 inhibition. Also, DEPTOR knockdown resulted in an increase in pERK1/2 levels in the absence or presence of rapamycin (Figure 3C), even though mTORC1-S6K1 activation was inhibited. Treatment of DEPTOR siRNA-transfected cells with Torin1, an established ATP competitive inhibitor of both mTORC1 and mTORC2,37 further confirmed that DEPTOR regulates ERK1/2 in the absence of mTOR signaling.
We also observed an increase in the levels of total STAT1, pSTAT1 (Y701), and pSTAT1 (S727) in DEPTOR siRNA-treated cells vs controls. In addition, we found that overexpression of pSTAT1 (Y701) was unchanged in siRNA-transfected ECs that were treated with rapamycin (Figure 3C) or Torin 1 (Figure 3D), and levels of expression were minimally reduced in ECs treated with U0126 (Figure 3C-D). However, when DEPTOR siRNA-transfected ECs were treated with both Torin1 and U0126 in combination, the increase in pSTAT1 (Y701) was almost completely inhibited, and levels of pSTAT1 (S727) and total STAT1 were markedly reduced. We interpret these observations to suggest that DEPTOR interacts with the mTOR and ERK1/2 signaling pathways through independent mechanisms and that it regulates STAT1 signaling in part via cross-talk among intermediaries within both the ERK1/2 and the mTOR pathways.
Effects of DEPTOR on EC activation and proinflammation
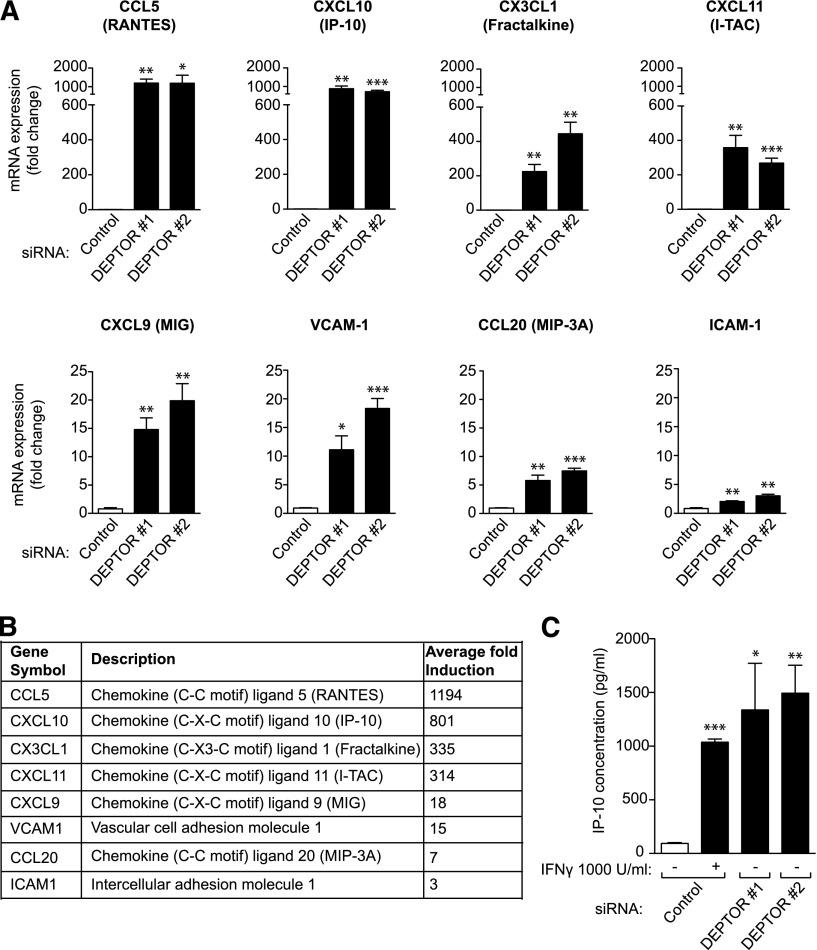
We next evaluated the effect of DEPTOR on the regulation of functional EC activation responses using 2 commercial PCR-based arrays. Of a total of 155 genes examined, 47 genes were strongly modulated in DEPTOR siRNA-transfected vs control siRNA-transfected HUVECs. Of these, a total of 27 genes were strongly induced (up to ∼360-fold) and 20 genes were moderately down-regulated following DEPTOR siRNA transfection (supplemental Table 2). There was a most notable effect of DEPTOR knockdown on the induction of T-cell chemoattractant chemokines, including CXCL9 (MIG), CXCL10 (IP-10), CXCL11 (I-TAC), CCL5 (RANTES), CX3CL1 (Fractalkine), and CCL20 (MIP3A) in the array (supplemental Table 2), as well as in subsequent real-time quantitative PCR analyses (Figure 4A-B). The mRNA expression of the adhesion molecules intercellular adhesion molecule-1 and vascular cell adhesion molecule (VCAM)-1 was also significantly up-regulated in DEPTOR siRNA-transfected EC vs controls (Figure 4A-B). By ELISA (Figure 4C), we also found that CXCL10 was dramatically increased in the supernatant of DEPTOR siRNA-transfected cells (mean = 1415 ± 562 pg/mL) vs control siRNA-transfected HUVECs (mean = 94 ± 13 pg/mL, P < .05), and was in the same range as that observed in HUVECs treated with interferon γ for 24 hours as a positive control (mean = 1037 ± 50 pg/mL). These potent effects of DEPTOR on activation responses were similar in ECs derived from different vascular beds (skin, lung, and coronary artery; data not shown). To test whether DEPTOR regulates EC