Abstract
Nodularia spumigena is a filamentous diazotrophic cyanobacterium that forms blooms in brackish water bodies. This cyanobacterium produces linear and cyclic peptide protease inhibitors which are thought to be part of a chemical defense against grazers. Here we show that N. spumigena produces structurally novel members of the aeruginosin family of serine protease inhibitors. Extensive chemical analyses including NMR demonstrated that the aeruginosins are comprised of an N-terminal short fatty acid chain, L-Tyr, L-Choi and L-argininal and in some cases pentose sugar. The genome of N. spumigena CCY9414 contains a compact 18-kb aeruginosin gene cluster encoding a peptide synthetase with a reductive release mechanism which offloads the aeruginosins as reactive peptide aldehydes. Analysis of the aeruginosin and spumigin gene clusters revealed two different strategies for the incorporation of N-terminal protecting carboxylic acids. These results demonstrate that strains of N. spumigena produce aeruginosins and spumigins, two families of structurally similar linear peptide aldehydes using separate peptide synthetases. The aeruginosins were chemically diverse and we found 11 structural variants in 16 strains from the Baltic Sea and Australia. Our findings broaden the known structural diversity of the aeruginosin peptide family to include peptides with rare N-terminal short chain (C2–C10) fatty acid moieties.
Introduction
N. spumigena is a filamentous diazotrophic cyanobacterium that forms extensive summer blooms in brackish water bodies. The ability to fix atmospheric nitrogen confers a competitive advantage on N. spumigena in nitrogen-poor and iron-limited brackish water ecosystems [1], [2], [3]. N. spumigena is responsible for a large part of the new nitrogen input in the Baltic Sea and is a source of environmental concern [1]. The consumption of water containing N. spumigena is associated with the death of wild and domestic animals [4], [5], [6]. These blooms are toxic through the production of nodularin, a cyclic pentapeptide toxin [4], [5]. Nodularin is the end-product of a complex hybrid non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) biosynthetic pathway [5]. N. spumigena produces other non-ribosomal peptides in addition to nodularin, including spumigin and nodulapeptin [7], [8], [9].
Spumigins are linear peptides which contain an N-terminal hydroxyphenyl lactic acid, almost exclusively D-homotyrosine, proline or 4-methylproline (mPro), and a C-terminal lysine or arginine derivative [7], [8], [9]. Spumigins are assembled on a NRPS enzyme complex which offloads the peptides as reactive aldehydes [8]. The enzymatic steps necessary for the synthesis of the unusual mPro are encoded together with the peptide synthetases in the 21-kb spumigin gene cluster [8]. Spumigins are potent inhibitors of serine proteases, active in the micromolar to nanomolar range [7], [8], [10].
Aeruginosins are another family of serine protease inhibitors that have been described from aquatic bloom-forming genera of cyanobacteria [11]. This family of linear peptides contain the rare 2-carboxy-6-hydroxyoctahydroindole (Choi) moiety as well as the C-terminal arginine derivatives argininal, argininol, agmatine, 1-amidino-2-ethoxy-3-aminopiperidine and more rarely 1-amino-2-(N-amidino-Δ3-pyrrolinyl)-ethyl moiety [11]. The N-terminus typically consists of either hydroxyphenyl lactic acid in Microcystis or phenyl lactic acid in Planktothrix [11]. Aeruginosins are the end-products of highly variable NRPS biosynthetic pathways and may be modified to contain chlorine, sulfate or sugars [12], [13]. Chemical variation in the aeruginosin structure is achieved by the action of tailoring enzymes and variation in the loading and release mechanisms [12], [13], [14]. A close relationship between aeruginosins and spumigins has been suspected for some time [12], [13], [15], [16].
N. spumigena encodes a number of cryptic NRPS clusters for which end-products have not been characterized [3], [17]. It was proposed based on the presence of Choi biosynthetic genes that N. spumigena may produce aeruginosins [3]. A recent study reported an incomplete peptide structure which contains Choi from N. spumigena [18]. Here we show that N. spumigena produces new members of the aeruginosin family of protease inhibitors using extensive chemical analyses including NMR studies (Figure 1) and demonstrate that N. spumigena strains produce two classes of similar non-ribosomal peptides, aeruginosins and spumigins, simultaneously using separate peptide synthetases. These results broaden the structural diversity of the aeruginosin family of peptides to include peptides with fatty acid side chains.
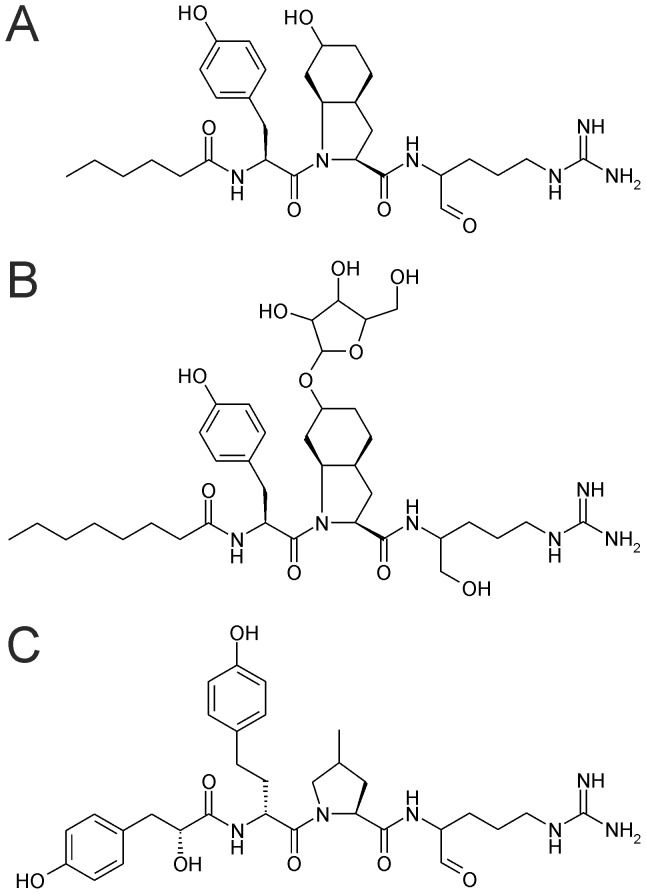
Figure 1. Representatives of the aeruginosin and spumigin families.
These tetrapeptides are produced by strains of Nodularia spumigena isolated from brackish water bodies in Australia and the Baltic Sea. (A) aeruginosin NAL2, (B) an aeruginosin NOL6 containing O-linked pentose, (C) spumigin E.
