Abstract
Background
The study of coffee polysaccharides-degrading enzymes from the coffee berry borer Hypothenemus hampei, has become an important alternative in the identification for enzymatic inhibitors that can be used as an alternative control of this dangerous insect. We report the cloning, expression and biochemical characterization of a mannanase gene that was identified in the midgut of the coffee berry borer and is responsible for the degradation of the most abundant polysaccharide in the coffee bean.
Methods
The amino acid sequence of HhMan was analyzed by multiple sequence alignment comparisons with BLAST (Basic Local Alignment Search Tool) and CLUSTALW. A Pichia pastoris expression system was used to express the recombinant form of the enzyme. The mannanase activity was quantified by the 3,5-dinitrosalicylic (DNS) and the hydrolitic properties were detected by TLC.
Results
An endo-1,4-β-mannanase from the digestive tract of the insect Hypothenemus hampei was cloned and expressed as a recombinant protein in the Pichia pastoris system. This enzyme is 56% identical to the sequence of an endo-β-mannanase from Bacillus circulans that belongs to the glycosyl hydrolase 5 (GH5) family. The purified recombinant protein (rHhMan) exhibited a single band (35.5 kDa) by SDS-PAGE, and its activity was confirmed by zymography. rHhMan displays optimal activity levels at pH 5.5 and 30°C and can hydrolyze galactomannans of varying mannose:galactose ratios, suggesting that the enzymatic activity is independent of the presence of side chains such as galactose residues. The enzyme cannot hydrolyze manno-oligosaccharides such as mannobiose and mannotriose; however, it can degrade mannotetraose, likely through a transglycosylation reaction. The Km and kcat values of this enzyme on guar gum were 2.074 mg ml-1 and 50.87 s-1, respectively, which is similar to other mannanases.
Conclusion
This work is the first study of an endo-1,4-β-mannanase from an insect using this expression system. Due to this enzyme’s importance in the digestive processes of the coffee berry borer, this study may enable the design of inhibitors against endo-1,4-β-mannanase to decrease the economic losses stemming from this insect.
Keywords: Endo-mannanase, Coffee berry borer, Hypotenemus hampei, Glycosyl hydrolase
Background
As an extension of previous study on different glycosyl hydrolase (GHs) enzymes that are secreted in the digestive system from coffee berry borer Hypothenemus hampei, this investigation is concerned with an specific GHs, an endo-1,4-β-mannanase (EC 3.2.1.78) [1]. This enzyme plays an important role in the random internal hydrolysis of β-mannosidic links in the main structure of mannans, releasing small manno-oligosaccharides (MOS) that are absorbed by the berry borer and used for development and growth. Thus, mannanases are an important inhibitory target in the design of control strategies for the coffee berry borer. Previous studies have relied in synthetic inhibitors evaluation of this enzyme [2].
Mannans are a group of polysaccharides that include four types: linear mannans, galactomannans, glucomannans and galactoglucomannans [3]. These polysaccharides are structural components of the cell walls and intracellular matrices of terrestrial and marine plants. In the coffee bean (Coffea sp.), a crop that provides sustenance to approximately half a million families of growers in Colombia [4], galactomannans constitute 25% of total polysaccharides, which in turn represents 50% of the coffee bean’s dry weight [5].
This seed is the only food source for the coffee berry borer (Hypothenemus hampei), which is the most detrimental insect for coffee growers around the world [6]. Throughout its life cycle, the berry borer remains inside the fruit, thus making it difficult to control with insecticides or natural enemies. Searching for mechanisms that interfere with the insect’s normal physiology may be the foundation for developing alternative biotechnologies for controlling this insect population.
Other studies have focused on bacterial endo-1,4-β-mannanases [7-9], fungi [10-12], superior plants [13] and mollusks [14,15]. In this article, the cloning and biochemical characterization of a recombinant endo-1,4-β-mannanase (rHhMan) is described for the first time for the H. hampei insect. Additionally, the investigation of this enzyme’s hydrolytic properties allows characteristics associated with the mechanism of this enzyme to be inferred to facilitate the design of inhibitors for use in future control strategies.