activation through mTORC1- and/or ERK1/2-dependent signals, we also evaluated EC activation responses in DEPTOR siRNA-transfected HUVECs pretreated for 24 hours with rapamycin (10 ng/mL) and/or U0126 (10 μM). As illustrated in supplemental Figure 4, we found that each inhibitor had different effects on the overexpression of individual chemokines and adhesion molecules, suggesting that the DEPTOR-mediated regulation of EC activation likely results from the simultaneous modulation of multiple signaling pathways.
Figure 4.
DEPTOR regulates the expression of proinflammatory chemokines and adhesion molecules in ECs. mRNA array analysis (supplemental Table 2) identified 27 genes that were induced in expression in DEPTOR siRNA-transfected EC vs controls. Of these, 8 represented established EC activation response genes, members of the chemokine and adhesion molecule families. (A) qPCR was performed on control or 2 DEPTOR siRNA-transfected ECs after 48 hours to validate the function of DEPTOR on the expression of the 8 genes, CXCL9 (MIG), CXCL10 (IP-10), CXCL11 (I-TAC), CX3CL1 (Fractalkine), CCL5 (RANTES), and CCL20 (MIP-3A), and the adhesion molecules VCAM-1 and intercellular adhesion molecule-1. Graphs represent the mean fold change in mRNA expression (±SEM) in 3 independent experiments. (B) Average fold induction (high to low) in mRNA expression of each gene, as indicated, in DEPTOR siRNA-transfected EC vs controls. (C) HUVECs were transfected with control or 2 DEPTOR siRNAs, and the concentration of IP-10 protein was evaluated by ELISA in culture supernatants after 96 hours. Control siRNA-transfected HUVECs treated with interferon γ (1000 U/mL) for 24 hours were used as positive control. The bar graphs illustrate the mean IP-10 concentration (±SEM) from 3 experiments (*P < .05, **P < .01, ***P < .001).
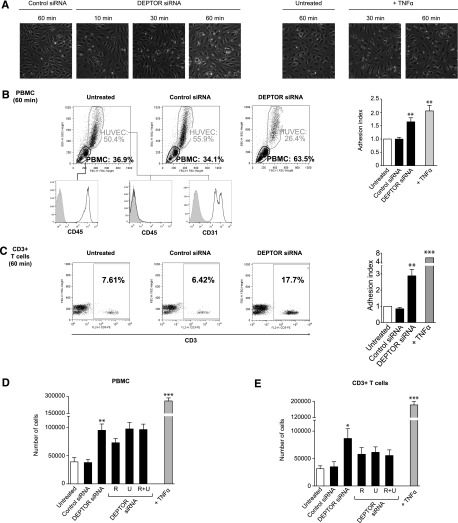
We next questioned whether these cell intrinsic effects of DEPTOR are of functional importance in proinflammation. We cocultured freshly isolated human PBMCs with confluent cultures of control siRNA- or DEPTOR siRNA-transfected HUVECs for 10 to 60 minutes at 37°C. Cocultures of PBMCs with TNFα-activated ECs served as a positive control. At each time point, nonadherent PBMCs were removed by washing, and adherent leukocytes were assessed by microscopy. Cultures of HUVECs and adherent leukocytes were also collected and analyzed by FACS. As illustrated in Figure 5A-B, we found higher numbers of adherent PBMCs in cocultures with DEPTOR siRNA-transfected ECs vs control siRNA-transfected ECs or untreated ECs at all time points examined, but the effect of DEPTOR knockdown on leukocyte adhesion was most prominent after 30 to 60 minutes of coculture (P < .005). We also evaluated the number of CD3+ T cells within each population of adherent PBMCs and found a significant effect (P < .005) of DEPTOR knockdown on CD3+ T cell–EC interactions (Figure 5C).
