Abstract
Heat shock proteins 90 (Hsp90) have an essential role in sarcomere formation and differentiation in skeletal muscle and also act as molecular chaperones during protein folding impacting a wide range of physiological processes. We characterised and provided a phylogenetically consistent nomenclature for the complete repertoire of six Hsp90 paralogues present in duplicated salmonid fish genomes (Hsp90α1a, Hsp90α1b, Hsp90α2a, Hsp90α2b, Hsp90ß1a and Hsp90ß1b). The expression of paralogues in fast skeletal muscle was investigated using in vivo fasting-feeding experiments and primary myogenic cultures. Fasted juvenile Atlantic salmon (Salmo salar) showed a transient 2 to 8-fold increase in the expression of all 4 Hsp90α paralogues within 24h of satiation feeding. Hsp90α1a and hsp90α1b also showed a pronounced secondary increase in expression after 10 days, concomitant with muscle differentiation and the expression of myogenin and sarcomeric proteins (mlc2, myhc). Hsp90ß1b was constitutively expressed whereas Hsp90ß1a expression was downregulated 10-fold between fasted and fed individuals. Hsp90α1a and Hsp90α1b were upregulated 10 to 15-fold concomitant with myotube formation and muscle differentiation in vitro whereas other Hsp90 paralogues showed no change in expression. In cells starved of amino acid (AA) and serum for 72h the addition of AA, but not insulin-like growth factor 1, increased phosphorylation of mTor and expression of all 4 hsp90α paralogues and associated co-chaperones including hsp30, tbcb, pdia4, pdia6, stga and fk504bp1, indicating a general activation of the protein folding response. In contrast, Hsp90ß1a expression in vitro was unresponsive to AA treatment indicating that some other as yet uncharacterised signal(s) regulate its expression in response to altered nutritional state.
Introduction
The cytosolic Hsp90s (Hsp90AA1 and Hsp90AB1) are some of the most abundant proteins in many cell types, representing around 2% of the soluble proteins in non-stressed cells. Hsp90α (Hsp90AA1) and Hsp90ß (Hsp90AB1) are considered to have originated by duplication in the vertebrate common ancestor around 500 million years (MY) ago [1,2]. Almost 200 different proteins are thought to require Hsp90s to achieve their final configuration including kinases [3], growth factor receptors [4], transcription factors [5] and a remarkable number of oncogenic proteins [6]. Hsp90s are therefore key regulators in cell physiology and are involved in diverse processes including signal transduction, protein folding, morphology and differentiation. Hsp90s function and client specificity is regulated by interaction with a group of non-client binding partners, known as co-chaperones, that includes Cdc37, p23, Fk506bp1, Chip, Sgta, Aha1 or Unc45 [7,8].
Three main functional domains have been described for Hsp90: the N-terminal domain, the flexible middle region and the C-terminal domain [9,10]. The N-terminal region contains the ATP binding domain with ATPase activity. The hydrolysis of ATP initiates a cycle that modifies Hsp90 configuration and acts as a molecular motor for the folding process (for a complete description see 9). Hsp90 chaperone function as a homodimer, with the C-terminal domain and middle region being responsible for dimerization [9,10].
Several studies have highlighted the importance of Hsp90 for skeletal muscle embryogenesis and myofibrillogenesis involving direct interaction with the myoblast determination factor MyoD [3,11]. Inhibition of Hsp90 function in C2C12 cells induced a significant decrease in myogenin and Akt expression, and caused disruption of the MyoD-Cdc37-Hsp90 complex, inhibiting myotube formation [12]. It is also known that Hsp90 regulates myosin heavy chain motor domain folding, one of the main components of the sarcomere, by interaction with the co-chaperone Unc45 [13,14].
Teleosts have undergone whole genome duplication (WGD) relative to the ancestor of tetrapods (Jaillon et al. 2004). The zebrafish (Danio rerio) genome contains Hsp90α1 and Hsp90α2 genes. Functional studies demonstrated that inhibition of hsp90α1 expression strongly perturbed embryonic muscle development, whilst blocking hsp90α2 had no effect, suggesting sub-functionalization between paralogues [15]. Several studies have described increased expression of hsp90 in response to temperature stress, although the issue of gene paralogues is generally ignored [16–18]. More recently, a dramatic and rapid increase in hsp90α expression has been reported in fish fed to satiation following a period of dietary restriction though the signals involved have not been investigated [19–22].
Atlantic salmon and other salmonid fish underwent a second lineage-specific whole genome duplication event around 88 million years ago followed by gene loss [23,24]. Potentially two copies of each zebrafish gene may be present in the Atlantic salmon genome [23]. The aim of the present study was to characterise the complete repertoire of Hsp90α and Hsp90β paralogues in salmonid genomes and determine their expression in a fasting-refeeding model using juvenile Atlantic salmon (Salmo salar L.). To provide further insight into the transcriptional regulation of this gene family we used primary myogenic cell cultures and investigated the effects of amino acid and IGF1 treatments on Akt and mTor phosphorylation and the expression of Hsp90 paralogues and co-chaperones.
Methods
Ethics statement
This study was conducted on Atlantic salmon (Salmo salar L.) of aquaculture origin. Fish were humanely killed by a blow to the head following Schedule 1 of the Animals (Scientific Procedures) Act 1986 (Home Office Code of Practice. HMSO: London January 1997), and all husbandry procedures and experimental protocols were approved by University of St Andrews Animal Ethics and Welfare Committee.
Hsp90 sequencing
Previously sequenced Atlantic salmon Hsp90α and Hsp90β (NM_001173702 and NM_01146473 respectively) were blasted against the Atlantic salmon genome draft deposited in the NCBI genome database (ASM23337v1). Four sequences similar to Hsp90α and two to Hsp90β were found and read trace quality was checked using the Trace Archive Nucleotide BLAST from NCBI. All sequences were experimentally confirmed as follows: primers against the start and stop regions (Table S1) were used to amplify Hsp90 paralogues from a cDNA mix of different Atlantic salmon tissues (fast muscle, liver, slow muscle, brain, skin, gills, heart, liver, fat, kidney and myoblast). Specific bands in the right size range (around 2.2kb) were cloned as a whole in TOPO® plasmid (Invitrogen), transformed by thermal shock into E. Coli Top10 (Invitrogen) and grown in LB-Agar plates with 75ng/ml ampicillin. 10-12 clones per product were sent for SANGER sequencing to the Dundee Sequencing Services. Hsp90 paralogues sequences were submitted to the GenBank database: KC150878, KC150879, KC150880, KC150881, KC150882 and KC150883 (Table S1). Hsp90 paralogues sequences were conceptually translated to amino acid using Virtual ribosome [25] and aligned with MAFFT version 6 [26] (Figure S1).
Rainbow trout transcriptome
Ten Rainbow trout specific 454-Titanium libraries from different sources were retrieved from the Sequence Reads Archive database (SRA): SRX041526-31 (skeletal fast muscle), SRX085156 (stressed Rainbow trout), DRX000493 (adipose tissue), SRX007396 (non-stressed Rainbow trout) and SRX041532-7 (larvae trunks). A total of 6,154,973 reads were de novo assembled by Newbler 2.5 assembly software (Roche, 454 Life Sciences). Assemblies were compiled using a Debain Linux system, IBM x3755 8877, with 8 CPU cores (4 x dual-core AMD Opteron), 64-bit, 2.8 GHz processor with 128 Gb of RAM maintained by the University of St Andrews. Hsp90 sequences were retrieved from the rainbow trout transcriptome by comparison with the Atlantic salmon paralogues using BioEdit free software [27].
Phylogenetic and synteny analysis
Teleost Hsp90α (Hsp90AA1) and Hsp90β (Hsp90AB1) sequences were retrieved for Oryzias latipes, Danio rerio, Oreochromis niloticus , Gadus moruha , Gasterosteus aculeatus , Takifugu rubripes and Tetraodon nigroviridis from the Ensembl database [28]. A total of 37 coding sequences, including Atlantic salmon and rainbow trout, were aligned using GUIDENCE webserver [29]. Only aligned regions with a GUIDENCE quality score over 0.93 were used for further analysis. Maximum Likelihood (ML) and Neighbour-Joining (NJ) trees with 500 bootstraps replications were constructed using MEGA5 [30] with Jone-Taylor-Thorton (JTT) and Gamma distribution as the evolutionary model. Trees generated were visualized and edited using Fig tree v 1.3.1.