Results and discussion
Sequence analysis of the HhMan
Pfam analysis (E-value: 1.4×10-34) indicated that the enzyme belongs to the glycosyl hydrolase GH5 family, an extensive group of enzymes that catalyze the cleavage of glycosidic bonds between two or more carbohydrates [16,17]. These results were demonstrated by multiple alignment of the deduced amino acid sequence against other mannanases of distinct origins within this family: Bacillus circulans (GenBank Accession Number AAX87003.1), Paenibacillus polymyxa (GenBank Accession Number ADO54643.1), Cellvibrio japonicus (GenBank Accession Number AA031760.1), Clostridium butyricum (GenBank Accession Number EEP53331.1) and Vibrio furnissii (GenBank Accession Number EEX39836.1), yielding percentage identities (PID) of 56%, 57%, 39%, 52% and 39% respectively. Figure 1 shows that all of the mannanases in the analysis exhibit features common to the GH5 family, including two strictly conserved glutamic acid residues that are important for catalytic activity (E147 and E242) by acting as nucleophiles or general acid/base catalysts, as well as six active site catalytic residues (R72, H108, N146, H212, Y214 and W272) [18-21].
Figure 1.

Alignment of the deduced amino acid sequence of HhMan of Hypothenemus hampei with those of other GH5 mannanases. Identical or similar amino acid residues are listed in black or gray, respectively. Glutamic acid residues and the residues highly conserved of the active site are indicated asterisk (*) below the alignment. The regions of HhMan with secondary structure α-helices or β-folded sheets shown on the alignment.
Cloning and expression of the HhMan gene
The HhMan gene was amplified by PCR with primers designed from the available GenBank sequence (ADF22325.1) and cloned in the pPICZαA vector. The coding sequence corresponds to a 900-bp open reading frame (ORF) for a 300-aa protein and does not include the native signaling sequence.
rHhMan was expressed as an extracellular protein in Pichia pastoris. The protein content in the supernatant was purified by affinity chromatography on Ni-NTA agarose with a one-step purification protocol. SDS-PAGE analysis demonstrated that an apparently homogeneous protein was obtained with an approximate molecular weight of 35.5 kDa, corresponding to 33 kDa from the protein sequence without the native signaling sequence and 2.5 kDa from the N-terminal polyhistidine tag (Figure 2A). The calculated molecular weight range for rHhMan (35–40 kDa) lies within those previously reported for other mannanases, such as mannanases from Solanum lycopersicon (39 kDa) [22], Bacillus subtilis (39.6 kDa) [9], Aplysia kurodai (40 kDa) [23] and Cryptopygus antarcticus (40 kDa) [24].
Figure 2.
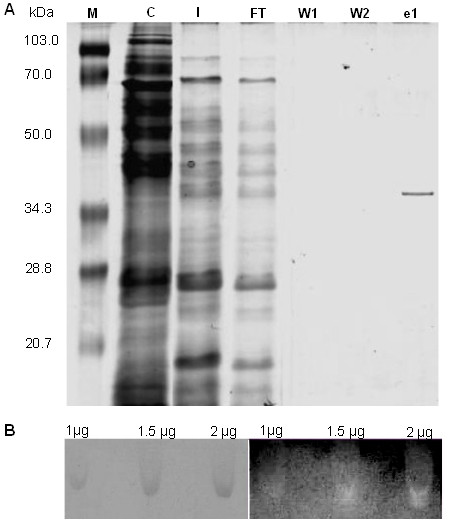
SDS- page, native- page and zymogram of the purified rHhMan. A) SDS-PAGE. M: Broad Range weight marker, C: Negative control for expression, I: Unpurified samples. FT: Fraction not retained in the matrix. W1: wash 1, W2: wash 2, e1: purified rHhMan. B) NATIVE- PAGE AND ZYMOGRAM Samples of pure protein (1, 1.5 y 2μg) were loaded on two native gels (12%). Left panel in absence of guar gum, the gel was stained with Coomassie brilliant blue R250 to visualize the band corresponing to mannanase (35.5 kDa). Right panel in the presence of 1% guar gum, the gel was stained with Congo red to verify the mannanase activity.
The expression protocol yielded 6 mg of pure protein per liter of culture medium with a specific activity on guar gum of 1075.3 U/mg under standard assay conditions.