Figure 5.
Function of DEPTOR in leukocyte-endothelial interactions in vitro. (A-C) Confluent cultures of untransfected and control siRNA- and DEPTOR siRNA-transfected HUVECs were cocultured with freshly isolated human PBMCs at 37°C for 10 to 60 minutes. Subsequently, the cultures were washed 3 times, and the number of adherent PBMCs were evaluated by microscopy or FACS analysis. (A) Representative photomicrographs of HUVECs transfected with control or DEPTOR siRNAs and cultured with human PBMCs for 10, 30, and 60 minutes. Cells cultured with TNFα-treated ECs (100 U/mL for 6 hours) are illustrated as a positive control. Microscopy was carried out (10× objective) using a digital inverted microscope (AMG Evos XL Core; Fisher Scientific). (B) Representative FACS dot plots of the patterns of forward scatter (FSC) and side scatter (SSC) for 10 000 cells per group, illustrating the percentage of adherent PBMCs to each group of HUVECs after 60 minutes. The expression of CD45 within the PBMC gate (lower left panel, open histogram), as well as CD45 and CD31 within the HUVEC gate (lower center and right panels, open histograms) are shown vs isotype antibody as a control (shaded histogram). The bar graph illustrates the mean adhesion index of PBMC (±SEM) to ECs from 5 independent experiments. (C) Representative FACS plots of CD3+ T cells within each PBMC gate shown in B. The bar graph shows the mean adhesion index of CD3+ T cells (±SEM) to ECs from 5 independent experiments. (D-E) PBMCs and freshly isolated CD3+ T cells were labeled with CFSE (5 μM) prior to coculture in adhesion assays with untransfected, control siRNA, or 2 DEPTOR siRNA-transfected HUVECs. ECs were cultured in the absence or presence of rapamycin (labeled R, 10 ng/mL) and/or U0126 (labeled U, 10 μM) for 24 hours prior to each assay. After 60 minutes of coculture, nonadherent cells were removed by washing, and adherent leukocytes were evaluated by measurement of fluorescence in each well. The number of adherent leukocytes was calculated based on a standard curve, as described in Materials and methods. The bar graphs show the mean number (±SEM) of adherent (D) PBMCs and (E) CD3+ T cells from 5 independent experiments. Data from ECs transfected with DEPTOR siRNA1 #1 or #2 were pooled for analysis in the bar graphs. *P < .05, **P < .005, ***P < .001.
To quantify leukocyte-EC adhesion, HUVECs were transfected with control or DEPTOR siRNAs and cocultured with freshly isolated and CFSE-labeled human PBMCs or CD3+ T cells for 60 minutes. After washing, the numbers of adherent PBMCs or CD3+ T cells in each experimental condition was evaluated by measuring fluorescent intensity; quantification of cell number was determined by comparison with a standard curve (supplemental Figure 2). As illustrated in Figure 5D-E, we found that DEPTOR knockdown in HUVECs resulted in a marked increase in the number of adherent PBMCs (∼2.5-fold, P < .01; Figure 5D) and CD3+ T cells (∼2.5-fold, P < .05; Figure 5E) compared with controls. In addition, we found that the pretreatment of DEPTOR siRNA-transfected HUVECs with rapamycin (10 ng/mL, to target mTORC1) and/or U0126 (10 μM, to target ERK1/2) for 24 hours before the assay did not inhibit the adhesion of PBMCs to ECs (Figure 5D). However, treatment of DEPTOR siRNA-transfected ECs with the inhibitors, alone or in combination, decreased the binding of CD3+ T cells to the EC monolayer, although the effect was not statistically significant (Figure 5E). Of note, pretreatment of control siRNA-transfected ECs with rapamycin and/or U0126 did not decrease, and tended to increase, adhesion of PBMCs and CD3+ T cells (data not shown). Collectively, these findings suggest that the endogenous expression of DEPTOR in ECs is of functional importance in leukocyte-EC adhesion and proinflammation.