Synteny surrounding Hsp90 genes was manually inferred by study of Ensembl genome assemblies of Danio rerio, Gasterosteus aculeatus , Oryzias latipes, Oreochromis niloticus , Tetraodont nigroviridis , Takifugu rubripes , Gadus morhua, Xiphophorus maculatus , Homo sapiens, Xenopus tropicalis and Meleagris gallopavo [28].
Atlantic salmon fasting refeeding experiment
A total of 150 Atlantic salmon juveniles (59.8 ± 7.9 g; mean ± SD) were obtained from Landcatch Natural Selection Ltd and maintained at ~10.6oC in replicate tanks at the Scottish Oceans Institute. Fish were fed a maintenance diet for 21 days and then fasted for 7 days and fed to satiation with commercial feed (EWOS) supplemented by bloodworms. Fast skeletal muscle was dissected from dorsal epaxial at 0.6 standard length at day 0 (after 7 days of fasting) and 1, 3, 5, 8, 15 and 21 days of satiation feeding.
Atlantic salmon primary myogenic cell culture
Myoblast cell culture was performed as previously described [31]. In brief, myogenic cells were extracted from fast skeletal muscle and maintained in 10% (v/v) Foetal Bovine Serum (SIGMA) DMEM (SIGMA) medium containing antibiotics (SIGMA) for 16 days. Cells RNA was extracted after 2, 4, 6, 8, 10, 12, 14 and 16 days after plating using RNeasy mini kit (QIAGEN). For immunofluorescence, cells were fixed in 4% (m/v) paraformaldehyde in PBS (SIGMA) at 2, 6, 10 and 14d following initiation of the culture.
Amino acid starvation and treatments
Myogenic cells at day 8 of culture were incubated for 72 hours with free amino acid media containing: Earle’s balance salt solution x1 (SIGMA), 20mM HEPES (SIGMA), 9mM NaCO3 (SIGMA), Vitamin mix x1 (SIGMA), antibiotics-antimycotic mix x1 (SIGMA) and 4g/L D-glucose (SIGMA), prepared using sterilised MilliQ water filtered at 0.22µm. Starved cells were then growth for 24 h in either free-amino acid media (-AA), media supplemented with recombinant salmon IGF1 (100ng/ml) (GroPep, Australia), medium with complete amino acid (DMEM) or a combination of both. RNA was collected 12 and 24h following treatments using RNeasy mini kit (QIAGEN). For immunofluorescence staining, cells were treated for 48 h and then fixed in 4% (m/v) paraformaldehyde in PBS (SIGMA). Control cells were maintained in DMEM (SIGMA) serum free media.
Immunofluorescence
Cells were washed in PBS (SIGMA), fixed with 4% (m/v) paraformaldehyde in PBS (SIGMA) for 20 min and made permeable with 0.5% (v/v) X-100 triton (SIGMA)-PBS for 5 min. After PBS washes, cells were blocked for 1h in 5% (v/v) normal goat serum (SIGMA), 1.5% (m/v) Bovine serum albumin (SIGMA), 0.1% (v/v) X-100 triton (SIGMA)-PBS. All antibodies were incubated overnight at 4oC in 0.1% (v/v) X-100 triton, 1.5% (m/v) BSA-PBS. Phalloidin-Texas Green antibody (Invitrogen) was diluted 1:1000 (v/v) and anti-desmin (SIGMA) was diluted 1:20 (v/v). For desmin detection cells where incubated with anti-rabbit Alexa Fluor 450 antibody (Invitrogen) for 4h at room temperature. When phalloidin was used, cells were counter-stained for nuclei with DAPI 1:10000 (v/v) (Invitrogen). When desmin was detected cells nuclei were counter-stained with SYTOX-green 1:1000 (v/v). Cells were viewed using a Leica TCS SP2 confocal microscope (Model TCS SP2, Leica Microsystems, Wetzlar, Germany).
Western Blot Analysis
A total of 20 µg of protein was added to 3 µl of 5X protein loading buffer and 1 µl of 20X reducing agent (Fermentas, Vilnius, Lithuania) and RIPA buffer to 15 µl. Samples were heated for 10 min at 95oC and loaded on to a NuPAGE® Novex 4-12% (m/v) poly-acrylamide gel (Invitrogen, Carlsbad, CA, USA). A pre-stained protein ladder ranging from 250 to 10 kDa (BioRad, Hemel, Hetfordshire, UK) was included in all gels to determine the molecular mass of bands. Samples were resolved by electrophoresis for 2h at 100V and room temperature (RT). Proteins were then transferred to a PDVF Immobilon-P Transfer Membrane (Millipore, Billerica, MA, USA) at 25V for 2h at RT. Membranes were washed twice with PBT (0.1% (v/v) Tween 20, SIGMA, in PBS) and blocked for non-specific binding with 5% (m/v) non-fat milk (AppliChem, Darmstadt, Germany) solution in PBT for 1h at RT. After washing in PBT three times for 10 min membranes were incubated at 4oC overnight with the following primary antibodies: phospho-Akt (Ser473) (Cell Signalling, #4060, Danvers, MA, USA), Akt (Cell Signalling, #2966), phospho-mTOR (Cell Signalling, #2971) and mTOR (Cell Signal, #2972). Akt and mTOR antibodies were diluted 1:1000 (v/v) and actin in 1:20000 (v/v) in PBT-0.01% (m/v) NaN3. Membranes were subsequently incubated with secondary anti-rabbit antibody linked to a horseradish peroxidase (HRP) (SIGMA) diluted 1:40000 (v/v) in 5% (m/v) non-fat milk PBT solution for 1h at RT. After washing in PBT three times for 15 min, membranes were incubated for 1 min with ECL Western Blot detection reagents (GE HealthCare, Amersham, Buckinghamshire, UK). Membranes were exposed to Hyperfilm ECL (GE HealthCare). The resulting films were scanned and band density evaluated with TotalLab Quant software (TotalLab, Newcastle, UK). Phospho-protein levels were normalized with respect to the total protein. A common pooled sample was loaded onto all gels for normalization.
RNA extraction and cDNA synthesis
Tissue RNA was obtained using TRIsure (Bioline, UK) phenol-chloroform extraction following manufacturer’s recommendations. Cellular RNA was extracted from individual wells using RNeasy plus minikit (Qiagen) following the manufacturer’s recommendations. In all cases RNA concentration, 260/280 and 260/230 ratios were determined using a Nanodrop 1000 Spectophotometer (Thermo, Fisher Scientific, Waltham, MA, USA) and their integrity was confirmed running a 1% (m/v) agarose gel. Only non-degraded RNA samples with 260/280 and 260/230 ratios over 2 were used for cDNA synthesis using a QuantiTec reverse transcription kit (Qiagen). After residual genomic DNA was removed using the DNA wipeout step provided in the kit, 500ng of total RNA were reversed transcribed for 30 minutes at 42oC following manufacturer’s recommendations. To detect any contamination of genomic DNA in the samples a reaction with no RT was added to the synthesis reaction (RT-).
Quantitative Real Time PCR (qPCR)
qPCR was compliant with the Minimum Information for Publication of Quantitative Real-Time PCR experiments MIQE guidelines [32]. Each qPCR mixture contained 6µl of cDNA 1:80 (v/v) diluted, 7.5µl of 2x Brilliant II (Stratagene) and 1.5µl of 500nM primer mix with the following protocol: 1 cycle 5min 95oC, 40 cycles 95oC 30sec 60oC 30sec 72oc 30sec and a final cycle of 7min 72oC performed in a Stratagene MX3005P real time PCR machine (Stratagene, La Jolla, CA, USA). A gradient was run from 60 to 95oC to confirm the presence of single amplification product following dissociation analysis of the PCR products. Products were sequenced to confirm their identity. PCR efficiencies were calculated from dilution series of pooled cDNA samples.