Results
Discovery of Aeruginosins
Two abundant peptides with a mass of m/z 587 and m/z 589 were identified from cell extracts of Nodularia spumigena AV1 by LC-MS analysis. They were initially suspected to be new variants of spumigin based on their mass and chromatographic behavior (Figure S1 in File S1). However, fragmentation of the protonated ions m/z 587 and m/z 589 did not produce sufficient information for overall substructure elucidation and only the presence of argininal and argininol could be postulated (Figures S1–S2 in File S1). Surprisingly, MDA and DNPH derivatization of the compounds and subsequent MS2 data suggested that the two peptides contained but differed by the presence of alcohol and aldehyde versions of arginine (Figure S3 in File S1). The moiety is a unique component of the aeruginosin family of linear peptides and we hypothesized that N. spumigena make members of the aeruginosin family of peptides.
Aeruginosin Gene Cluster
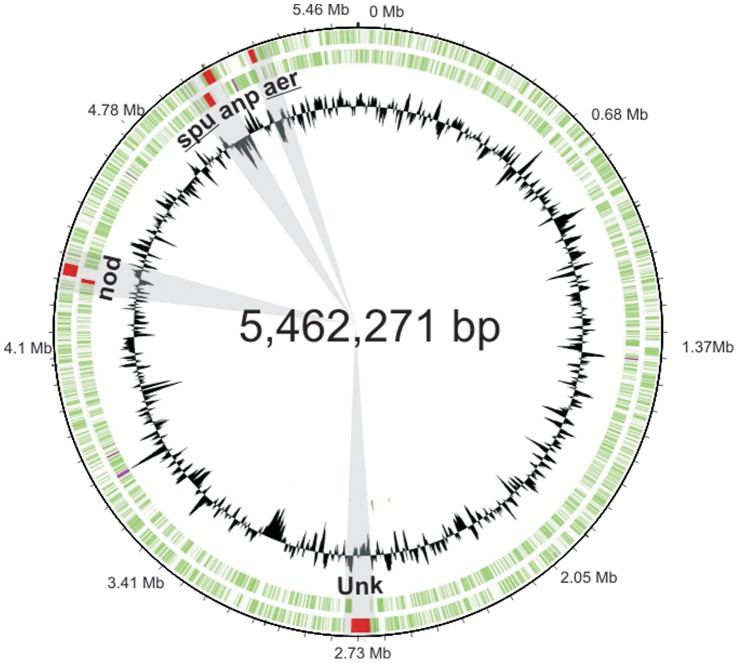
A 17.6 kb aeruginosin (aer) gene cluster was subsequently identified on a single contig in the genome of N. spumigena CCY9414 (GenBank accession number CM001793) through tBLASTn searches using AerD, AerE and AerF protein sequences involved in the synthesis of the Choi moiety (Figure 2). The aer gene cluster was located 133 kb apart from the spumigin gene cluster (Figure 2), which was located just 11 kb from the nodulapeptin gene cluster (Figure 2).
Figure 2. Location of the spumigin and aeruginosin gene clusters on the circular genome of N. spumigena CCY9414.
The N. spumigena CCY9414 genome encodes 5 non-ribosomal peptide gene clusters. The spumigin (spu) and aeruginosin (aer) gene clusters are encoded 133 kb apart. The spumigin and nodulapeptin (anp) gene clusters are encoded 11 kb apart. The nodularin (nda) gene cluster and a cryptic NRPS gene cluster (unk) are encoded at separate locations on the chromosome.
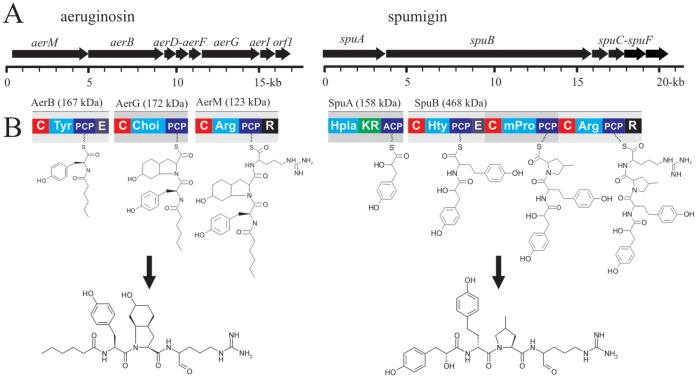
The aer gene cluster encodes 8 proteins organized in a single operon (Figure 3; Table 1). The predicted substrate specificities of the AerM, AerB and AerG peptide synthetases were L-Arg, L-Tyr and Choi through comparison with other aeruginosin biosynthetic pathways (Table 2). Aeruginosin biosynthesis was predicted to start by loading short-chain fatty acids using the AerB condensation domain, as in a number of other non-ribosomal biosynthetic pathways (Figure 3). In order to test this we performed phylogenetic analyses to assign the condensation domains of AerB, AerM and AerG to different condensation domain subtypes (Figure 4). Phylogenetic analysis of the AerB condensation domain showed a close relationship between the loading condensation domains of the nostopeptolide and cyanopeptolin biosynthetic pathways (Figure 4). Genes encoding the AerD, AerE and AerF enzymes were also located in the aer gene cluster (Figure 3) as was a gene encoding a putative glycosyltransferase, AerI (Table 1).
Figure 3. The biosynthesis of aeruginosins and spumigins (A) The organization of the genes in the aeruginosin (18 kb) and spumigin (21 kb) gene clusters in Nodularia spumigena CCY9414.
(B) The proposed biosynthetic routes and main end-product of each biosynthetic pathway. Adenylation domains and their predicted substrates are given in blue. C, condensation domain; PCP, peptidyl carrier protein; E, epimerase domain; R, reductase domain; KR, ketoreductase domain; ACP, acyl carrier protein.