Functional effects of DEPTOR on angiogenic responses
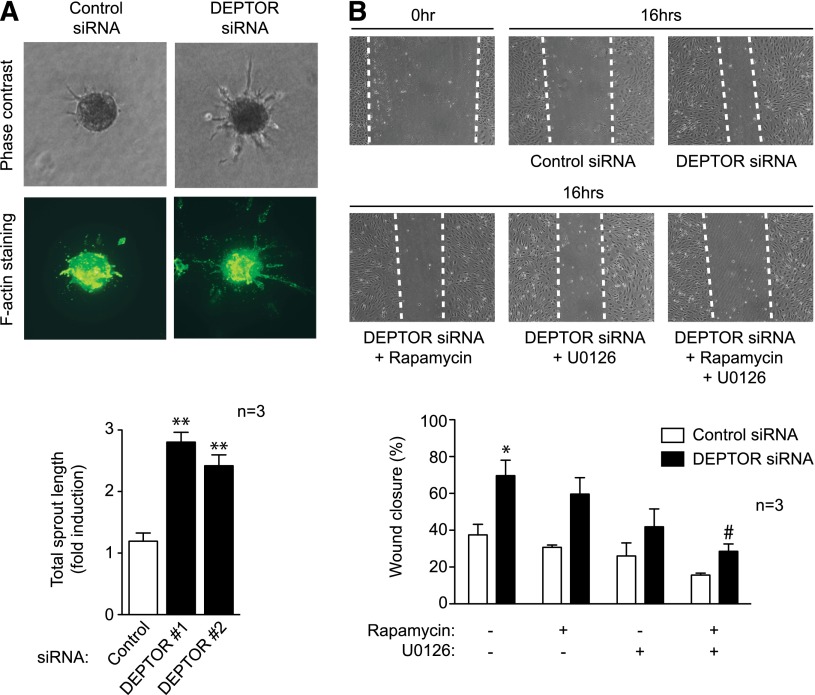
We also used 2 well-established in vitro models to evaluate the function of DEPTOR in angiogenic responses. In the tridimensional spheroid EC sprouting assay, DEPTOR siRNA-transfected HUVECs were seeded to form spheroids and embedded in a collagen type I matrix, as described.30 After 24 hours, total sprout length was quantified in >5 spheroids per condition. Consistently, we found a significant increase (P < .01) in total sprout length in DEPTOR siRNA-transfected EC vs controls, as visualized and quantified by direct phase microscopy (Figure 6A, upper) and following staining with phalloidin (Figure 6A, lower). In the wound healing assay, DEPTOR siRNA- or control siRNA-transfected ECs were grown to confluence, and a linear wound was subsequently created in the monolayer. The migration of EC into the wound was quantified over a 12- to 24-hour period, as described.7 Consistent with our findings in the spheroid assay, we found that DEPTOR siRNA-transfected ECs migrated into the wound at an increased rate (P < .01) vs controls (Figure 6B). In addition, to evaluate whether DEPTOR regulates EC migration through mTORC1- and/or ERK1/2-dependent signals, we treated siRNA-transfected EC with rapamycin or U0126, respectively, and we evaluated migration in the wound-healing assay. As illustrated in Figure 6B, each inhibitor alone partially attenuated the increase in migration induced by DEPTOR knockdown (P = not significant), but treatment with both rapamycin and U0126 in combination significantly reduced the EC migration response (P < .05; Figure 6B). This suggests that DEPTOR controls EC migration responses by regulating both mTORC1 and ERK1/2 activity. In contrast, EC proliferative responses (as assessed by [3H] thymidine incorporation after 72 hours) and rates of apoptosis (as assessed by annexin V and propidium iodide staining) were similar in DEPTOR siRNA-transfected EC vs controls (supplemental Figure 5).
Figure 6.