Primer design
Some of the primers used in the present study have been described previously elsewhere [31,33,34]. Primers were designed using Primer3 [35] with a melting temperature of 60oC and, where possible, in region that spanned an exon-exon junction. Primers possible hairpins or non-desirable primer-dimmers were investigated using NetPrimer [36]. Primers used in the present study are summarized in Table S1.
Data Analysis
GeNorm [37] was used to choose the best reference genes and to analyse the data from each experiment in the study. For the cell culture gene expression analysis ribosomal protein 19 (rps19) and Hypoxanthine phosphoribosyl transferase 1 (hprt1) were reported as the most stable genes. Elongation factor alpha-1 (ef1α) and ribosomal protein L13 (rspl13) were identified as the most stable for amino acid deprivation experiment and rspl13 and rps29 for the Atlantic salmon fasting re-feeding experiment. GeNorm normalization was performed using the geometric average of the housekeeping genes, and values are shown as arbitrary units.
For data analysis, when data conformed to parametric assumptions an ANOVA using Bonferroni post hoc test was used to detect significant differences. When parametric assumptions were not upheld a Kruskal-Wallis-H test was conducted. Statistical analysis of the data was performed using the SPSS-Statistics (IBM) package.
Results
Atlantic salmon possess 6 highly conserved paralogues
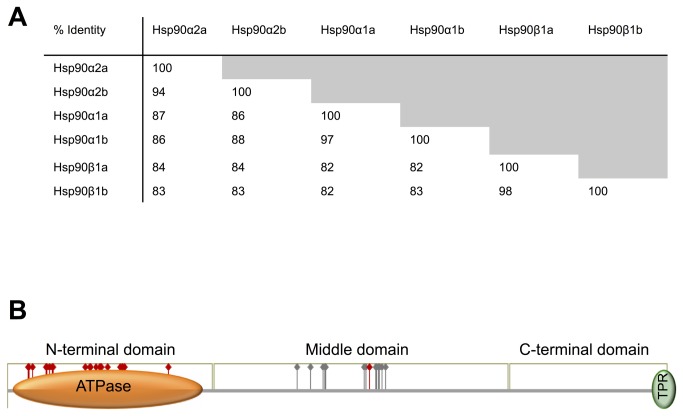
Previous to this study only two cytosolic Hsp90 were described for Atlantic salmon (Salmo salar): Hsp90α (NM_001173702) and Hsp90β (NM_001123532). A total of 6 Hsp90 paralogues, 4 Hsp90α (KC150878, KC150879, KC150880, KC150881) and 2 Hsp90β (KC150882, KC150883) were identified in the Atlantic salmon genome draft [38]. A high degree of amino acid identity was found between groups of paralogues, Hsp90α paralogues shared 87 to 97% identity (Figure 1A) and Hsp90β paralogues shared 98% identity. The percentage identity was reduced to 82-84% when Hsp90ß paralogues were compared to any of the Hsp90α paralogues (Figure 1A, Figure S1).
Figure 1. Atlantic salmon Heat shock protein 90 (Hsp90) paralogues.
A. Identity (%) between amino acid sequences of the Hsp90 paralogues identified for Atlantic salmon B. Functional domain organization of the different paralogues. Orange oval represents the extension of the N-terminal ATPase domain. Grey oval identifies the position of the tetratricopeptide motif MEEVD (TRP domain). Red flags highlight amino acids involved in ATP binding conserved in all paralogues. Grey flags highlight residues involved in protein binding conserved in all paralogues. Domains and residues identification are based on previous studies [8,9,47,48].
Characteristic domains of the Hsp90 family were conserved in all Atlantic salmon paralogues (Figure 1B). The N-terminal ATPase domain was localized between amino acid 14 to 300 (Figure 1B, orange domain) and all the residues and motifs previously identified in the literature as key components for the ATP binding and hydrolysis appeared conserved (Figure 1B red flags, Figure S1 red squares). The middle region (between amino acid 300 to 500) was the most variable between paralogues (Figure S1), but residues considered important for binding client proteins were also retained (Figure 1B grey flags, Figure S1 black squares). The MEEVD motif, responsible of the interaction with the tetratricopeptide-repeat domain (TPR)-containing co-chaperones, was also present (Figure 1 green domain, Figure S1 green square).
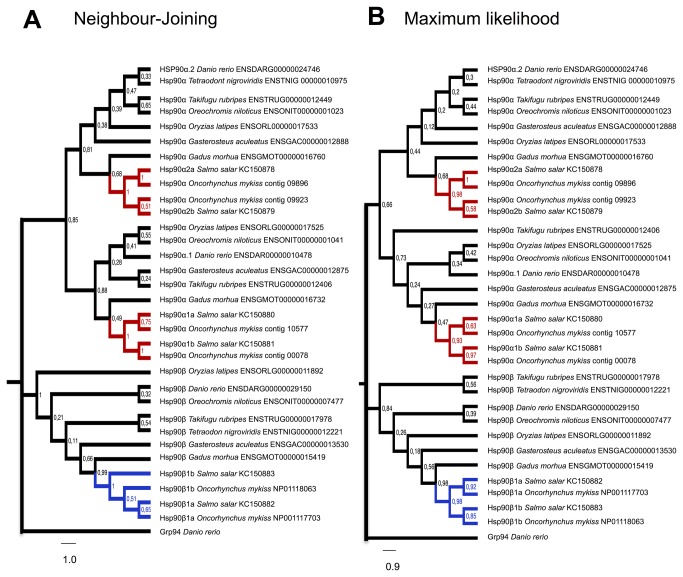
To confirm the Salmonidae specificity of the paralogues found in Atlantic salmon an extensive rainbow trout ( Oncorhychus mykiss ) transcriptome was constructed on the bases that both species would share the same number of paralogues. With this objective over 6 millions reads were used to generate a total of 279,336 isotigs (transcriptome details are described in Table S2). Six rainbow trout isotigs with similarity to the Atlantic salmon Hsp90 paralogues were found. Rainbow trout Hsp90 identity and Atlantic salmon relationship was further validated by phylogenetic analysis. Teleost Hsp90α and β orthologues were retrieved from: Atlantic cod (Gadus morhua), stickleback ( Gasterosteus aculeatus ), zebrafish (Danio rerio), tilapia ( Oreochromis niloticus ), medaka (Oryzias latipes), fugu ( Takifugu rubripes ) and green pufferfish (Tetraodon nigroviridis) genome sequences.
A total 32 Hsp90 sequences were used producing 674 aligned residues suitable for phylogenetic analysis. NJ and ML trees correctly grouped all sequences into two main clades for Hsp90α and β, confirming the nature of the trout and salmon sequences (Figure 2A and B). Hsp90α sequences were separated in two monophyletic branches with one sequence from each teleost species and two from rainbow trout and Atlantic salmon. Also, the Atlantic salmon Hsp90α paralogues were pairwise branched with individual rainbow trout orthologues (Figure 2, red branches). Based on this analysis, Atlantic salmon Hsp90α paralogues were named according to the zebrafish orthologue branching in the same monophyletic group: Hsp90α1a, Hsp90α1b, Hsp90α2a and Hsp90α2b. Tree topology for Hsp90β orthologues was not well resolved with different topologies between NJ and ML trees (Figure 2, blue branches). Beta paralogues were named as Hsp90ß1a and Hsp90ß1b as have been described for rainbow trout.
Figure 2. Teleost Hsp90 phylogenetic analysis.
Neighbour-Joining (A) and Maximum Likelihood (B) trees showing phylogenetic relationships between cytosolic Hsp90s from different teleost species. Branches containing Atlantic salmon Hsp90α paralogues are highlighted in red and those with Hsp90β in blue. Phylogenetic trees were constructed from a highly confidence alignment of 37 sequences. Trees were constructed using JJT+G as best fitted evolutionary model. Resulting trees were constructed using MEGA5 with 500 bootstrap repetitions. Bootstrap-posterior values are indicated on the node of each branch.