Enzyme activity of the commercial enzyme was checked (data not shown). The enzymatic activity of rHhMan was evaluated by zymography assays in 1% guar gum substrate. The enzymatic activity was directly proportional to the enzyme concentration. Figure 2B shows that the highest activity levels were obtained with 2 μg of enzyme, which was the highest concentration tested. The activity was reduced when a lower concentration of 1 μg was used.
Effects of pH and temperature on enzymatic activity
The optimal enzymatic activity of rHhMan occurred at pH 5.5, with 95% activity at pH 5.0 and 75% activity at pH 4.0, 6 and 7. The activity gradually decreased to 50% at pH 7.5 and 8 (Figure 3A). These results demonstrate that rHhMan remains stable under acidic pH conditions, which is in agreement with the physiological pH levels in the digestive tract of H. hampei, which lie between pH 4.5 and 5.2 [25]. Similarly, the intestinal pH in other coleoptera is between 4.5 and 5.5 [26]. Other mannanases from diverse fungi exhibit optimal activity at acidic pH, and mannanases from Aspergillus niger, A. fumigatus and A. oryzae are optimally active at pH 5.5, 4.5 and 4.5, respectively [10,27,28].
Figure 3.
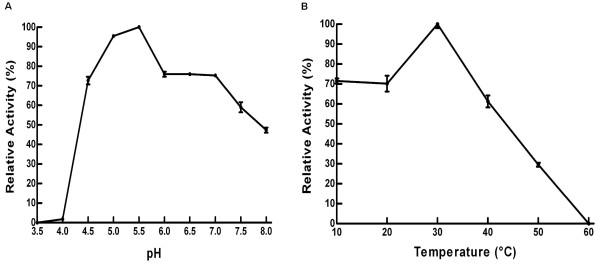
Characterization of recombinant rHhMan. A) The effect of pH on the activity of rHhMan. B) The effect of the temperature on the activity of rHhMan.
The maximum activity of rHhMan occurred at 30°C, and the activity was reduced to approximately 70% between 10 and 20°C. The activity decreased substantially at high temperatures. At 40°C, 60% of the activity was conserved, while only 30% activity remained at 50°C (Figure 3B). These findings coincide with the reported activity levels in the digestive tracts of insects, with the highest activity between 30 and 40°C and a rapid decrease in the maximum activity at higher temperatures [26].
Hydrolytic properties
Purified rHhMan protein can hydrolyze high-molecular weight polysaccharides with β-1,4-glycosidic bonds in their structure. Figure 4A-B shows that the main by products of the hydrolysis of both guar and locust bean gum are mannose and manno-oligosaccharides of different sizes (mannobiose, mannotriose and mannotetraose) after a 15-min reaction. Similarly, other Bacillus sp. mannanases hydrolyze different substrates with the same structural characteristics of the aforementioned galactomannans [9]. In those reactions, the hydrolysis by products obtained can be used as food additives because of the prebiotic and anti-obesity effects that MOS have in different regions of the digestive tract [29].
Figure 4.
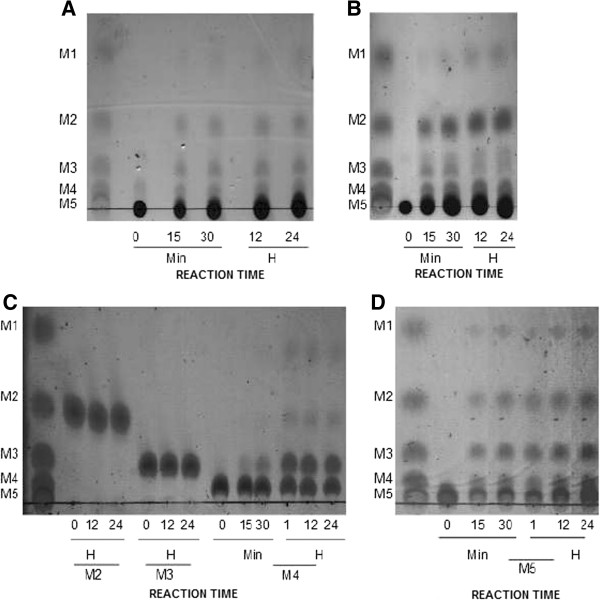
Thin layer chromatography (tlc) of the hydrolytic products generated by rhhman on Mannans and Manno-oligosaccharides. The reactions in the substrates respectives in 0,1 M sodium citrate buffer (pH 5.5) with 0,4 μg of rHhMan were incubated at 30°C for 24 hours. 2 μl of each reaction were analized by TLC. The substrates were A) Guar gum, B) Locust vean, C) Manno- oligosaccharides: mannobiose, mannotriose and mannotetraose and D) Manno-oligosaccharides: Mannopentose. The incubation time was shown (minutes and hours). The standar was a mix of mannose and manno- oligosaccharides where M1 is mannose; M2, mannobiose; M3, mannotriose; M4, mannotetraose y M5 mannopentose.