Function of DEPTOR in angiogenic responses in vitro. (A) Spheroids derived from either control or DEPTOR siRNA-transfected HUVECs were embedded in a collagen I matrix, cultured for 24 hours, and stained with Alexa Fluor 488–conjugated phalloidin. A representative image of each group is shown without (upper panels) and with (lower panels) staining. The bar graph shows quantitative analysis of the mean total sprout length (±SEM) performed on ≥5 spheroids per experimental group in 3 independent experiments (**P < .01). (B) HUVECs were transfected with control or 2 DEPTOR siRNAs and cultured for 48 hours until confluent. Subsequently, linear scratch/wounds were created in the monolayers with a pipet tip, and the migration of cells into the wound was measured after 16 hours in the absence or presence of rapamycin (10 ng/mL) and/or U0126 (10 μM). Illustrated are representative photomicrographs of wounds at 0 hours and after 16 hours; dotted lines highlight the linear scratch/wound for each group of cells (representative of 3 experiments). The bar graph shows the mean percentage wound closure in pooled DEPTOR siRNA-transfected cells vs controls (±SEM; *P < .05 vs untreated control siRNA-transfected ECs; #P < .05 vs untreated DEPTOR siRNA-transfected ECs).
Discussion
Microvascular ECs participate in all aspects of acute and chronic inflammation, from their initial encounter with leukocytes in the course of recruitment and transmigration, to the immune angiogenesis reaction that is characteristic of delayed-type hypersensitivity.1-3,38 Activation responses and proinflammatory intracellular signals in ECs are initiated and coordinated by cytokines,38 growth factors,2,28 and cell-cell surface receptor-mediated interactions.38-40 In contrast, EC-dependent signals mediating inflammation resolution are poorly defined and less well understood. In these studies, we find that a reduction in the level of expression of cell intrinsic DEPTOR has in striking effects on mTORC1-dependent signaling and ERK1/2 and STAT1 activity in ECs. DEPTOR knockdown also results in EC activation, including the induced expression of several T-cell chemoattractant chemokines and adhesion molecules. Also, DEPTOR-regulated EC activation is associated with leukocyte-EC adhesion and a marked increase in EC migratory responses in vitro. Our studies are suggestive that DEPTOR has potent regulatory effects within ECs and that the level of DEPTOR expression is likely of importance for the maintenance of a quiescent EC phenotype.
Similar to other cell types,21,23-25 we find that DEPTOR expression in EC regulates mTORC1-dependent activation responses, but in these studies, we also identify an important regulatory role for DEPTOR in ERK1/2 activity. Although there is a well-established intracellular cross-talk among the mTOR signaling pathway (and intermediaries) and ERK1/2 signaling,41-43 we find that the treatment of ECs with pharmacological inhibitors of mTOR activity (either rapamycin or Torin1) fails to alter the effect of DEPTOR knockdown on the augmentation of ERK1/2 activity. Furthermore, we find that the treatment of ECs with a pharmacological inhibitor of MEK-ERK1/2 activity (U0126) fails to alter the effect of DEPTOR knockdown on mTORC1-induced signals. Thus, our findings indicate that DEPTOR selectively regulates the endogenous activity of ERK1/2 and mTORC1 via independent mechanisms. Although it is known that DEPTOR binds to mTOR,21 this observation is consistent with a report suggesting that DEPTOR contains a putative ERK-binding site31; it also associates with ERK1/2 in cells that have been cotransfected with a Flag-tagged DEPTOR construct and HA-tagged ERK1/2.32 However, we did not find any association between endogenous DEPTOR and ERK1/2 in our ECs (data not shown). Thus, it is possible that DEPTOR may interact with another intermediary to regulate endogenous ERK1/2 signaling.
In these studies, we also identify a critical role for DEPTOR in the regulation of STAT1 expression and activity through a mechanism that is dependent in part on both mTOR and ERK1/2. Indeed, pharmacological inhibition of either pathway alone in DEPTOR siRNA-transfected EC failed to suppress phosphorylation of STAT1, but combined pharmacological inhibition (using U0126 and Torin1) was associated with a significant reduction in pSTAT1 levels. Interestingly, in contrast to Torin1, rapamycin had no effect on the phosphorylation of STAT1, suggesting that the ability of mTOR to regulate STAT1 activity is rapamycin resistant. In addition, we found that DEPTOR regulates levels of both pSTAT1(Tyr701) and pSTAT1(Ser727). Because pSTAT1(Tyr701) is Janus kinase dependent44 and pSTAT1(Ser727) is in part ERK1/2 dependent,45 this observation is consistent with the possibility that DEPTOR has a direct effect(s) on STAT1 itself. Indeed, in general, DEPTOR knockdown resulted in an increase in total STAT1 levels within ECs, indicating that it may function via a direct STAT1-dependent autoregulation loop.