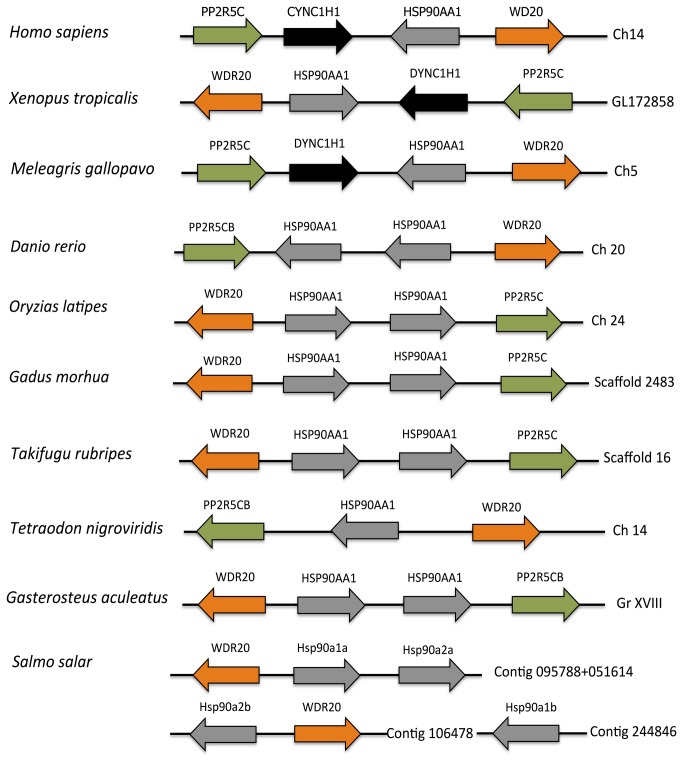
Synteny analysis showed that, with exception of Tetraodon , all basal teleost genomes had two copies of hsp90α and one copy of hsp90β (Figure 3). Also, in all teleost genomes analysed the Hsp90α paralogues had WD repeat-containing protein 20 (wdr20) and serine/threonine-protein phosphatase 2A 56kDa regulatory subunit gamma (pp2r5c) as neighbours. Despite the fragmented Atlantic salmon genome it was possible to perform partial synteny analysis by fusing two of the genome-contigs where Hsp90 paralogues were identified. The analysis of these “super-contigs” revealed that hsp90α1a and hsp90α2a are located next to one another with a copy of wdr20 next to hsp90α1a (Figure 3). We also found a second copy of wdr20 next to the hsp90α2b paralogue (Figure 3). After WGD events, neighbouring genes at the duplicated chromosomes tend to maintain their relative positions or orders during evolutionary time. The finding that two copies of wdr20 are found next to two hsp90α genes suggests the likely WGD origin of the second set of salmon hsp90α paralogues.
Figure 3. Hsp90 synteny analysis.
Hsp90AA1 genetic environment in different vertebrate genomes: Homo sapiens, Meleagris gallopavo , Xenopus tropicalis , Danio rerio, Gasterosteus aculeatus , Takifugu rubripes , Gadus morhua, Tetraodont nigroviridis and Salmo salar. Arrow direction refers the orientation of the reading frame. Common genes between genome sections are label with the same colour. Abbreviations are as follow: WD repeat-containing protein 20 (WD20), dynein cytoplasmatic 1 heavy chain 1 (CYNC1H1), serine/threonine-protein phosphatase 2A 56kDa regulatory subunit gamma (PP2R5C), chromosome (Ch), group (Gr).
Hsp90 paralogues expression is regulated by food intake
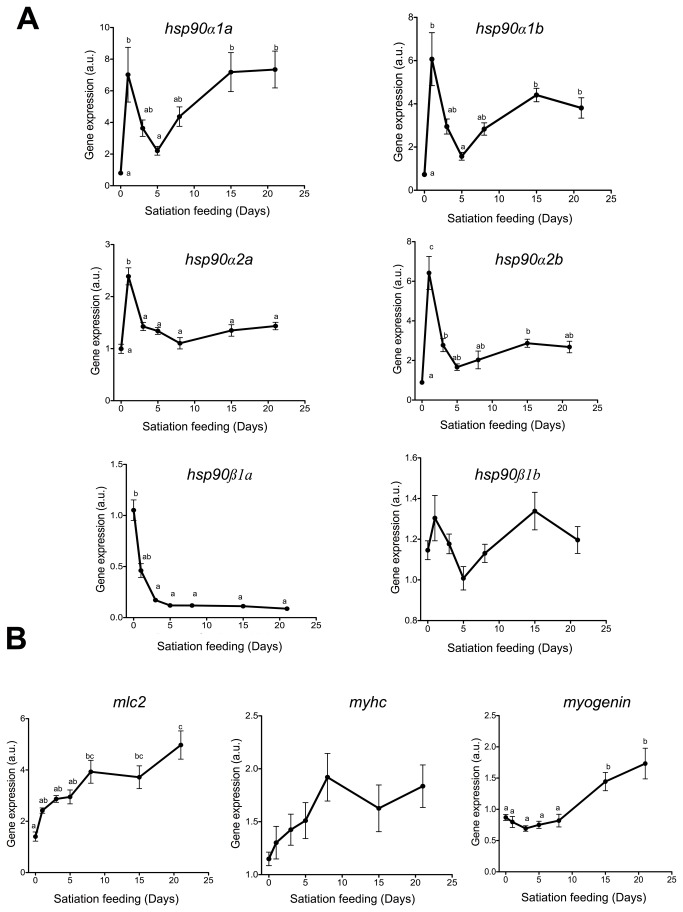
We investigated the expression of hsp90 paralogues after manipulating nutritional status in juvenile Atlantic salmon involving a transition from fasting (7d) to satiation feeding (up to 21d). The expression of all hsp90α paralogues showed a transient 2 to 8-fold increase in expression peaking 24 h after the initiation of feeding (Figure 4A; P < 0.01). This was followed by a recovery to pre-refeeding levels after 5d. Hsp90α2a and 2b expression remained stable over the remainder of the experiment. In contrast, both hsp90α1 paralogues transcripts increased after 8d and reached a new peak of expression after 15d (4-fold for hsp90α1b (P<0.01) and 8-fold for hsp90α1a (P<0.01)). This second delayed increase in hsp90α1a and hsp90α1b expression may reflect the activation of myogenesis and fibre growth induced by refeeding, as previously suggested [19–22]. In support of this idea, and as an indicator of skeletal muscle response, the expression of sarcomeric proteins (myosin light chain 2 (mlc2) and myosin heavy chain (myhc) and the differentiation factor myogenin increased over a similar timescale (Figure 4B).
Figure 4. Gene relative expression in Atlantic salmon fast skeletal muscle in response to changes in nutritional status.
Effect of fasting for 7d and subsequent refeeding to satiation for 1, 3, 5, 8, 15 and 21 days on gene expression in fast skeletal muscle. Values represent mean ± SE (N=6). Different letters indicate significant differences between means (P<0.05) A. Hsp90α1a, Hsp90α1b, Hsp90α2a, Hsp90α2b, Hsp90ß1a and Hsp90ß1b B. Myosin light chain 2 (mlc2), myosin heavy chain (myhc) and myogenin.