Table 1. Proposed functions of genes in the suspected aeruginosin gene cluster.
| Protein | Amino acids | Proposed function | Sequence similarity | Organism | Identity (%) | Accession number |
| ORF | 60 | MFS-1 protein | Cyanothece sp. PCC 8801 | 43 | YP_002373798 | |
| ORF | 78 | MFS-1 protein | Anabaena sp. 90 | 51 | YP_006996107 | |
| AerM | 1469 | NRPS | Peptide synthetase | M. aeruginosa NIES-843 | 70 | BAG05472 |
| AerB | 1451 | NRPS | Peptide synthetase | M. aeruginosa NIES-843 | 66 | BAG05480 |
| AerD | 196 | Decarboxylase | AerD protein | P. rubescens NIVA-CYA 98 | 73 | CAQ48269 |
| AerE | 211 | Unknown | AerE protein | P. agardhii NIVA-CYA 126 | 64 | CAM59604 |
| AerF | 266 | Reductase | AerF protein | M. aeruginosa NIES-98 | 83 | ACM68688 |
| AerG | 1093 | NRPS | AerG protein | M. aeruginosa NIES-843 | 75 | BAG05474 |
| AerI | 133 | glycosyltransferase | Glycosyl-transferase | P. agardhii NIVA-CYA 126 | 82 | CAM59604 |
| ORF1 | 259 | oxidoreductase | M. aeruginosa NIES-98 | 80 | ACM68692 | |
| ORF2 | 394 | Signal transduction | N. punctiforme PCC 73102 | 87 | ACC84291 | |
| ORF3 | 497 | Two-component hybridsensor and regulator | Nostoc sp. PCC 7120 | 87 | BAB73236 |
The assignment of proposed functions to gene products is based upon BLASTp searches.
Table 2. The substrate specificity of aeruginosin NRPS adenylation domains.
| Protein | Strain | Residue | Proposed substrate | |||||||||
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | |||
| AerB | CCY 9414 | D | A | S | T | I | A | A | V | C | K | Tyr |
| NIES-843 | – | – | – | – | – | – | – | – | – | – | Tyr | |
| PCC 7806 | – | – | – | – | – | – | – | – | – | – | Tyr | |
| NIES-98 | – | – | F | F | L | G | V | T | F | – | Ile | |
| CYA126-8 | – | – | W | F | L | G | N | – | V | – | Leu | |
| AerG | CCY 9414 | D | V | H | I | C | A | F | L | V | K | Choi |
| NIES-843 | – | – | – | – | – | – | – | – | – | – | Choi | |
| PCC 7806 | – | – | – | – | – | – | Y | – | – | – | Choi | |
| NIES-98 | – | – | – | – | – | – | Y | – | – | – | Choi | |
| CYA 126-8 | – | – | – | – | – | – | – | L | – | – | Choi | |
| AerM | CCY 9414 | D | V | E | N | V | G | A | I | T | K | Arg |
| NIES-843 | – | – | – | – | I | – | – | – | – | – | Arg | |
| PCC 7806 | – | – | – | – | G | A | V | V | – | – | Arg | |
The aeruginosin producers include Nodularia spumigena CCY9414, Microcystis aeruginosa strains NIES-843, PCC 7806, NIES-98, and Planktothrix agardhii NIVA-CYA 126/8.
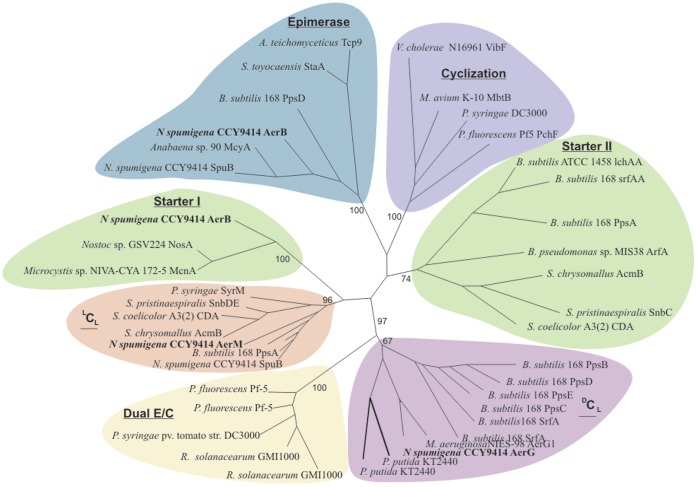
Figure 4. Phylogenetic analyses of the condensation and epimerase domains from the Nodularia spumigena CCY9414 aeruginosin gene cluster.
The condensation domain of AerB clusters with the starter condensation domains of nostopeptolide (NosA) and cyanopeptolin (McnA) which load short chain fatty acids and forms a well-supported group. The phylogenetic tree includes all condensation subtypes, including condensation (LCL, DCL), condensation and heterocyclization catalyzed by heterocyclization domains, epimerization followed by condensation catalyzed by a Dual E/C domain, and loading condesation domains which are found on initiation modules [29]. The phylogeny was reconstructed using phyml, employing the JTT model of amino acid substitution and a gamma-distributed rate variation with four categories. The support values are based on 100-fold bootstrapping.
Preliminary MS2 fragmentation was not sufficient to resolve the first two sub-structural elements and suggested that the second amino acid could be Leu or Tyr while bioinformatic analyses suggested that the substrate could be L-Tyr (Table 2). In order to gain more information on the substructure of the aeruginosins an ATP-PPi exchange assay was performed which demonstrated that L-Tyr was activated by the AerB adenylation domain in vitro (Figure 5). The presence of an epimerase domain suggested that the substrate of AerB was L-Tyr, which would be epimerized to D-Tyr as found in other aeuginosins.
Figure 5. The substrate specificity of the AerB adenylation domain.
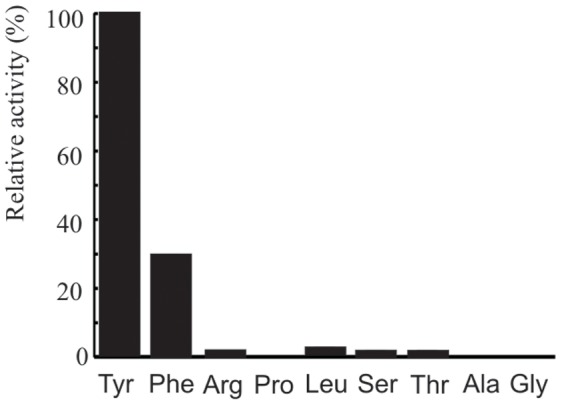
ATP-PPi exchange assay results for the AerB adenylation domain, showing preferential activation of L-Tyr, a predicted substrate of AerB.
Aeruginosin Chemical Structure
The main aeruginosin variant (m/z 587) was hydrolyzed in order to confirm these biochemical and bioinformatic predictions and to obtain further information on the structure of the new peptides. However, despite the presence of a full-length and apparently functional epimerase domain in AerB, the second amino acid was unambiguously determined to be L-Tyr according to chromatographic amino acid analysis of aeruginosins subjected to acid hydrolysis. The amino acids found at this position in other aeruginosins are almost exclusively D-amino acids. Reanalysis of the main aeruginosin (m/z 589) product ion spectrum also showed the presence of Tyr in position 2 (Figure S4 in File S1). The third amino acid, Choi, was in L configuration based to the configuration of Choi in aeruginosin 298-A. The N-terminal moiety could not be detected by amino acid analysis. However, GC-MS analysis of the hydrolyzed aeruginosin allowed unequivocal identification of the N-terminal moiety as hexanoic acid based on retention time and spectra (Figure 6).