The mode of action of rHhMan was analyzed by thin layer chromatography (TLC) with different manno-oligosaccharides (Figure 4C). The enzyme cannot hydrolyze mannobiose and mannotriose in a 24-hr incubation. However, mannotetraose hydrolysis produces mannose, mannobiose and mannotriose. This results suggest that probably it is a process that include a transglycosylation reaction in the mechanism that allows the enzyme to hydrolyze this manno-oligosaccharide [14,27]. In these types of enzymes, mannans must have a degree of polymerization of at least 4 units to achieve significant hydrolysis, as occurs with mannotetraose [30].
Additionally the knowledge of hydrolytic properties of this enzyme and the methodology described has been used to evaluate five analogues that mimic the HhMan substrate and function as inhibitors of the enzyme. The compounds were evaluated by TLC and enzymatic inhibiton test. One of the tested compounds, the 4-nitrophenyl-thio-β-D-mannopyranoside was identified as an inhibitor of HhMan. Thus, this strategy is a starting point for the development of new molecules to be used in pest control strategies to reduce the serious damage caused by the coffee berry borer during coffee cultivation [2].
Kinetic parameters
rHhMan displayed Michaelis-Menten reaction behavior in guar gum substrate assays, as shown in Figure 5. This polysaccharide has a mannose:galactose ratio of 1.6:1 and, therefore, has a higher content of galactose than other polysaccharides in which galactose is substituted by other monosaccharides, such as locust bean gum (4:1) and carob gum (3.76:1). Our results indicate that the affinity of rHhMan for the guar gum substrate (Km= 2.074 mg ml-1) is similar to that reported for other mannanases for locust bean gum (Km=2.0 mg ml-1) and carob gum (Km= 2.2 mg ml-1) substrates. These data suggest that enzymatic activity is not affected by the presence of side chains such as galactose residues. In addition, rHhMan displays a higher affinity for guar gum than the mannanases of Aspergillus niger and Bacillus subtilis WY34 (Km= 7.7 mg ml-1 and 27.4 mg ml-1, respectively) [9,17].
Figure 5.
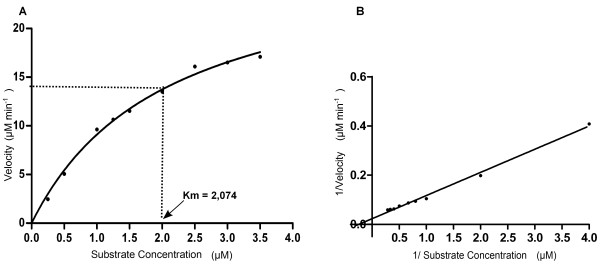
Michaelis- menten and lineweaver- burk plots of rHhMan using guar gum as substrate. A) Michaelis-Menten plot. B) Lineweaver-Burk plot. The measures were performed in optimum pH and temperature conditions.
The catalytic rate, kcat, for rHhMan is 50.87 s-1, which is greater than that of the mannanase of Bacillus licheniformis, which has a lower catalytic potential for glucomannan substrates (kcat= 21.00 s-1), locust bean gum (kcat= 31,20 s-1) and D-mannan (kcat= 18.2 s-1) [31].