Hsp90 paralogues are differently expressed during maturation of primary myogenic cell cultures
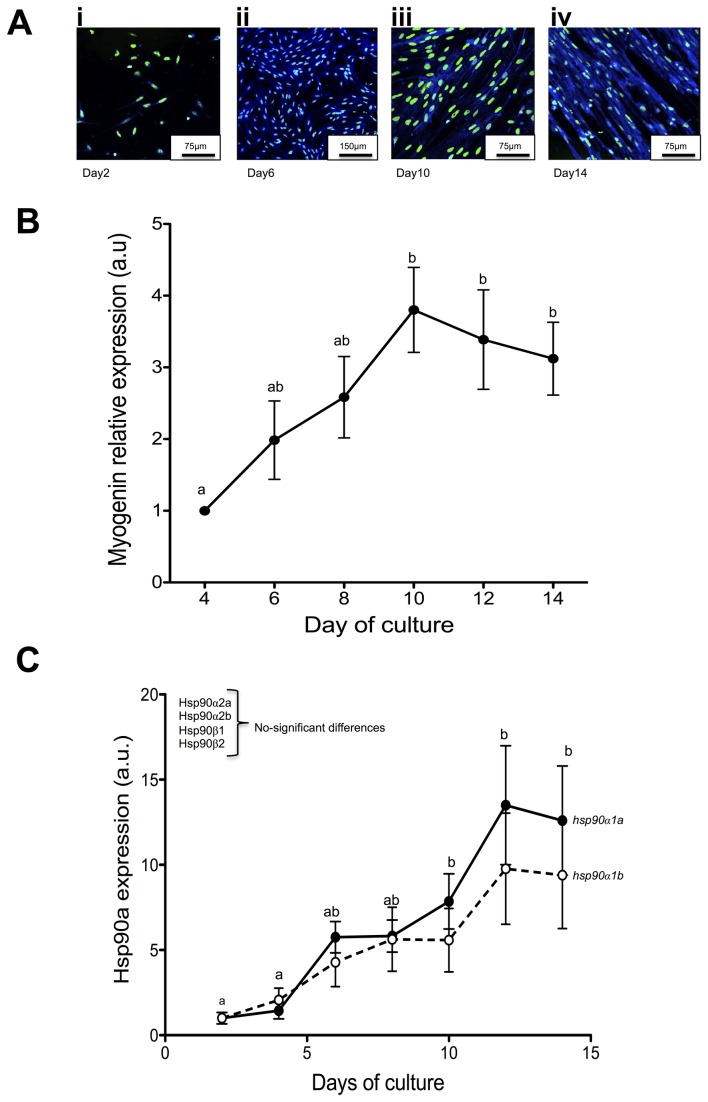
We then used primary myogenic cell cultures from fast muscle to test the hypothesis that hsp90α1, but not hsp90α2 paralogues, showed increased expression concomitant with myotube formation. Shortly after myoblasts were plated (Figure 5Ai), cells started proliferating until day 5-6 (Figure 5Aii), with the first signs of cell fusion observed between days 8 to 10 (Figure 5Aiii) and fully developed myotubes at day 14 (Figure 5Aiv). Myoblast differentiation was monitored by measuring the expression of the Myogenic Regulatory Factor myogenin which significantly increased it abundance after 10 days of culture (Figure 5B). Hsp90α1a and hsp90α1b expression increased 10 to- 15 fold (P<0.05) from day 10 concomitant with myotube formation and muscle differentiation (Figure 5C) whereas the expression of other hsp90 paralogues was unchanged (data not show). It is interesting to note that in both in vivo and in vitro the Ct values of hsp90α1a/b were always much lower than hsp90α2a/b (data not show), suggesting higher expression of these paralogues.
Figure 5. Hsp90 expression in Atlantic salmon myoblast cell culture.
A. Atlantic salmon fast skeletal muscle myoblasts at days 4 (i), 6 (ii), 10 (iii) and 14 (iv) of culture. Cells nuclei were stained with SYTOX green and myogenic cells were identified by staining for desmin (blue). B. myogenin expression in myoblast cell culture at days 4, 6, 8, 10, 12 and 14. C. hsp90α1a and hsp90α1b expression in myoblast cell culture at days 2, 4, 6, 10, 12 and 14. To facilitate interpretation of the data, values at day 2 (and 4 for myogenin) were transformed so the mean value = 1. Values represent mean ± SE from 5 different cultures. Different letters indicate significant differences between means (P<0.05).
Hsp90α paralogues are regulated by amino acid
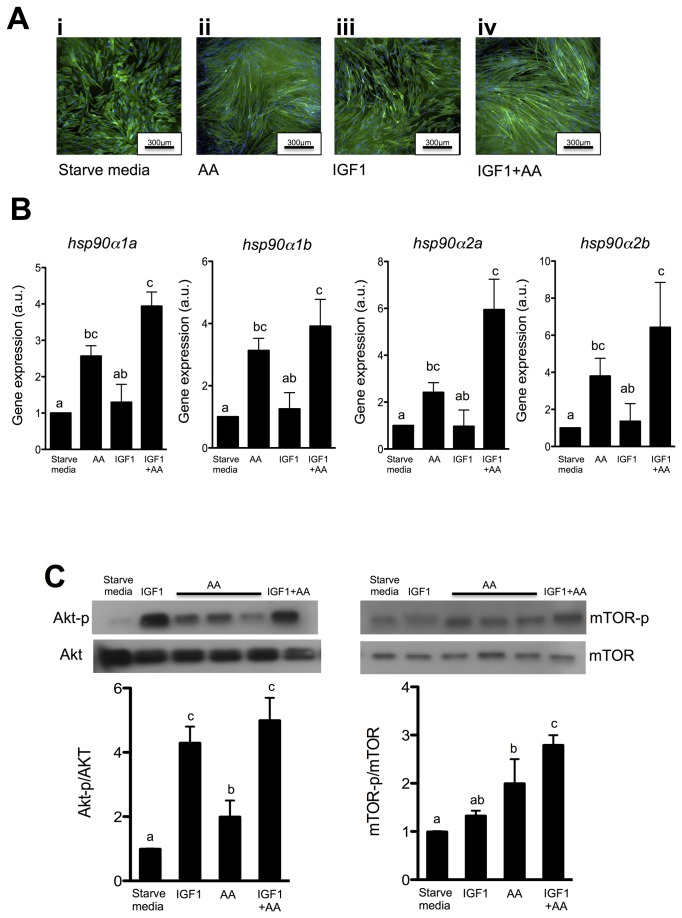
To further explore the transcriptional regulation of hsp90 paralogues by nutrition their expression was analysed in 8 d cultures following treatment with amino acids and IGF1. Cultures were transferred to serum/amino acid free medium (“starved media”) for 72h and then incubated with medium containing either amino acid (AA), IGF1 or a combination of both (IGF1+AA). Akt and mTOR phosphorylation was also measured to provide evidence for the activation of the PI3K and mTor pathways respectively (Figure 6C). Myoblasts incubated with amino acid/serum-free medium stopped their progression through myogenesis and failed to fuse to form myotubes (Figure 6Ai). Myotubes formed again after 48h in the presence of amino acid alone or in combination with IGF1 (Figure 6Aii and Aiv). IGF1 alone failed to recover myotube formation (Figure 6Aiii). Phosphorylation of Akt increased 4-5 fold in the presence of IGF1 indicating activation of PI3K signalling. In contrast, phosphorylation of mTor was unchanged by IGF1 treatment, but increased 2-3 fold when amino acid were added alone or in combination with IGF1, consistent with the activation of mTor signalling (Figure 6C).
Figure 6. Hsp90 paralogues expression in Atlantic salmon myoblasts incubated with amino acid and IGF1 after 72h of amino acid deprivation.
A. Actin was stained with green phalloidin and nuclei were counterstained with DAPI (blue) in myoblast incubated with free amino acid medium to simulated starvation (Starve media) (i), with amino acid (AA) (ii), insulin-like growth factor 1 (IGF1) (iii) or amino acid combined with IGF1 (IGF1+AA) (iv). B. Expression profiles of hsp90α1a, hsp90α1b, hsp90α2a and hsp90α2b in 10 day myogenic cell cultures starved for 72h (Starve media) and then treated with amino acid (AA), insulin-like growth factor 1 (IGF1) or a combination of both (AA+IGF1) for 24h. Values represent mean ± SE from 5 independent cell cultures. For ease of interpretation, values of starved cells (Starve media) were transformed so the mean value = 1. C. Akt and mTOR phosphorylation in response to Starve media, AA, IGF1 and AA+IGF1 treatments. Relative units were obtained by normalizing the phosphorylated-protein to the total protein.