In our studies, knockdown of DEPTOR in ECs resulted in the overexpression of several proinflammatory molecules that are both STAT1 dependent (eg, IP-10/CXCL10) and STAT1 independent (eg, VCAM-1), and there was an associated increase in the adhesion of leukocytes to DEPTOR siRNA-transfected ECs. Also, we found that the treatment of DEPTOR siRNA-transfected ECs with rapamycin (to inhibit mTORC1) or U0126 (to inhibit ERK1/2), alone or in combination, had variable effects on the overexpression of individual chemokines and adhesion molecules and leukocyte-EC adhesion. These findings suggest that the effects of DEPTOR on the regulation of EC activation responses is likely related to its broad functions as a cell intrinsic inhibitor of several intracellular signaling pathways. In addition, knockdown of DEPTOR resulted in enhanced EC migration and angiogenesis responses that are well established to be associated with mTOR and ERK1/2 signaling; consistently, we found that the treatment of DEPTOR siRNA-transfected ECs with the combination of rapamycin and U0126 dramatically decreased the induced EC migratory response. Some DEPTOR-regulated chemokines (eg, IP-10/CXCL-10) have well-established antiangiogenic effects,9 further suggesting that these promigratory effects of DEPTOR knockdown are associated with cell intrinsic signaling. Collectively, these data are suggestive that the potent regulatory effects of DEPTOR on mTORC1 and ERK1/2 in ECs translate into its ability to sustain EC quiescence and/or inflammation resolution mechanisms.
DEPTOR expression has been shown to be down-regulated by mTOR-mediated signals at the transcriptional and post-translational levels.21 mTOR signals result in the phosphorylation of DEPTOR, which leads in turn to its binding to the F box protein βTrCP and its subsequent ubiquitination and degradation.31-33 We found that cytokine-mediated stimulation of ECs (with TNFα) results in a significant decrease in DEPTOR mRNA expression after 6 hours. Thus, following cytokine-mediated activation of EC in association with inflammation, amplification loops may sustain a reduced expression of DEPTOR, such that regulatory responses are inhibited. Therefore, we speculate that agents known to sustain DEPTOR activity,46 and/or agents that target physiological DEPTOR degradation,47-50 will have anti-inflammatory and antiangiogenic properties.
Collectively, these studies define DEPTOR as a key endogenous regulator of EC activation and EC-dependent proinflammatory responses. Our findings are consistent with a new paradigm whereby the maintenance of anti-inflammation and/or inflammation resolution involves the expression and activity of endogenous regulators of EC activation. The development of therapeutics that target DEPTOR degradation will have implications for the prevention and treatment of chronic inflammation and angiogenesis-dependent diseases.
Supplementary Material
Acknowledgments
The authors thank Dr Alex Toker (Beth Israel Deaconess Medical Center, Boston, MA), Dr David Sabatini (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology, Boston, MA), and Dr Michael Klagsbrun (Boston Children’s Hospital) for consultation and advice regarding these studies. The authors also thank Drs Dipak Datta and Brian Wilson (Boston Children's Hospital) for technical support with assays and Dr Soumitro Pal (Boston Children's Hospital) for his constructive critique. Finally, the authors thank Ms Kay Case and the Vascular Biology Core at Brigham and Women’s Hospital (Boston, MA) for the generation of human ECs for these studies.
This work was supported by National Institutes of Health grants R01 AI046756 and R01 AI092305 (National Institute of Allergy and Infectious Diseases), T32 DK007726 (National Institute of Diabetes and Digestive and Kidney Diseases), and an Advancing Research in Transplantation Science Investigator Initiated Grant from Pfizer, Inc. S.B. was also supported by a fellowship grant from the American Society of Transplantation. Microscopy imaging was performed at the Boston Children's Hospital Intellectual and Developmental Disabilities Research Center Imaging Core (NIH-P30-HD-18655).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.B. participated in research design, performed the studies, analyzed and interpreted the data, and wrote the manuscript; H.N., C.B.W., and E.A.F assisted with experiments; and D.M.B. conceived and designed the study, analyzed and interpreted the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David M. Briscoe, Transplant Research Program, Division of Nephrology, Boston Children's Hospital, 300 Longwood Ave, Boston, MA 02115; e-mail: david.briscoe@childrens.harvard.edu.