Conclusions
This study is the first report on an insect endo-1,4-β-mannanase expressed in Pichia pastoris. This enzyme has a molecular weight of 35.5 kDa without its native signaling sequence, belongs to the glycosyl hydrolase GH5 family and contains the eight strictly conserved residues found in proteins within this family. The enzyme exhibits optimal activity under conditions similar to those of the digestive tract of the coffee berry borer. The rHhMan protein can hydrolyze different galactomannans and manno-oligosaccharides that can be used in the pharmaceutical and food industries because of their beneficial effects on human health. Coffee beans are the only food source for coffee berry borer insects, and the role that this enzyme performs in the utilization of galactomannan, the main polysaccharide found in coffee beans, makes these results a good starting point for designing rHhMan inhibitors. According with previous studies made for evaluate synthetic inhibitors of the enzyme, the results of this investigations may be used in the development of new molecules that could be used in pest control strategies that would help reduce the economic losses resulting from this insect.
Methods
Endo-1,4-β-mannanase (HhMan) protein sequence analysis
The amino acid sequence of HhMan available (GenBank Accession Number ADF22325.1) was analyzed by multiple sequence alignment comparisons with BLAST (Basic Local Alignment Search Tool) and CLUSTALW, respectively [32,33]. Secondary structure prediction was performed with the PSIPRED server [34], and queries on protein family and domain information were performed with the Pfam database [35].
Cloning, expression of the endo-1,4-β-mannanase (HhMan) gene and purification of the recombinant protein (rHhMan)
The cloning and heterologous expression of the P. pastoris system as well as the purification of the recombinant protein were performed under the methodology described previously [1]. The HhMan gene was amplified without its native signaling sequence with the primers Pp_Man_Fw (5′-CCGCTCGAGAAAAGAGTACCCGGATTCACGGTTTC-3′) and Pp_Man_Rv (5′-TGCTCTAGACCATTGAATATTGAACAGATTG -3′). Pp_Man_Fw includes a XhoI restriction site, and Pp_Man_Rv includes the site for XbaI (italics).
SDS-PAGE and zymography analysis
An aliquot of the purified recombinant protein and the commercial enzyme: Rohalase®GMP used like a positive control of mannanase activity were analyzed by SDS- PAGE (12%) as described by Laemmli [36]. The bands were visualized by Coomassie Blue R-250 staining. A low-range molecular weight standard (BIORAD, Hercules, California, USA) was used as a reference marker.
Another aliquot of protein and the positive control were used to assay enzymatic activity in a native PAGE, containing 1% (w/v) guar gum (Sigma, St Louis, Missouri, USA) as the substrate; activity was evaluated with 1, 1.5 and 2 μg of rHhMan, at 4°C. To evaluate enzyme activity, the gels were incubated in 0.1 M sodium citrate buffer (pH 5.5) for 20 min at 30°C. Enzymatic activity was visualized by Red Congo staining as described previously [1].
Enzymatic assays
The activity of the rHhMan enzyme was determined by the 3,5-dinitrosalicylic acid (DNS) method [37]. The substrate, guar gum (1%), was dissolved in 500 μl of 0.1 M sodium citrate buffer (pH 5.5) and incubated with 0.05 μg of rHhMan enzyme at 30°C for 30 min. Rohalase®GMP was used like a positive control of mannanase activity. The amount of reducing sugars released during the reaction was measured by mixing 5 μl of the enzymatic reaction and 5 μl of DNS solution, according to the methodology described by Padilla [1]. One unit of endo-1,4-β-mannanase activity is defined as the amount of enzyme required to release 1 μmol of reducing sugar per minute under the experimental conditions described with D-mannose as the standard substrate.
Effect of pH and temperature on enzymatic activity
The optimal pH for rHhMan activity was determined over a pH range of 3.5 to 8.0 under standard assay conditions with two buffering systems: 0.1 M sodium citrate (pH 3.0 – 6.0) and 0.1 M potassium phosphate (pH 6.0 – 8.0).
The effect of temperature on rHhMan activity was measured by incubating 0.05 μg of enzyme with 1% guar gum substrate in 0.1 M sodium citrate buffer (pH 5.5) at different temperatures between 10 and 60°C. The enzymatic activity was evaluated under standard assay conditions.
Hydrolytic properties
Fractions of 0.4 μg rHhMan were added to 0.5% guar gum and locust bean gum solutions (Sigma, St Louis, Missouri, USA) in 0.1 M sodium citrate buffer (pH 5.5). Reactions were incubated at 30°C for 24 hr. Aliquots were collected at 0, 15, and 30 min time points as well as at 12 and 24 hr, and they were then heated at 100°C for 5 min. Hydrolysis byproducts were separated on 60F 254 silica plates (Merck, Darmstadt, Germany) with chloroform: ethyl acetate: n-propanol: water (0.2:1:1.5:0.5 v/v) and detected by sulfuric acid aspersion in 5% ethanol, followed by heating at 100°C for 5 min.