Values represent mean ± SE from 5 different cultures. Different letters indicate significant differences between means (P<0.05).
Significant changes in gene expression were observed 24h after treatments. When amino acid were added to the medium all hsp90α paralogues showed an almost 3-fold increase in expression (P<0.05). Hsp90α expression was unchanged in myoblasts treated with IGF1 alone. However, when amino acids were combined with IGF1, all 4 hsp90α paralogues significantly increased their expression (Figure 6B; P < 0.05 for hsp90α2a/b; P<0.01 for hsp90α1a/b) to levels greater than observed with amino acids alone. Hsp90β expression was not affected by any of the treatments (data not show).
Our working hypothesis is that amino acid influx to the muscle stimulates the mTor pathway leading to increased peptide translation and concomitant activation of the protein folding response pathway. To investigate this further the expression of other chaperones, co-chaperones and protein editing genes including hsp70, hsp30, tbcb, sgta, fkbp4, pdia4 and pdia6 were analysed under the same in vitro conditions (Table 1). Hsp30, sgta, tbcb and fkbp4 increased their expression 2 to 3-fold (P<0.05) in response to amino acid combined with IGF1 after 24h (Table 1). Expression of the disulfide-isomerases pdia4 and pdia6 showed a significant response after 12h to amino acid alone (2-4 fold; P<0.05) or when were combined with IGF1 (3-5 fold; P<0.05) (Table 1). In contrast, there were no changes in rab9a expression and, although hsp70 levels were significantly altered, expression did not follow a consistent pattern (Table 1).
Table 1. Expression profiles of folding and protein-editing genes in Atlantic salmon myoblasts incubated with amino acid and insulin-like growth factor 1 (IGF1) after 72h of amino acid deprivation.
| Time |
12h
|
24h
|
||||||
|---|---|---|---|---|---|---|---|---|
| Condition | Starvation media | AA | IGF1 | AA+IGF1 | Starvation media | AA | IGF1 | AA+IGF1 |
| Gene ID | ||||||||
| sgta | nd | nd | nd | nd | 1.0 ± 0.0 | 1.98 ± 0.3 | 0.84 ± 0.1 | 3.36 ± 0.3* |
| fkbp4 | nd | nd | nd | nd | 1.0 ± 0.0 | 1.89 ±0.1 | 0.79 ± 2.1 | 2.17 ± 0.1* |
| tbcb | nd | nd | nd | nd | 1.0 ± 0.0 | 1.79 ± 0.2 | 0.97 ± 0.1 | 2.66 ± 0.2* |
| hsp30 | nd | nd | nd | nd | 1.0 ± 0.0 | 1.54 ± 0.4 | 0.70 ± 0.2 | 2.89 ± 0.1* |
| pdia4 | 1.0 ± 0.0 | 2.68 ± 0.9 | 1.06 ± 0.3 | 3.37 ± 0.9* | nd | nd | nd | nd |
| pdia6 | 1.0 ± 0.0 | 4.43 ± 1.8* | 1.55 ± 0.9 | 5.67 ± 1.5* | nd | nd | nd | nd |
| hsp70 | 1.0 ± 0.0 | 5.53 ± 1.7* | 2.25 ± 0.7 | 4.03 ± 1.0* | 1.0 ± 0.0 | 2.65 ± 1.4 | 5.3 ± 1.1* | 5.02 ± 1.3* |
Values are expressed as mean ± SE from 5 independent Atlantic salmon myoblast cell cultures. Significant differences between treatments for each gene with respect to the cells in “starved media” are indicated with an asterisk (*) (P<0.05).
nd: no statistical difference
Starved media: cells incubated with amino acid free medium
AA: cells incubated with medium containing amino acid
IGF1: cells incubated with medium containing 100ng/ml recombinant Atlantic salmon IGF1
AA+IGF1: cells incubated with a combination of amino acid and IGF1
Discussion
Our synteny analysis of teleost genomes has shown that the majority of studied species apart from Tetraodon nigriviridis have two copies of hsp90α and one copy of hsp90ß (Figure 3). Hsp90α paralogues were always found next to each other on the same chromosome/contig and flanked by wdr20 and pp2r5c genes (Figure 3). These results indicate that Hsp90α paralogues originated by tandem duplication at the base of the teleost linage (Figure 3). Salmonid genomes contain four paralogues of Hsp90α and two Hsp90ßs which were named hsp90α1a, hsp90α1b, hsp90α2a, hsp90α2b, hsp90ß1a and hsp90ß1b based on a phylogenetic analysis (Figure 2). Despite fragmented synteny an examination of the available contigs allowed us to make two conclusions; 1: that hsp90α1a and hsp90α2a are found next to one another as observed in other teleost genomes, indicating they originated by tandem duplication and 2: the presence of two copies of wd20 adjacent to hsp90α1a and hsp90α2b (Figure 3) suggests that the second pair of Hsp90α salmon paralogues probably originated during the salmonid-specific WGD [24] since neighbouring genes on the duplicated chromosomes tend to maintain their relative positions or orders over evolutionary time [39]. It has previously been suggested that proteins which form homodimers, such as Hsp90, are more likely to be retained after WGD [40].
Research on Hsp90s in teleosts has mainly focused on their roles in temperature stress [16–18] and embryogenesis [14,41,42] including skeletal muscle formation [15]. Studies in Atlantic salmon [20], zebrafish [21] and gilthead sea bream [22], have reported that hsp90α expression is regulated by feeding in agreement with our findings (Figure 4). In the present study in vitro experiments have provided direct evidence for the stimulation of hsp90α1a and hsp90α1b expression by amino acids (Figure 6). Furthermore the growth factor IGF1, which stimulates protein synthesis via the PI3K pathway in muscle [43], was not able to stimulate hsp90α expression unless amino acids were also present (Figure 6B). Our data suggests that hsp90α regulation by amino acids acts in synergy with the PI3K/Akt pathway, but is independent of Akt phosphorylation (Figure 6). Amino acids (alone or combined with IGF1) were able to increase mTor phosphorylation (Figure 6) in agreement with the general idea that mTor acts as an intracellular nutrient sensor [44]. mTor has been reported to regulate hsp90 expression in response to stress through the phosphorylation of Heat shock factor 1 (HSF1) [45]. Other in vitro studies with salmon myoblasts similarly reported that, in absence of amino acids, IGF1 was unable to increase the expression of several genes related with muscle growth including pcna, pax7, myod1b, myod1c, igf1 itself, igf2, igfbp4, igfbp5.1 and igfbp5.2 [31,33]. In the present study other chaperones (hsp30, hsp70, tcbc), co-chaperones (sgta, fkbp4) and protein-disulphide isomerases (pdia4, pdia6) associated with Hsp90 and protein editing also showed increased expression with amino acid treatment, indicating a general activation of the protein folding response (Table 1).
Our results suggest that all four hsp90α share regulatory elements that are activated in response to changes in intracellular amino acid concentration (Figures 4 and 6), whereas only hsp90α1a and hsp90α1b were associated with myotube formation and muscle differentiation (Figures 4 and 5). Hsp90α1 paralogues are required for the correct formation of the skeletal muscle in zebrafish embryos [15]. It is therefore likely that salmon hsp90α1 paralogues retains analogous myogenesis-related functions to those describes for the zebrafish hsp90α1 orthologue.
It has been widely accepted that Hsp90α is the facultative form of cytosolic Hsp90 and that hsp90ß is constitutive expressed [46]. We found that hsp90ß1a and ß1b did not change their expression during maturation of primary myogenic cultures, whereas hsp90ß1a was significantly downregulated between the fasting and feeding states in vivo (Figure 4A). In gilthead sea bream hsp90ß was also elevated during fasting and returned to lower values after re-feeding [22]. Since hsp90ß1a was not affected when cells were incubated in a free amino acid/hormone medium it seems that is not the absence of nutrients or serum that regulates hsp90ß1a expression, but rather some as yet uncharacterised signal(s) from fasting, such as glucagon, ghrelin or other hormones.