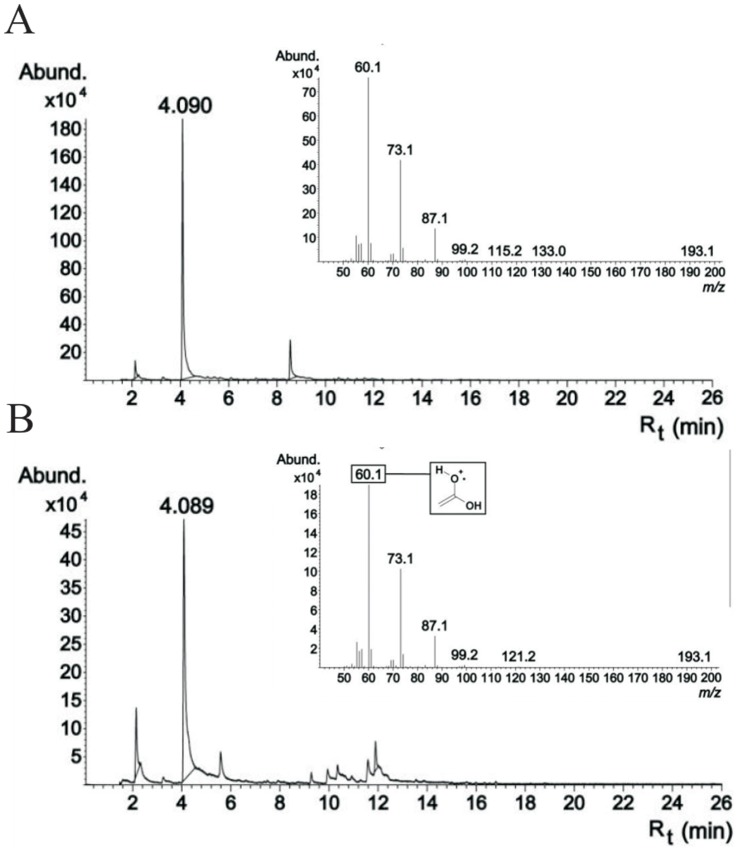
Figure 6. The identity of the C-terminal fatty acid determined using GC-MS.
Chromatographic behavior of the hexanoic acid standard and the hydrolysed aeruginosin NOL3. Commercial hexanoic acid and hydrolysis product from NOL3 had identical retention times, 4.089 and 4.090 min, respectively, and identical mass spectra which unambiguously prove the first moiety of the NOL3 aeruginosin. (A) hydrolysed aeruginosin NOL3 (B) hexanoic acid standard.
The complete structure of the main aeruginosin (m/z 587) was confirmed by NMR analysis. Separation and purification of this peptide from the other aeruginosins and spumigins produced by the N. spumigena AV1 strain was hindered by the reactive aldehydic nature of the compound (Figure S1 in File S1). NaBH4 reduction of the peptides in the methanol extract converted the aldehyde to an alcohol which made it possible to purify reduced aeruginosin by HPLC. 1H and 13C NMR signals yielded four partial structures confirming the aeruginosin structure (Figure 1A). The NMR spectral data are presented in the supplementary material (Table S1 in File S1; Figures S5–S7 in File S1). Accurate mass measurement of the protonated molecules together with the 15N-labeling of the aeruginosins NAL2 and NOL3 were in full agreement with the other results.
Chemical Variation
Detailed inspection of N. spumigena AV1 and CH307 strains allowed the identification of 11 structural variants of aeruginosins (Table 3; Table S2 in File S1). These could be divided into aeruginosins containing either an aldehyde (NAL1-NAL4) or alcohol (NOL1-NOL7) functionality. A range of short straight-chained carboxylic acids were found at the N-terminus (Table 1). Approximately 83% of the variants contained hexanoic acid in AV1 while 93% of the variants contained octanoic acid in CH307 but both strains produced small amounts of aeruginosins with shorter and longer chained carboxylic acids at this position. An O-linked pentose was identified in 4 of the 11 aeruginosins and located on the Choi moiety (Figure 1B, Figure S8 in File S1). Inspection of 16 strains of N. spumigena revealed just a single strain which lacked detectable levels of aeruginosins (Figure 7 and Table S3 in File S1). Glycosylated aeruginosins could be detected in just 3 of the 16 strains (Table S3 in File S1). Three strains, CH307, CH311 and P38, produced glycosylated aeruginosins, of which CH307 produced variants containing octanoic acid as the main fatty acid.
Table 3. Chemical variation of aeruginosins detected from Nodularia spumigena.
| Aeruginosinvariant | Structural subunits | Rt | [M+H]+ (m/z) | Relative amount (%) | ||||||
| 1 | 2 | 3 | 4 | (min) | Experimentala | Calculated | AV1 | CCY9414 | CH307 | |
| NAL1 | Bu | Tyr | Choi | Argininal | 15.0 | 559.3236 | 559.3239 | 1 | <1 | |
| NAL2 | Hex | Tyr | Choi | Argininal | 21.9 | 587.3553 | 587.3552 | 64 | 66 | |
| NAL3 | Oct | Tyr | Choi | Argininal | 32.0 | 615.3877 | 615.3865 | 13 | 1 | |
| NAL4 | Oct | Tyr | Choi-P | Argininal | 27.8 | 747 | 15 | |||
| NOL1 | Ac | Tyr | Choi | Argininol | 12.4 | 533 | <1 | <1 | ||
| NOL2 | Bu | Tyr | Choi | Argininol | 15.1 | 561.3381 | 561.3395 | 1 | 1 | |
| NOL3 | Hex | Tyr | Choi | Argininol | 23.3 | 589.3708 | 589.3708 | 19 | 29 | |
| NOL4 | Oct | Tyr | Choi | Argininol | 32.4 | 617.4050 | 617.4021 | 1 | <1 | |
| NOL5 | Hex | Tyr | Choi-P | Argininol | 18.2 | 721 | 4 | |||
| NOL6 | Oct | Tyr | Choi-P | Argininol | 27.9 | 749 | 81 | |||
| NOL7 | Dec | Tyr | Choi-P | Argininol | 38.2 | 777 | <1 | |||
Structure, ion mass, retention time and abundance of aeruginosin identified from N. spumigena AV1, CCY9414 and CH307.