To determine the mode of action of the enzyme, 0.4 μg of rHhMan was added to 25 mM mannobiose, mannotriose and mannotetraose mannooligosaccharide solutions in 0.1 M sodium citrate buffer (pH 5.5). The reactions were incubated at 30°C for 24 hr, and aliquots were collected at different time points and heated at 100°C for 5 min. The hydrolysis products were separated on silica plates as previously described. A mannooligosaccharide mix of mannose, mannobiose, mannotriose, mannotetraose and mannopentose (Megazyme, Co., Wicklow, Ireland) was used as a standard.
Kinetic parameters
To determine the kinetic parameters Km and Vmax, guar gum substrate was used in a concentration range of 2.5 to 35 mg ml-1 in 0.1 M sodium citrate buffer (pH 5.5). The reaction velocity was determined in triplicate for each substrate concentration. The data were fitted to a nonlinear regression model of the Michaelis-Menten equation with Prism software (GraphPad Software, San Diego, California, USA).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CAG and JVO: Performed the identification, amplification and cloning of HhMan as well as the subcloning in a Pichia vector and expression. CAG: purification and biochemical characterization of HhMan, performed the HhMan enzymatic assays and drafted the manuscript. PGS: Helped in enzymatic assays design. RAZ: Conceived the study and participated in its design, coordination and helped draft the manuscript. All authors read and approved the final version of this manuscript.
Contributor Information
Carolina Aguilera-Gálvez, Email: Carolina.Aguilera@unisarc.edu.co.
Juan J Vásquez-Ospina, Email: juan.vasquez_ospina@uni-wuerzburg.de.
Pablo Gutiérrez-Sanchez, Email: paguties@unal.edu.co.
Ricardo Acuña-Zornosa, Email: Ricardo.Acuna@cafedecolombia.com.
Acknowledgments
This work was financed by the Ministry of Agriculture and Rural Development of Colombia and the National Federation of Coffee Growers of Colombia (Contract # 074/2007). The authors thank Beatriz E. Padilla Hurtado and Jefferson Medina Olaya from the Plant Breeding Program of CENICAFE for their contributions in the development of this research, as well as Edison Osorio Durango and Carlos Muskus López for text revisions and valuable contributions in the production of the final manuscript.
References
- Padilla-Hurtado B, Florez-Ramos C, Aguilera-Gálvez C, Medina-Olaya J, Ramirez-Sanjuan A, Rubio-Gomez J, Acuna-Zornosa R. Cloning and expression of an endo-1,4-beta-xylanase from the coffee berry borer. Hypothenemus hampei. BMC Res Notes. 2012;5(1):23. doi: 10.1186/1756-0500-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera-Gálvez C, Gutierrez- Sanchez P, Acuña-Zornosa R. Modelado molecular e interacción enzima-ligando de potenciales inhibidores de la endo-1,4-β-mananasa de la broca del café Hypothenemus hampei. Boletin de Investigaciones Unisarc. 2012;10(1):17–23. [Google Scholar]
- Moreira L, Filho E. An overview of mannan structure and mannan-degrading enzyme systems. Appl Microbiol Biotechnol. 2008;79(2):165–178. doi: 10.1007/s00253-008-1423-4. [DOI] [PubMed] [Google Scholar]
- Bustillo Pardey AE. Una revisión sobre la broca del café, Hypothenemus hampei (Coleoptera: Curculionidae: Scolytinae), en Colombia. Revista Colombiana de Entomología. 2006;32:101–116. [Google Scholar]
- Redgwell R, Fischer M. Coffee carbohydrates. Braz J Plant Physiol. 2006;18(1):165–174. doi: 10.1590/S1677-04202006000100012. [DOI] [Google Scholar]
- Jaramillo J, Borgemeister C, Baker P. Coffee berry borer Hypothenemus hampei (Coleoptera: Curculionidae): searching for sustainable control strategies. Bull Entomol Res. 2006;96(03):223–233. doi: 10.1079/BER2006434. [DOI] [PubMed] [Google Scholar]