Conclusions
In the present study we found that Atlantic salmon possess an expanded set of Hsp90 paralogues that probably originated during the salmonidae-specific WGD. We have demonstrated that the hsp90α paralogues together with other chaperones and co-chaperones increase their expression in response to amino stimulation during feeding, likely involving enhanced mTor signalling. The present study suggests specific roles for Hsp90α1a/b in muscle differentiation based on in vivo and in vitro expression patterns. We also show that Hsp90ß1b expression is regulated by signals other than amino acids whereas Hsp90ß1a is constitutively expressed as is the single Hsp90ß in mammals and zebrafish.
Supporting Information
Hsp90 amino acid sequence alignments. The ATPase domain is indicated with an underscored red line, middle-domain with a low blue line and C-terminal domain with an underscored green line. Motifs and amino acid involved in ATP binding and hydrolysis are highlighted in red. Residues implicated in protein binding are highlighted in black. The MEEVD motif is highlighted in green.
(TIFF)
Quantitative PCR primer sequences, PCR efficiencies and correlation coefficients of standard curves for genes. E, PCR efficiency; Tm, melting temperature; F, forward; R, reverse. Genes are as follow: heat shock protein 90 kDa (hsp90), Heat shock protein 70kDan (hsp70), heat shock protein 30kDa (hsp30), small glutamine-rich tetratricopeptide repeat-containing protein alpha (sgta), protein disulfide-isomerase A (pdia), myosin heavy chain (Myhc), myosin light chain 2 (mlc2), beta actin (β-actin), RNA polymerase 2 (rnapol II), elongation factor alpha (ef1a), hypoxanthine-guanine phosphoribosyltransferase 1 (hprt1), 40S ribosomal protein S19 (rps19), 60S ribosomal protein L13 (rpl13), tubulin folding cofactor b (tbcb) and fk506 binding protein 4 (fkbp4).
(DOCX)
Rainbow trout de novo transcriptome metrics. bp: base pair.
Singletons: reads not contained in the final assembly.
Isotig: contigs consistently connected by a set of reads.
N50: The value was computed by sorting all contigs from largest to smallest and by determining the minimum set of contigs whose sizes total 50% of the entire transcriptome.
(DOCX)
Acknowledgments
The authors want to thank Dr Neil Bower, University of Queensland (Australia), for the provision of Atlantic salmon cDNA. The authors also thank Dr Daniel MacQueen, University of Aberdeen, for his help in the design of the Hsp90 primers and advice.
Funding Statement
This research was funded by the European Community’s Seventh Framework program (FP7/2007-2013) under grant agreement No 222719- LIFECYCLE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stechmann A, Cavalier-Smith T (2004) Evolutionary origins of Hsp90 chaperones and a deep paralogy in their bacterial ancestors. J Eukaryot Microbiol 51: 364-373. doi:10.1111/j.1550-7408.2004.tb00580.x. PubMed: 15218707. [DOI] [PubMed] [Google Scholar]
- 2. Chen B, Zhong D, Monteiro A (2006) Comparative genomics and evolution of the Hsp90 family of genes across all kingdoms of organisms. BMC Genomics, 7: 156-. doi:10.1186/1471-2164-7-156. PubMed: 16780600. PubMed: 16780600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yun BG, Matts RL (2005) Differential effects of Hsp90 inhibition on protein kinases regulating signal transduction pathways required for myoblast differentiation. Exp Cell Res 307: 212-223. doi:10.1016/j.yexcr.2005.03.003. PubMed: 15922741. [DOI] [PubMed] [Google Scholar]
- 4. Sawai A, Chandarlapaty S, Greulich H, Gonen M, Ye Q et al. (2008) Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer Res 68: 589-596. doi:10.1158/0008-5472.CAN-07-1570. PubMed: 18199556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dittmar KD, Demady DR, Stancato LF, Krishna P, Pratt WB (1999) Folding of the Glucocorticoid Receptor by the Heat Shock Protein (hsp) 90-based Chaperone Machinery. J Biol Chem 272: 21213-21220. [DOI] [PubMed] [Google Scholar]
- 6. da Silva VC, Ramos CH (2012) The network interaction of the human cytosolic 90 kDa heat shock protein Hsp90: A target for cancer therapeutics. J Proteomics 75: 2790-2802. doi:10.1016/j.jprot.2011.12.028. PubMed: 22236519. [DOI] [PubMed] [Google Scholar]
- 7. Johnson JL (2012) Evolution and function of diverse Hsp90 homologs cochaperones proteins. BBA-Mol. Cell Res 1823: 607-613. [DOI] [PubMed] [Google Scholar]
- 8. Li J, Soroka J, Buchner J (2012) The Hsp90 machinery: Conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta 1823: 624-635. doi:10.1016/j.bbamcr.2011.09.003. PubMed: 21951723. [DOI] [PubMed] [Google Scholar]
- 9. Pearl LH, Prodromou C (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75: 271-294. doi:10.1146/annurev.biochem.75.103004.142738. PubMed: 16756493. [DOI] [PubMed] [Google Scholar]
- 10. Street TO, Lavery LA, Verba KA, Lee CT, Mayer MP et al. (2012) Cross-monomer substrate contacts reposition the Hsp90 N-terminal domain and prime the chaperone activity. J Mol Biol 415: 3-15. doi:10.1016/j.jmb.2011.10.038. PubMed: 22063096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hawkins TA, Haramis AP, Etard C, Prodromou C, Vaughan CK et al. (2008) The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development 135: 1147-1156. doi:10.1242/dev.018150. PubMed: 18256191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yun BG, Matts RL (2005) Hsp90 function to balance the phosphorylation state of Akt during C2C12 myoblast differentiation. Cell Signal 17: 1477-1485. doi:10.1016/j.cellsig.2005.03.006. PubMed: 15935620. [DOI] [PubMed] [Google Scholar]
- 13. Liu L, Srikakulam R, Winkelmann DA (2008) Unc45 activates Hsp90-dependent folding of the myosin motor domain. J Biol Chem 283: 13185-13193. doi:10.1074/jbc.M800757200. PubMed: 18326487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wohlgemuth SL, Crawford BD, Pilgrim DB (2007) The myosin co-chaperone UNC45 is required for skeletal and cardiac muscle function in zebrafish. Dev Biol 303: 483-492. doi:10.1016/j.ydbio.2006.11.027. PubMed: 17189627. [DOI] [PubMed] [Google Scholar]
- 15. Du SJ, Li H, Bian Y, Zhong Y (2008) Heat-shock protein 90α1 is required for organized myofibril assembly in skeletal muscle of zebrafish embryos. Proc Natl Acad Sci USA 105: 554-559. doi:10.1073/pnas.0707330105. PubMed: 18182494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Basu N, Todgham AE, Ackerman PA, Bibeau MR, Nakano K et al. (2002) Heat shock protein genes and their functional significance in fish. Gene 295: 173-183. doi:10.1016/S0378-1119(02)00687-X. PubMed: 12354651. [DOI] [PubMed] [Google Scholar]
- 17. Wu CX, Zhao FY, Zhang Y, Zhu YJ, Ma MS et al. (2012) Overexpression of Hsp90 from grass carp (Ctenopharyngodon idella) increases thermal protection against heat stress. Fish Shelfish Immunol 33: 42-47. doi:10.1016/j.fsi.2012.03.033. PubMed: 22510210. [DOI] [PubMed] [Google Scholar]
- 18. Manchado M, Salas-Leiton E, Infante C, Ponce M, Asensio E et al. (2008) Molecular characterization, gene expression and transcriptional regulation of cytosolic HSP90 genes in the flatfish Senegalese sole (Solea senegalensis Kaup). Gene 16: 77-84. PubMed: 18442885. [DOI] [PubMed] [Google Scholar]
- 19. Rescan PY, Montfort J, Rallière C, Le Cam A, Esquerré D et al. (2007) Dynamic gene expression in fish muscle during recovery growth induced by a fasting-refeeding schedule. BMC Genomics 8: 438. doi:10.1186/1471-2164-8-438. PubMed: 18045468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bower NI, Johnston IA (2010) Discovery and characterization of nutritionally regulated genes associated with muscle growth in Atlantic salmon. Physiol Genomics 42: 114-130. PubMed: 20663983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Amaral IPG, Johnston IA (2011) Insulin-like growth factor (IGF) signalling and genome-wide transcriptional regulation in the fast muscle of zebrafish following a single-satiating meal. J Exp Biol 214: 2125-2139. doi:10.1242/jeb.053298. PubMed: 21653807. [DOI] [PubMed] [Google Scholar]
- 22. de la Garcia serrana D, Vieira VLA, Andree KB, Darias M, Estévez A, et al (2012) Development temperature has persistent effects on muscle growth responses in gilthead sea bream. PLOS ONE 12: e51884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Macqueen DJ, Garcia de la serrana D, Johnston IA (2013) Evolution of ancient functions in the vertebrate insulin-like growth factor system uncovered by study of duplicated salmonid fish genomes. Mol Biol Evol 30:1060-1076 [Google Scholar]
- 24. Allendorf FW, Thorgaard GH (1984) Tetraploidy and the evolution of salmonid fishes. Turner BJ. Evolutionary Genetics of Fishes. New York: Plenum Press; pp. 55-93. [Google Scholar]
- 25. Wernersson R (2006) Virtual Ribosome- a comprehensive translation tool with support for sequence feature integration. Nucleic Acids Res 34: 385-388. doi:10.1093/nar/gkl252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.http://mafft.cbrc.jp/alignment/server/ MAFFT version 7 Available: . Accessed 2013 June 25th.