References
- 1.Cotran R. Inflammation and repair. In: Cottran RS, Kumar V, Robbins SL, editors. Pathologic Basis of Disease. Philadelphia, PA: W. B. Saunders; 1994:51-92. [Google Scholar]
- 2.Bruneau S, Woda CB, Daly KP, et al. Key features of the intragraft microenvironment that determine long-term survival following transplantation. Front Immunol. 2012;3(54):1-13. [DOI] [PMC free article] [PubMed]
- 3.Auerbach R, Sidky YA. Nature of the stimulus leading to lymphocyte-induced angiogenesis. J Immunol. 1979;123(2):751–754. [PubMed] [Google Scholar]
- 4.Recchiuti A, Serhan CN. Pro-resolving lipid mediators (SPMs) and their actions in regulating miRNA in novel resolution circuits in inflammation. Front Immunol. 2012;3(298):1-23. [DOI] [PMC free article] [PubMed]
- 5.Serhan CN. The resolution of inflammation: the devil in the flask and in the details. FASEB J. 2011;25(5):1441–1448. doi: 10.1096/fj.11-0502ufm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serhan CN, Brain SD, Buckley CD, et al. Resolution of inflammation: state of the art, definitions and terms. FASEB J. 2007;21(2):325–332. doi: 10.1096/fj.06-7227rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruneau S, Datta D, Flaxenburg JA, Pal S, Briscoe DM. TRAF6 inhibits proangiogenic signals in endothelial cells and regulates the expression of vascular endothelial growth factor. Biochem Biophys Res Commun. 2012;419(1):66–71. doi: 10.1016/j.bbrc.2012.01.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu A, Banerjee P, Contreras AG, Flynn E, Pal S. Calcineurin inhibitor-induced and Ras-mediated overexpression of VEGF in renal cancer cells involves mTOR through the regulation of PRAS40. PLoS ONE. 2011;6(8):e23919. doi: 10.1371/journal.pone.0023919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulday G, Haskova Z, Reinders ME, Pal S, Briscoe DM. Vascular endothelial growth factor-induced signaling pathways in endothelial cells that mediate overexpression of the chemokine IFN-gamma-inducible protein of 10 kDa in vitro and in vivo. J Immunol. 2006;176(5):3098–3107. doi: 10.4049/jimmunol.176.5.3098. [DOI] [PubMed] [Google Scholar]
- 10.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormond O, Madsen JC, Briscoe DM. The effects of mTOR-Akt interactions on anti-apoptotic signaling in vascular endothelial cells. J Biol Chem. 2007;282(32):23679–23686. doi: 10.1074/jbc.M700563200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guba M, von Breitenbuch P, Steinbauer M, et al. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat Med. 2002;8(2):128–135. doi: 10.1038/nm0202-128. [DOI] [PubMed] [Google Scholar]
- 13.Phung TL, Ziv K, Dabydeen D, et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. 2006;10(2):159–170. doi: 10.1016/j.ccr.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinders ME, Rabelink TJ, Briscoe DM. Angiogenesis and endothelial cell repair in renal disease and allograft rejection. J Am Soc Nephrol. 2006;17(4):932–942. doi: 10.1681/ASN.2005121250. [DOI] [PubMed] [Google Scholar]
- 15.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 17.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 18.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17(6):596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 20.Facchinetti V, Ouyang W, Wei H, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27(14):1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proud CG. Dynamic balancing: DEPTOR tips the scales. J Mol Cell Biol. 2009;1(2):61–63. doi: 10.1093/jmcb/mjp012. [DOI] [PubMed] [Google Scholar]
- 23.Foster H, Coley HM, Goumenou A, Pados G, Harvey A, Karteris E. Differential expression of mTOR signalling components in drug resistance in ovarian cancer. Anticancer Res. 2010;30(9):3529–3534. [PubMed] [Google Scholar]
- 24.Pei L, Xie P, Zhou E, Yang Q, Luo Y, Tang Z. Overexpression of DEP domain containing mTOR-interacting protein correlates with poor prognosis in differentiated thyroid carcinoma. Mol Med Rep. 2011;4(5):817–823. doi: 10.3892/mmr.2011.503. [DOI] [PubMed] [Google Scholar]