Unit mass from ion trap, accurate mass from Q-TOF. Ac = acetic acid, Bu = butyric acid,
Hex = hexanoic acid, Oct = octanoic acid, Dec = decanoic acid, P = pentose.
Figure 7. The production of aeruginosins, spumigins, nodularin, nodulapeptin and suomilide by N. spumigena, N. sphaerocarpa, and N. harveyana.
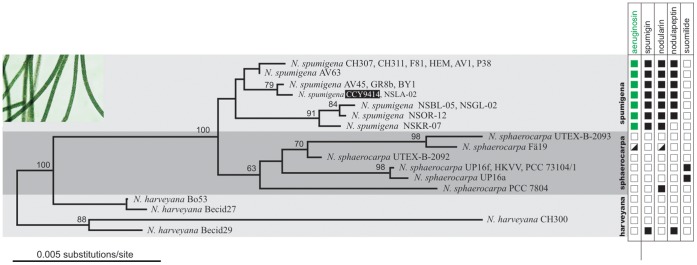
A maximum-likelihood tree based on the 16S rRNA gene with bootstrap values based on 1000 bootstrap replicates. The taxonomy of the strains follows Lyra et al. 2005 [30] and Lehtimäki et al. 2000 [31]. See also Table S3 in File S1.
The presence of aeruginosins as well as the distribution of genes encoding AerM, AerB, AerG, and AerI was determined in 16 strains of N. spumigena, 4 strains of N. harveyana, and 8 strains of N. sphaerocarpa (Table S3 in File S1). All 16 strains of N. spumigena contained aeruginosin biosynthetic genes and 15 of these strains produced aeruginosins (Figure 7). Aeruginosins were not found from N. spumigena AV45, which seems to encode only a partial aer gene cluster. All strains of N. spumigena from the Baltic Sea contained aer clusters encoding the AerI putative glycosyltransferase. The gene encoding this enzyme was not detected in any of the strains isolated from Australia. Neither aeruginosins nor aeruginosin biosynthetic genes could be found in any strains of the benthic N. harveyana or N. sphaerocarpa tested (Table S3 in File S1). The majority of the 16 strains of N. spumigena produced both spumigin and aeruginosin (Figure 7). Some strains produced more aeruginosins and spumigins than others (Figure 7). About half of the 16 strains produced mPro containing spumigins while the remainder produced spumigins containing Pro. Using UV (280 nm) to quantify the amount of aeruginosins in the AV1 strain demonstrated that spumigins comprise 7 ‰ and aeruginosins 3–4 ‰ of the dry weight. All 16 strains of Nodularia spumigena produced spumigins and the nodularin toxin. Almost all of the 16 strains of Nodularia spumigena produced nodulapeptins. Suomilide was identified only in N. sphaerocarpa strains (Table S3 in File S1).
Discussion
Aeruginosins are a chemically diverse family of peptides known to date from just the bloom-forming cyanobacterial genera Microcystis and Planktothrix [11], [12], [13], [14]. The relationship between aeruginosins and spumigins has been subject to speculation for some time [12], [13], [15], [16]. Phylogenetic analyses suggest that the spumigin gene cluster of N. spumigena and the aeruginosin gene clusters of Microcystis and Planktothrix are unrelated [8]. Recent studies demonstrated that N. spumigena encodes a number of cryptic NRPS clusters for which the products were unknown [3], [17]. It was suggested based on the presence of Choi biosynthetic genes that it may produce an aeruginosin [3]. Interestingly a recent study has shown that N. spumigena isolated from variety of geographic locations produce a diversity of peptides including a partial peptide which contains Choi and may be assigned to the aeruginosin family [18]. This study suggests that N. spumigena produces bona fide aeruginosins in addition to spumigin [18]. However, the aeruginosin structures presented by Mazur-Marzec and coworkers were based on MS2 data and are incomplete [18]. The identities of the N-terminal moiety and amino acid at position 2 were not resolved in their analyses [18]. Here we show that the complete structure of the main aeruginosin variant is comprised of an N-terminal short fatty acid chain, L-Tyr, L-Choi and L-argininal.
We found 11 structural variants of the aeruginosins in 16 strains from the Baltic Sea and Australia. The aeruginosins produced by N. spumigena contain fatty acids at the N-terminus including acetic acid, butyric acid, hexanoic acid, octanoic acid and decanoic acid. The N-termini of previously reported aeruginosins consists of either hydroxyphenyl lactic acid in Microcystis or phenyl lactic acid in Planktothrix [11]. Aeruginosins may also be modified to contain chlorine, sulfate or sugars [12] [13]. Glycosylation of aeruginosides, members of the aeruginosin family reported from P. agardhii, is catalyzed by the AerI glycosyltransferase [12]. However, the majority of aeruginosins detected here lacked the O-linked pentose despite the presence of the AerI glycosyltransferase in the genome of the producing strain.
It has been anticipated that aeruginosins and spumigins might be related compounds based on their structural similarities [12], [13], [15], [16]. Our results show that these two peptides are assembled on separate peptide synthetases (Figure 2). The organization of catalytic domains in AerB, AerG and AerM suggests an orthodox model for aeruginosin assembly (Figure 2). N. spumigena CCY9414 lacks the reductive loading mechanism of the Microcystis and Planktothix aeruginosin biosynthetic pathways entirely [12], [13]. The organization of catalytic domains in the aer gene cluster (Figure 3) and phylogenetic analyses (Figure 4) suggests that the condensation domain of AerB is responsible for lipidation of the aeruginosins. N-terminal condensation domains have been proposed to prime the synthetase with short-chain carboxylic acids in lichenysin [19], daptomycin [20], nostopeptolide [21] and cyanopeptolin [22] [23] biosynthesis. Our results suggest substrate specificities ranging from C2 to C10 fatty acid moieties. However, the exact lipidation mechanism remains unknown. The reductase domain of AerM releases the C-terminal arginine as a reactive aldehyde.
Members of the aeruginosin family of natural products commonly have strong inhibitory activity against serine proteases [11], [15]. Serine proteases are involved in a number of important physiological processes, and their importance in the complex blood coagulation cascade is well established [11]. Planktonic bloom-forming cyanobacteria produce a range of protease inhibitors [15]. The function of these peptides is unclear but they are widely believed to be part of a chemical defense system, acting as a grazing deterrent [24], [25]. Our results show that N. spumigena strains produce a complex cocktail of protease inhibitors comprising up to 1% of the dry weight of the organism, which may explain in part its ecological success.