- 27. BioEdit Available: http://www.mbio.ncsu.edu/bioedit/bioedit.html. Accessed 2013 April 2nd.
- 28.http://www.ensembl.org/index.html Ensembl Available: . Accessed 2012 December 10th.
- 29. Penn O, Privman E, Landan G, Graur D, Pupko T (2010) An alignment confidence score capturing robustness to guide-tree uncertainty. Mol Biol Evol. doi:10.1093/molbev/msq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamura K, Peterson D, Peterson N, Stecher G, Nei M et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28: 2731-2739. doi:10.1093/molbev/msr121. PubMed: 21546353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bower NI, Johnston IA (2010) Paralogues of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferation and differentiating myogenic cells. Am J Physiol Regul Integr Comp Physiol 298: 1615-1626. doi:10.1152/ajpregu.00114.2010. [DOI] [PubMed] [Google Scholar]
- 32. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J et al. (2009) The MIQUE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem 55: 611-622. doi:10.1373/clinchem.2008.112797. PubMed: 19246619. [DOI] [PubMed] [Google Scholar]
- 33. Bower NI, Johnston IA (2010) Transcription Regulation of the IGF Signalling Pathway by Amino Acid and Insulin-Like Growth Factors during Myogenesis in Atlantic Salmon. PLOS ONE 5: e11100. doi:10.1371/journal.pone.0011100. PubMed: 20559434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valente LM, Bower NI, Johnston IA (2012) Postpandrial expression of growth related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. Br J Nutr 108: 2148-2157. doi:10.1017/S0007114512000396. PubMed: 22464448. [DOI] [PubMed] [Google Scholar]
- 35.http://biotools.umassmed.edu/bioapps/primer3_www.cgi Primer3: WWW primer tool Available: . Accessed 2012 October 24th.
- 36. NetPrimer Available: http://www.premierbiosoft.com/netprimer/index.html. Accessed 2012 October 24th.
- 37. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 0034.1-0034.11 PubMed: 12184808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmohttp://www.ncbi.nlm.nih.gov/genome/?term=salmo%20salar salar (Atlantic salmon) Available: . Accessed 2012 November 22nd.
- 39. Catchen JM, Conery JS, Postlethwait JH (2009) Automated identification of conserved synteny after whole-genome duplication. Genome Res 19: 1497-1505. doi:10.1101/gr.090480.108. PubMed: 19465509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Presser A, Elowitz MB, Kellis M, Kishony R (2008) The evolutionary dynamics of the Saccharomyces cerevisiae protein interaction network after duplications. Proc Natl Acad Sci U S A, 105: 950-954. doi:10.1073/pnas.0707293105. PubMed: 18199840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krone PH, Sass JB (1994) Hsp90α and Hsp90β genes are present in the zebrafish and are differently regulated on developing embryos 204 Biochem Biophys Res Co. pp. 746-752. PubMed: 7980538. [DOI] [PubMed] [Google Scholar]
- 42. Lele Z, Hartson SD, Martin CC, Whitesell L, Matts RL et al. (1999) Disruption of zebrafish somite development by pharmacologic inhibition of Hsp90. Dev Biol 210: 56-70. doi:10.1006/dbio.1999.9262. PubMed: 10364427. [DOI] [PubMed] [Google Scholar]
- 43. Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L et al. (2001) Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3: 1009-1013. doi:10.1038/ncb1101-1009. PubMed: 11715022. [DOI] [PubMed] [Google Scholar]
- 44. Lian J, Yan XH, Peng J, Jiang SW (2008) The mammalian target of rapamycin pathway and its role in molecular nutrition regulation. Mol Nutr Food Res 52: 393-399. doi:10.1002/mnfr.200700005. PubMed: 18306429. [DOI] [PubMed] [Google Scholar]
- 45. Chou SD, Prince T, Gong J, Calderwood SK (2012) mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLOS ONE 7: e39679. doi:10.1371/journal.pone.0039679. PubMed: 22768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Millson SH, Truman AW, Rácz A, Hu B, Panaretou B et al. (2007) Expressed as the sole Hsp90 of yeast, the alpha and beta isoforms of human Hsp90 differ with regard to their capacities for activation of certain client proteins, whereas only Hsp90beta generates sensitivity to the Hsp90 inhibitor radicicol. FEBS 274: 4453-4463. doi:10.1111/j.1742-4658.2007.05974.x. PubMed: 17681020. [DOI] [PubMed] [Google Scholar]
- 47. Sreeramulu S, Jonker HR, Langer T, Richter C, Lancaster CR et al. (2009) The human Cdc27-Hsp90 complex studied by heteronuclear NMR spectroscopy. J Biol Chem 284: 3885-3896. PubMed: 19073599. [DOI] [PubMed] [Google Scholar]
- 48. Meyer P, Prodromou C, Liao C, Hu B, Mark Roe S et al. (2004) Structural basis of recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J 23: 511-519. doi:10.1038/sj.emboj.7600060. PubMed: 14739935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hsp90 amino acid sequence alignments. The ATPase domain is indicated with an underscored red line, middle-domain with a low blue line and C-terminal domain with an underscored green line. Motifs and amino acid involved in ATP binding and hydrolysis are highlighted in red. Residues implicated in protein binding are highlighted in black. The MEEVD motif is highlighted in green.
(TIFF)
Quantitative PCR primer sequences, PCR efficiencies and correlation coefficients of standard curves for genes. E, PCR efficiency; Tm, melting temperature; F, forward; R, reverse. Genes are as follow: heat shock protein 90 kDa (hsp90), Heat shock protein 70kDan (hsp70), heat shock protein 30kDa (hsp30), small glutamine-rich tetratricopeptide repeat-containing protein alpha (sgta), protein disulfide-isomerase A (pdia), myosin heavy chain (Myhc), myosin light chain 2 (mlc2), beta actin (β-actin), RNA polymerase 2 (rnapol II), elongation factor alpha (ef1a), hypoxanthine-guanine phosphoribosyltransferase 1 (hprt1), 40S ribosomal protein S19 (rps19), 60S ribosomal protein L13 (rpl13), tubulin folding cofactor b (tbcb) and fk506 binding protein 4 (fkbp4).
(DOCX)
Rainbow trout de novo transcriptome metrics. bp: base pair.
Singletons: reads not contained in the final assembly.
Isotig: contigs consistently connected by a set of reads.
N50: The value was computed by sorting all contigs from largest to smallest and by determining the minimum set of contigs whose sizes total 50% of the entire transcriptome.
(DOCX)