- 25.Kazi AA, Hong-Brown L, Lang SM, Lang CH. Deptor knockdown enhances mTOR Activity and protein synthesis in myocytes and ameliorates disuse muscle atrophy. Mol Med. 2011;17(9-10):925–936. doi: 10.2119/molmed.2011.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laplante M, Horvat S, Festuccia WT, Birsoy K, Prevorsek Z, Efeyan A, Sabatini DM. DEPTOR cell-autonomously promotes adipogenesis, and its expression is associated with obesity. Cell Metab. 2012;16(2):202–212. doi: 10.1016/j.cmet.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimbrone MA, Jr, Cotran RS, Folkman J. Human vascular endothelial cells in culture. Growth and DNA synthesis. J Cell Biol. 1974;60(3):673–684. doi: 10.1083/jcb.60.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edelbauer M, Datta D, Vos IH, et al. Effect of vascular endothelial growth factor and its receptor KDR on the transendothelial migration and local trafficking of human T cells in vitro and in vivo. Blood. 2010;116(11):1980–1989. doi: 10.1182/blood-2009-11-252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Korff T, Augustin HG. Tensional forces in fibrillar extracellular matrices control directional capillary sprouting. J Cell Sci. 1999;112(Pt 19):3249–3258. doi: 10.1242/jcs.112.19.3249. [DOI] [PubMed] [Google Scholar]
- 31.Gao D, Inuzuka H, Tan MK, et al. mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44(2):290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44(2):304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol Cell. 2011;44(2):317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen CH, Lu YC, Li CH, et al. Functional characterization of glycine N-methyltransferase and its interactive protein DEPDC6/DEPTOR in hepatocellular carcinoma. Mol Med. 2012;18(1):286–296. doi: 10.2119/molmed.2011.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dibble CC, Asara JM, Manning BD. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol. 2009;29(21):5657–5670. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66(3):1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7(10):803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 39.Melter M, Reinders ME, Sho M, et al. Ligation of CD40 induces the expression of vascular endothelial growth factor by endothelial cells and monocytes and promotes angiogenesis in vivo. Blood. 2000;96(12):3801–3808. [PubMed] [Google Scholar]
- 40.Dormond O, Dufour M, Seto T, Bruneau S, Briscoe DM. Targeting the intragraft microenvironment and the development of chronic allograft rejection. Hum Immunol. 2012;73(12):1261–1268. doi: 10.1016/j.humimm.2012.07.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langlais P, Yi Z, Mandarino LJ. The identification of raptor as a substrate for p44/42 MAPK. Endocrinology. 2011;152(4):1264–1273. doi: 10.1210/en.2010-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dormond-Meuwly A, Roulin D, Dufour M, Benoit M, Demartines N, Dormond O. The inhibition of MAPK potentiates the anti-angiogenic efficacy of mTOR inhibitors. Biochem Biophys Res Commun. 2011;407(4):714–719. doi: 10.1016/j.bbrc.2011.03.086. [DOI] [PubMed] [Google Scholar]
- 43.Guan KL, Figueroa C, Brtva TR, Zhu T, Taylor J, Barber TD, Vojtek AB. Negative regulation of the serine/threonine kinase B-Raf by Akt. J Biol Chem. 2000;275(35):27354–27359. doi: 10.1074/jbc.M004371200. [DOI] [PubMed] [Google Scholar]
- 44.Mowen K, David M. Regulation of STAT1 nuclear export by Jak1. Mol Cell Biol. 2000;20(19):7273–7281. doi: 10.1128/mcb.20.19.7273-7281.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 46.Liu M, Wilk SA, Wang A, et al. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. J Biol Chem. 2010;285(47):36387–36394. doi: 10.1074/jbc.M110.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo Z, Yu G, Lee HW, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72(13):3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, Sun Y. Targeting the mTOR-DEPTOR pathway by CRL E3 ubiquitin ligases: therapeutic application. Neoplasia. 2012;14(5):360–367. doi: 10.1593/neo.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo Z, Pan Y, Jeong LS, Liu J, Jia L. Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells. Autophagy. 2012;8(11):1677–1679. doi: 10.4161/auto.21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012;3,e386:1-13. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.