Materials and Methods
Strain Growth
Sixteen strains of N. spumigena, 4 strains of N. harveyana and 8 strains of N. sphaerocarpa (Table S3 in File S1) were grown at a photon irradiance of 15 µmol m−2 s−1 in saline Z8 medium lacking a source of combined nitrogen for 21 days [8]. 15N-labeling of N. spumigena AV1 was performed in similar way, except that medium was buffered with 10 mM HEPES (pH 8.0). 15N-urea (98+ % 15N, ISOTEC, USA) was used as nitrogen source and nitrogen-free argon (with 20.9% O2 and 0.45% CO2; quality 5.7; AGA Gas Ab, Sweden) was bubbled into the medium to prevent nitrogen fixation from air.
Gene Cluster Annotation
The aer gene cluster was identified on a single 5,462,271 bp scaffold in the genome of N. spumigena CCY9414 (GenBank accession number CM001793) through BLASTp searches using AerD, AerE and AerF proteins. The genes in the aer gene cluster were predicted with Artemis using Glimmer. The starting sites were refined manually. The amino acid sequences of the genes were used to query the non-redundant database at NCBI in order to predict a function for the genes (Table 2). The substrate specificity of the activated adenylation domains in the NRPS modules was predicted by using the 10 amino acid binding pocket signature [26].
Frequency of aer gene Clusters in N. spumigena Strains
Genomic DNA was extracted from the cultivated strains as previously described [8]. We amplified four genes from the aer gene cluster, aerM, aerB, aerG and aerI, by PCR using oligonucleotide primers designed from the N. spumigena CCY9414 genome sequence (Table S4 in File S1). The PCR reactions were performed in a 20 µl final volume containing 1 µl of DNA, 1× DyNAzyme II PCR buffer, 100 mM of each deoxynucleotide, 0.4 mM of each oligonucleotide primer, and 0.4 units of DyNAzyme II DNA polymerase (Finnzymes, Espoo, Finland). The following protocol was used: 94°C, 3 min; 25 cycles of 94°C, 30 s; 63°C, 30 s; 72°C, 1 min; and 72°C, 10 min. PCR to confirm the deletion of the aerI gene was performed as before but with an annealing temperature of 58°C. PCR products were visualized on 1.5% agarose gels containing 0.5× TAE run at 120 V for 20–25 min and scored for the presence or absence of PCR products of the expected length. The 16S rRNA gene was amplified and sequenced from N. spumigena CH307, P38 and AV45 as previously described [27] and the sequence data was deposited in GenBank (KF360086-KF360088). An alignment of 16 strains of N. spumigena, 8 strains of N. sphaerocarpa and 4 strains of N. harveyana was made using Bioedit. Gaps and ambiguous regions were excluded and a total of 1340 bp of sequence was considered for phylogenetic analysis. A neighbor-joining tree was constructed using DNADIST and NEIGHBOR as implemented in the PHYLIP package. The tree was midpoint rooted using RETREE. 1000 bootstrap replicates were constructed using SEQBOOT, DNADIST, NEIGHBOR and CONSENSE in the PHYLIP package. The production of aeruginosin, spumigin, nodularin, nodulaopeptin and suomilide was mapped to this tree.
LC-MS
LC-MS analyses were performed with an Agilent 1100 Series LC/MSD Ion Trap XCT Plus System (Agilent Technologies, Palo Alto, CA, USA) using a Phenomenex Luna C8 (150×2.0 mm, 5 µm, Phenomenex, Torrance, CA, USA) LC-column. Between 7 and 31 mg of freeze-dried cells of Nodularia strains were extracted for 20 s with 1 ml of methanol in 2 ml plastic tubes containing approximately 200 µl of 0.5 mm glass beads (Scientific Industries, New York) using Fast Prep homogenizer (FP120, Bio 101, Savant) at speed value of 6 m s−1. Extracts were centrifuged for 5 min at 10 000 g prior to LC-MS analysis. High accuracy mass of the aeruginosins of N. spumigena AV1 was measured by UPLC-ESI-QTOF mass spectrometry performed on Synapt G2 HDMS (Waters, MA, USA) in high resolution mode and m/z 500–850 mass range. In MS/MS analysis the mass range was m/z 50–531.
Derivatization
Aeruginosins were derivatized with malondialdehyde (MDA) using 100 µl of methanol extract from N. spumigena AV1 evaporated to dryness in vacuum centrifuge. The resultant residue was dissolved in 100 µl of 12 M H3PO4 and 2.4 µl of 1,1,3,3-tetraethoxypropane (Sigma) was added. The sample was evaporated in a vacuum centrifuge after 1 h at room temperature and dissolved in 100 µl of methanol. DNPH derivatives were prepared as previously described [8].
Amino Acid Hydrolysis
Isolated aeruginosin NOL3 (100 µg) were dried in a 300-µl glass vial which was then transferred to a 4-ml glass vial containing 1 ml of 6 M HCl. The vial was flushed with argon prior to being closed. Acid hydrolysis was performed by incubating overnight at 110°C. After hydrolysis, the inner vial was dried for 30 min with a vacuum centrifuge. Hydrolyzed and reference amino acids were derivatized by the Marfey method using L-FDAA (1-fluoro-2,4-dinitrophenyl-5)-L-alaninamide) reagent. Reaction mixtures were analyzed with a Luna C18(2) column (150×2, 5 µm; Phenomenex) eluted with an acetonitrile (solvent B) and 0.1% aqueous formic acid (solvent A) gradient (20% B to 75% B in 45 min and then 5 min at the reached level) at a flow rate of 0.2 ml min−1 at a temperature of 40°C. The detection wavelength for the Marfey derivatives was 340 nm. Acid hydrolysate of aeruginosin 298-A from Microcystis aeruginosa NIES-298 was used as a reference for L-Choi [28].