- Yan X-X, An X-M, Gui L-L, Liang D-C. From structure to function: insights into the catalytic substrate specificity and thermostability displayed by Bacillus subtilis mannanase BCman. J Mol Biol. 2008;379(3):535–544. doi: 10.1016/j.jmb.2008.03.068. [DOI] [PubMed] [Google Scholar]
- Yang P, Li Y, Wang Y, Meng K, Luo H, Yuan T, Bai Y, Zhan Z, Yao B. A novel β-mannanase with high specific activity from Bacillus circulans CGMCC1554: gene cloning, expression and enzymatic characterization. Appl Biochem Biotechnol. 2009;159(1):85–94. doi: 10.1007/s12010-008-8364-3. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Wei Y, Li D, Li L, Chai P, Kusakabe I. High-level production, purification and characterization of a thermostable β-mannanase from the newly isolated Bacillus subtilis WY34. Carbohydr Polym. 2006;66(1):88–96. doi: 10.1016/j.carbpol.2006.02.030. [DOI] [Google Scholar]
- Naganagouda K, Salimath PV, Mulimani VH. Purification and characterization of endo-beta-1,4 mannanase from Aspergillus niger gr for application in food processing industry. J Microbiol Biotechnol. 2009;19(10):1184–1190. [PubMed] [Google Scholar]
- Johnson KG, Ross NW. Enzymic properties of β-mannanase from Polyporus versicolor. Enzyme Microb Technol. 1990;12(12):960–964. doi: 10.1016/0141-0229(90)90117-9. [DOI] [Google Scholar]
- Malheiros Ferreira H, Ximenes Ferreira Filho E. Purification and characterization of a β-mannanase from Trichoderma harzianum strain T4. Carbohydr Polym. 2004;57(1):23–29. doi: 10.1016/j.carbpol.2004.02.010. [DOI] [Google Scholar]
- Marraccini P, Rogers WJ, Allard C, André M-L, Caillet V, Lacoste N, Lausanne F, Michaux S. Molecular and biochemical characterization of endo-ß-mannanases from germinating coffee (Coffea arabica) grains. Planta. 2001;213(2):296–308. doi: 10.1007/s004250100541. [DOI] [PubMed] [Google Scholar]
- Xu B, Hägglund P, Stålbrand H, Janson J-C. Endo-β-1,4-Mannanases from blue mussel, Mytilus edulis: purification, characterization, and mode of action. J Biotechnol. 2002;92(3):267–277. doi: 10.1016/S0168-1656(01)00367-4. [DOI] [PubMed] [Google Scholar]
- Ootsuka S, Saga N, Suzuki K-i, Inoue A, Ojima T. Isolation and cloning of an endo-β-1,4-mannanase from Pacific abalone Haliotis discus hannai. J Biotechnol. 2006;125(2):269–280. doi: 10.1016/j.jbiotec.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37(suppl 1):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Cuong D, Thi-Thu D, Berrin J-G, Haltrich D, Kim-Anh T, Sigoillot J-C, Yamabhai M. Cloning, expression in Pichia pastoris and characterization of a thermostable GH5 mannan endo-1,4-β-mannosidase from Aspergillus niger BK01. Microb Cell Fact. 2009;8(1):1–12. doi: 10.1186/1475-2859-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SD, Hefford MA, Johnson DA, Sung WL, Yaguchi M, Seligy VL. The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-β-1,4-glucanases is essential for enzymatic activity. Biochem Biophys Res Commun. 1990;169(3):1035–1039. doi: 10.1016/0006-291X(90)91998-8. [DOI] [PubMed] [Google Scholar]
- Guiseppi A, Cami B, Aymeric JL, Ball G, Creuzet N. Homology between endoglucanase Z of Erwinia chrysanthemi and endoglucanases of Bacillus subtilis and alkalophilic Bacillus. Mol Microbiol. 1988;2(1):159–164. doi: 10.1111/j.1365-2958.1988.tb00017.x. [DOI] [PubMed] [Google Scholar]
- Macarron R, van Beeumen J, Henrissat B, de la Mata I, Claeyssens M. Identification of an essential glutamate residue in the active site of endoglucanase III from Trichoderma reesei. FEBS Lett. 1993;316(2):137–140. doi: 10.1016/0014-5793(93)81202-B. [DOI] [PubMed] [Google Scholar]