GC-MS
100 µg of aeruginosin NOL3 was incubated with 100 µl of 5 M NaOH in a closed vial for 24 h at 110°C. The entire 100 µl solution was transferred to a 20 ml brown vial with 18 mm magnetic caps with silicon/Teflon disks (VWR International, USA) containing 4 ml of MilliQ water, 1.5 g of NaCl and 50 µl of 17.5 M H3PO4, and the vial was immediately sealed. The vial was agitated for 5 min at 70°C and 500 rpm in a GC-MS autosampler (combiPAL, CTC Analytics). A SPME (Solid Phase Micro Extraction) needle penetrated 12 mm through the vial cap, exposing 10 mm of the fiber (DVB/CAR/PDMS, Supelco, Sigma-Aldrich Co., USA) inside the vial. After a 30 min extraction the fiber was retracted and sample was injected in the GC column with an injection needle penetration of 32 mm and the fiber exposure of 10 mm. After 10 min of desorption time, the fiber was removed from the injection port and the sample was injected with a split ratio of 5∶1. HP 6890 gas chromatograph with an Agilent 5973 Network mass selective detector (Agilent Technologies, Wilmington, DE, USA) with a split/splitless injector and a SPB™-624 capillary column (30 m×0.25 mm, 1.4 um; Supelco, Sigma-Aldrich Co., USA) were used as follows: Injection and detector at 250°C, oven started at 150°C for 2 min from which temperature increased 5°C min−1 during 10 min. The final temperature was 220°C after a total run time of 26 min. Helium was used as carrier gas with a flow rate of 1 ml min−1. A standard 0.14 mM hexanoic acid (Sigma-Aldrich Co., USA) solution was prepared. The full scan electron impact mass spectra were obtained at a range of 50–200 m/z.
NMR Analysis
Aldehydes were converted to alcohols in the methanol extract using NaBH4 reduction which made it possible to purify aeruginosin NOL1 by HPLC. One gram of freeze dried AV1 cells was extracted with 70 ml of methanol using a tip homogenizer (SilentCrusher M, Heidolph, Germany) in three 30 sec cycles at ambient temperature with a speed of 16000 rpm 3×30 sec. The suspension was centrifuged (10000 g, 5 min) and dichloromethane and water was added to the supernatant in volume ratio of 1∶1:1. Phases were separated by centrifugation (5000 g, 5 min). The upper water/methanol phase was collected and vacuum evaporated to dryness. The residue was dissolved in 4 ml of methanol, 50 mg of NaBH4 was added and after 5 min reaction time the solution was vacuum evaporated to dryness. The residue was dissolved in 1 ml of 15% acetonitrile. 100 µl portions of the solution were injected 10 times into a Luna C8 (2) (10×150 mm, 5 µm, 100 Å, Phenomenex) column which was eluted isocratically with 0.1% TFA in 15% acetonitrile. Pooled fractions containing aeruginosin NOL1 were evaporated in a vacuum and dissolved in CD3OD for NMR. 1H and 13C NMR spectra were obtained with a Varian Unity Inova 600 MHz NMR spectrometer equipped with cryogenically cooled triple-resonance 1H, 13C, 15N probe head and actively shielded z-gradient system. DQF-COSY, TOCSY (120 ms mixing time) experiments were collected using 2048 and 512 data points in F1 and F2 dimensions, corresponding to acquisition times of 0.34 and 0.085 s, respectively. The corresponding acquisition times in 13C HSQC and 13C HMBC experiments were 0.02 (13C dimension) and 0.17 (1H dimension), and 0.014 (13C dimension) and 0.34 (1H dimension), respectively. The average one- and three-bond 1H-13C couplings were estimated to be 140 Hz and 8 Hz, and 1H-13C transfer delays for HSQC and HMBC were set to 3.57 and 62.5 ms, respectively. All spectra were collected at 25°C. Spectra were processed and analyzed using VNMRJ 2.1 version B and ACD/SpecManager version 11.03 software packages.
ATP-pyrophosphate Exchange Assay
The region of the aerB gene encoding the adenylation domain was amplified by PCR from N. spumigena CCY9414 using oligonucleotide primers designed to anneal to the substrate-conferring portion of each adenylation domain. Primer design and PCR reactions were performed as described previously [8]. PCR products were digested with NcoI and PmeI restriction enzymes, gel excised and ligated to pFN18A (HaloTag® 7) T7 Flexi® vector (Promega, WI, USA) opened with the same enzymes. Ligation mix was transformed into Escherichia coli (KRX) competent cells following the manufacturer’s instructions. Colonies were grown in shaker (160 rpm) at 37°C overnight in 3 ml of LB medium supplemented with 100 mg ml−1 ampicillin. In the following day, 400 µl was used to inoculate 20 ml of TB medium containing 50 mg ml−1 carbenicillin and incubated with shaking at 37°C (160 rpm) for 1.5 h and then induced by the addition of 0.1% of rhamnose and the culture was grown overnight (16–18 h) in shaker (100 rpm) at 24°C. E. coli cells were collected and sonicated as described previously [8]. The expression of soluble protein was observed in 10% SDS PAGE gel. The soluble adenylation domains were purified using HaloTag® Protein Purification System (Promega). Protein concentration of the preparations was measured with the BCA protein assay kit (Pierce). ATP-pyrophosphate exchange assay was performed as described previously [9].
Supporting Information
The Combined Supporting Information File S1 contains detailed data on the discovery and identification of aeruginosins by LC-MS (Figures S1–S4) and NMR (Figures S5–S7, Table S1), chemical variation of aeruginosins (Figure S8 and Table S2) and the results of the screening of individual Nodularia strains for various peptides and their biosynthetic genes (Table S3) and the PCR primers used (Table S4).
(DOCX)
Acknowledgments
We are indebted to Lyudmila Saari for her valuable assistance in handling the cultures. We are grateful to Lucas Stal and Wolfgang Hess for providing us access to an unpublished version of the Nodularia spumigena CCY9414 genome.
Funding Statement
This work was funded by a research Center of Excellency (118637) and Academy project grant (258827) both from the Academy of Finland to KS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stal LJ, Albertano P, Bergman B, von Bröckel K, Gallon JR, et al. (2003) BASIC: Baltic Sea cyanobacteria: An investigation of the structure and dynamics of water blooms of cyanobacteria in the Baltic Sea-responses to a changing environment. Cont Shelf Res 23: 1695–1714. [Google Scholar]
- 2. Jonasson S, Vintila S, Sivonen K, El-Shehawy R (2008) Expression of the nodularin synthetase genes in the Baltic Sea bloom-former cyanobacterium Nodularia spumigena strain AV1. FEMS Microbiol Ecol 65: 31–39. [DOI] [PubMed] [Google Scholar]
- 3. Voß B, Bolhuis H, Fewer DP, Kopf M, Möke F, et al. (2013) Insights into the physiology and ecology of the brackish-water-adapted cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis. PLoS One 8: e60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sivonen K, Kononen K, Carmichael WW, Dahlem AM, Rinehart KL, et al. (1989) Occurrence of the hepatotoxic cyanobacterium Nodularia spumigena in the Baltic Sea and structure of the toxin. Appl Environ Microbiol 55: 1990–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moffitt MC, Neilan BA (2004) Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl Environ Microbiol 70: 6353–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simola O, Wiberg M, Jokela J, Wahlsten M, Sivonen K, et al. (2012) Pathologic findings and toxin identification in cyanobacterial (Nodularia spumigena) intoxication in a dog. Vet Pathol 49: 755–759. [DOI] [PubMed] [Google Scholar]
- 7. Fujii K, Sivonen K, Adachi K, Noguchi K, Sano H, et al. (1997) Comparative study of toxic and non-toxic cyanobacterial products: Novel peptides from toxic Nodularia spumigena AV1. Tetrahedron Lett 38: 5525–5528. [Google Scholar]
- 8. Fewer DP, Jokela J, Rouhiainen L, Wahlsten M, Koskenniemi K, et al. (2009) The non-ribosomal assembly and frequent occurrence of the protease inhibitor spumigin in the bloom-forming cyanobacterium Nodularia spumigena . Mol Microbiol 73: 924–937. [DOI] [PubMed] [Google Scholar]
- 9. Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K (2010) Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria). Chem Biol 17: 265–273. [DOI] [PubMed] [Google Scholar]
- 10. Anas AR, Kisugi T, Umezawa T, Matsuda F, Campitelli MR, et al. (2012) Thrombin inhibitors from the freshwater cyanobacterium Anabaena compacta . J Nat Prod 75: 1546–1552. [DOI] [PubMed] [Google Scholar]
- 11. Ersmark K, Del ValleJR, Hanessian S (2008) Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew Chem Int Ed 47: 1202–1223. [DOI] [PubMed] [Google Scholar]
- 12. Ishida K, Christiansen G, Yoshida WY, Kurmayer R, Welker M, et al. (2007) Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem Biol 14: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishida K, Welker M, Christiansen G, Cadel-Six S, Bouchier C, et al. (2009) Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Appl Environ Microbiol 75: 2017–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cadel-Six S, Dauga C, Castets AM, Rippka R, Bouchier C, et al. (2008) Halogenase genes in non-ribosomal peptide synthetase gene clusters of Microcystis (cyanobacteria): sporadic distribution and evolution. Mol Biol Evol 25: 2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Welker M, von Döhren H (2006) Cyanobacterial peptides – nature’s own combinatorial biosynthesis. FEMS Microbiol Rev 30: 530–563. [DOI] [PubMed] [Google Scholar]
- 16. Schindler CS, Stephenson CRJ, Carreira EM (2008) Enantioselective synthesis of the core of banyaside, suomilide, and spumigin HKVV. Angew Chem Int Ed 47: 8852–8855. [DOI] [PubMed] [Google Scholar]
- 17. Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, et al. (2012) Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA 110: 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mazur-Marzec H, Kaczkowska MJ, Agata Blaszczyk A, Akcaalan R, Spoof L, et al. (2013) Diversity of peptides produced by Nodularia spumigena from various geographical regions. Mar Drugs 11: 1–19 doi:10.3390/md11010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Konz D, Doekel S, Marahiel MA (1999) Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J Bacteriol 181: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miao V, Coëffet-Legal MF, Brian P, Brost R, Penn J, et al. (2005) Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151: 1507–1523. [DOI] [PubMed] [Google Scholar]
- 21. Hoffmann D, Hevel JM, Moore RE, Moore BS (2003) Sequence analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene 311: 171–180. [DOI] [PubMed] [Google Scholar]
- 22. Bister B, Keller S, Baumann HI, Nicholson G, Weist S, et al. (2004) Cyanopeptolin 963A, a chymotrypsin inhibitor of Microcystis PCC 7806. J Nat Prod 67: 1755–1757. [DOI] [PubMed] [Google Scholar]
- 23. Nishizawa T, Ueda A, Nakano T, Nishizawa A, Miura T, et al. (2011) Characterization of the locus of genes encoding enzymes producing heptadepsipeptide micropeptin in the unicellular cyanobacterium Microcystis . J Biochem 149: 475–485. [DOI] [PubMed] [Google Scholar]
- 24. Rohrlack T, Christoffersen K, Kaebernick M, Neilan BA (2004) Cyanobacterial protease inhibitor microviridin J causes a lethal molting disruption in Daphnia pulicaria . Appl Environ Microbiol 70: 5047–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davis TW, Koch F, Marcoval MA, Wilhelm SW, Gobler CJ (2012) Mesozooplankton and microzooplankton grazing during cyanobacterial blooms in the western basin of Lake Erie Harmful Algae. 15: 26–35. [Google Scholar]
- 26. Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6: 493–505. [DOI] [PubMed] [Google Scholar]
- 27. Halinen K, Fewer DP, Sihvonen LM, Lyra C, Eronen E, et al. (2008) Genetic diversity in strains of the genus Anabaena isolated from planktonic and benthic habitats of the Gulf of Finland (Baltic Sea). FEMS Microbiol Ecol. 64: 199–208. [DOI] [PubMed] [Google Scholar]
- 28. Murakami M, Okita Y, Matsuda H, Okino T, Yamaguchi K (1994) Aeruginosin 298-A, a thrombin and trypsin inhibitor from the blue-green alga Microcystis aeruginosa (NIES-298). Tetrahedron Lett 35: 3129–3132. [Google Scholar]
- 29. Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH (2007) Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol. 7: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyra C, Laamanen M, Lehtimäki JM, Surakka A, Sivonen K (2005) Benthic cyanobacteria of the genus Nodularia are non-toxic, without gas vacuoles, able to glide and genetically more diverse than planktonic Nodularia. Int J Syst Evol Microbiol. 55: 555–568. [DOI] [PubMed] [Google Scholar]
- 31. Lehtimäki J, Lyra C, Suomalainen S, Sundman P, Rouhiainen L, et al. (2000) Characterization of Nodularia strains, cyanobacteria from brackish waters, by genotypic and phenotypic methods. Int J Syst Evol Microbiol. 3: 1043–1053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Combined Supporting Information File S1 contains detailed data on the discovery and identification of aeruginosins by LC-MS (Figures S1–S4) and NMR (Figures S5–S7, Table S1), chemical variation of aeruginosins (Figure S8 and Table S2) and the results of the screening of individual Nodularia strains for various peptides and their biosynthetic genes (Table S3) and the PCR primers used (Table S4).
(DOCX)