- Gilbert HJ, Stålbrand H, Brumer H. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol. 2008;11(3):338–348. doi: 10.1016/j.pbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Schröder R, Wegrzyn T, Sharma N, Atkinson R. LeMAN4 endo-β-mannanase from ripe tomato fruit can act as a mannan transglycosylase or hydrolase. Planta. 2006;224(5):1091–1102. doi: 10.1007/s00425-006-0286-0. [DOI] [PubMed] [Google Scholar]
- Zahura UA, Rahman MM, Inoue A, Tanaka H, Ojima T. An endo-β-1,4-mannanase, AkMan, from the common sea hare Aplysia kurodai. Comp Biochem Physiol, Part B: Biochem Mol Biol. 2010;157(1):137–143. doi: 10.1016/j.cbpb.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Song JM, Nam K-W, Kang SG, Kim C-G, Kwon S-T, Lee Y-H. Molecular cloning and characterization of a novel cold-active β-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus. Comp Biochem Physiol, Part B: Biochem Mol Biol. 2008;151(1):32–40. doi: 10.1016/j.cbpb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Valencia A, Bustillo AE, Ossa GE, Chrispeels MJ. α-Amylases of the coffee berry borer (Hypothenemus hampei) and their inhibition by two plant amylase inhibitors. Insect Biochem Mol Biol. 2000;30(3):207–213. doi: 10.1016/S0965-1748(99)00115-0. [DOI] [PubMed] [Google Scholar]
- Silva EM, Valencia A, Grossi-de-Sá MF, Rocha TL, Freire É, de Paula JE, Espindola LS. Inhibitory action of Cerrado plants against mammalian and insect α-amylases. Pestic Biochem Phys. 2009;95(3):141–146. doi: 10.1016/j.pestbp.2009.08.003. [DOI] [Google Scholar]
- Puchart V, Vršanská M, Svoboda P, Pohl J, Ogel ZB, Biely P. Purification and characterization of two forms of endo-β-1,4-mannanase from a thermotolerant fungus, Aspergillus fumigatus IMI 385708 (formerly Thermomyces lanuginosus IMI 158749) Biochimica et Biophysica Acta (BBA) - General Subjects. 2004;1674(3):239–250. doi: 10.1016/j.bbagen.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Regalado C, García-Almendárez BE, Venegas-Barrera LM, Téllez-Jurado A, Rodríguez-Serrano G, Huerta-Ochoa S, Whitaker JR. Production, partial purification and properties of β-mannanases obtained by solid substrate fermentation of spent soluble coffee wastes and copra paste using Aspergillus oryzae and Aspergillus niger. J Sci Food Agric. 2000;80(9):1343–1350. doi: 10.1002/1097-0010(200007)80:9<1343::AID-JSFA651>3.0.CO;2-#. [DOI] [Google Scholar]
- Smith DL, Nagy TR, Wilson LS, Dong S, Barnes S, Allison DB. The effect of mannan oligosaccharide supplementation on body weight gain and fat accrual in C57Bl/6J mice. Obesity. 2010;18(5):995–999. doi: 10.1038/oby.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ademark P, Varga A, Medve J, Harjunpää V, Torbjörn D, Tjerneld F, Stålbrand H. Softwood hemicellulose-degrading enzymes from Aspergillus niger: Purification and properties of a β-mannanase. J Biotechnol. 1998;63(3):199–210. doi: 10.1016/S0168-1656(98)00086-8. [DOI] [PubMed] [Google Scholar]
- Songsiriritthigul C, Buranabanyat B, Haltrich D, Yamabhai M. Efficient recombinant expression and secretion of a thermostable GH26 mannan endo-1,4-beta-mannosidase from Bacillus licheniformis in Escherichia coli. Microb Cell Fact. 2010;9(1):20. doi: 10.1186/1475-2859-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DWA, Ward SM, Lobley AE, Nugent TCO, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38(suppl 2):W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R. et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34(suppl 1):D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Peter B. In: Methods in Enzymology. Colowick SP, Kaplan NO, editor. Vol. 1. New York: Academic Press; 1955. Amylases, α and β; pp. 149–158. [Google Scholar]


