Abstract
Background
Many SXT/R391-like enterobacterial Integrative Conjugative Elements (ICEs) have been found to express an atypical, recA-dependent, UV-inducible, cell-sensitising phenotype observed as a reduction in post-irradiation cell survival rates in host cells. Characterisation of a complete deletion library of the prototype ICE R391 identified the involvement of three core ICE genes, orfs90/91 encoding a putative transcriptional enhancer complex, and orf43, encoding a putative type IV secretion system, outer membrane-associated, conjugative transfer protein.
Results
In this study, expression analysis of orf43 indicated that it was up-regulated as a result of UV irradiation in an orfs90/91-dependent manner. Induced expression was found to be controlled from a site preceding the gene which required functional orfs90/91. Expression of orfs90/91 was in turn found to be regulated by orf96, a λ cI-like regulator. Targeted construction of ICE R391 deletions, RT-PCR and qRT-PCR analysis confirmed a regulatory link between orfs90/91 and orf43 while site-directed mutagenesis of orf43 suggested an association with the cell membrane was a prerequisite for the cytotoxic effect.
Conclusions
Because of the recA-dependence of the effect, we hypothesise that UV induction of RecA results in cleavage of the cI-like ICE-encoded repressor protein, the product of orf96. This in turn allows expression of the transcriptional enhancer complex encoded by orfs90/91, which we conclude stimulates transcription of orf43, whose product is directly responsible for the effect.
Keywords: SXT/R391 ICE, Type IV secretion system, Conjugative transfer, UV-induced sensitisation and cytotoxicity
Background
Integrative Conjugative Elements (ICEs) are a class of bacterial mobile genetic elements that encode features necessary for their site-specific integration and excision from host genomes, self-circularisation and transfer by conjugation [1,2]. ICEs are divided into families based on similarity between core genes (specifically the integrase gene) and the site of integration they utilise within host chromosomes. The SXT/R391 family share a highly similar integrase gene and integrate into the prfC gene of enterobacterial hosts [1,3]. In addition to encoding host beneficial traits such as antibiotic resistance determinants [3-5], many SXT/R391 family ICEs express an unusual cell-sensitising function [6-8]. Preliminary characterisation of the UV-inducible, cell-sensitising function of the prototype, ICE R391, determined the effect to be recA-dependent [6], while further analysis based on construction of a deletion library of ICE R391 found that three core ICE genes, namely orfs90/91 and orf43 were involved [8]. Deletion analysis also revealed that orf96, which encodes a putative λ cI-like repressor protein [9], could only be deleted in strains where orfs90/91 had previously been removed suggesting that the repressor protein may prevent lethal expression of orfs90/91. Additionally, cloning and controlled expression of both orfs90/91 and orf43 revealed that expression of orf43 alone was cytotoxic to wild type E. coli while expression of orfs90/91 was only cytotoxic to wild type E. coli cells harbouring the ICE R391. This indicated that orf43 was responsible for the observed UV-inducible cytotoxicity [8].
RecA is a well-documented regulatory protein involved in UV-induced proteolysis of repressor proteins associated with the SOS response [10]. Induction of RecA (some 50 fold) following UV irradiation, results in cleavage of phage λ and phage λ -like cI repressors resulting in phage induction and indeed cleavage of other SOS repressors [10-13]. Bioinformatic analysis of the ICE R391 encoded orf96 has shown it encodes a cI-like repressor protein with homology to phage λ434 cI [9], while analysis of the ICE R391-encoded orfs90/91 has indicated that these genes may act as a putative transcriptional enhancer complex. It has been demonstrated that orfs90/91 stimulate the expression of ICE specific genes such as orf4 (jef, Figure 1) [14], which is an element-encoded excisionase, resulting in formation of increased levels of a circular form of the ICE, presumably as a transfer intermediate. The ICE R391-encoded orf43, has previously been found to be essential for ICE R391 transfer [8]. Indeed, based on bioinformatic homology, orf43 is predicted to encode a putative TraV homolog, an outer membrane protein involved in the ICE type IV secretion system and thought to function in the construction and stabilisation of the outer-membrane portion of the mating pore required for ICE transfer by conjugation [15]. Deletion of the ICE R391-encoded orf43 was recently shown to abolish the UV-inducible sensitising effect of this ICE while clones expressing orf43 under arabinose control were shown to compliment for the transfer deficiency but additionally mimic the cell toxicity associated with UV induction [8].
Figure 1.
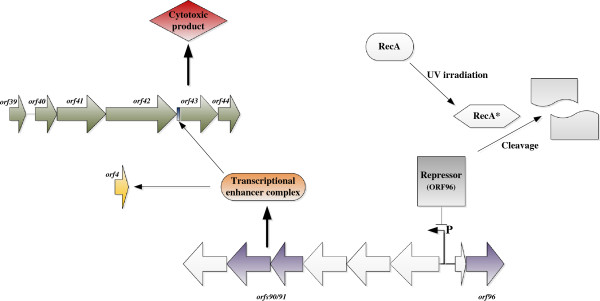
Proposed induction pathway for the UV-inducible cell-sensitising function of ICE R391. Stimulation of RecA to its active form (RecA*) by UV irradiation results in the cleavage of the putative orfs90/91 repressor protein (orf96) allowing the transcription of orfs90/91 which putatively encode a transcriptional enhancer complex that activates/increases expression of the orf43 gene product as well as the previously documented UV-inducible orf4 (jef) [14]. Expression of orf43 is then cytotoxic to E. coli host cells. Evidence to support this hypothesised pathway includes: RecA has been well documented to be stimulated to its active form (RecA*) by single-stranded DNA generated from exposure to UV irradiation [16], the observation that the cell-sensitising function of ICE R391 requires the presence of recA in the host genome [6], the deletion of orf96 encoding a putative repressor protein cannot be achieved without the previous deletion of orfs90/91[8], and orfs90/91 have previously been documented to enhance the transcription of other ICE R391 genes after host cell exposure to UV irradiation, specifically orf4 (jef), proposed to promote element excision from the host genome [14]. Additionally the ICE SXT homologs setR (orf96) and setC/D (orfs90/91) have been documented to have a similar recA-dependent, stress-inducible relationship [17].
Here, a model is proposed (Figure 1) for the control of this unusual ICE R391 UV-inducible sensitising effect based on expression data examining the key genes involved and supported by a number of directed ICE R391 deletions.
Results and discussion
orfs90/91 stimulate orf43 transcription after exposure to UV irradiation
We previously demonstrated that over-expression of orf43 when cloned into the arabinose inducible pBAD33-orf43 construct was responsible for the UV-inducible sensitisation observed in ICE R391 and other ICEs of the SXT/R391 family [8]. Mutagenesis data also suggested that the putative transcriptional controller encoded by orfs90/91 was also involved, although not directly. To investigate the relationship between orfs90/91 and orf43, we utilised both qualitative and quantitative RT-PCR targeting these genes in different mutant backgrounds and with and without UV irradiation.
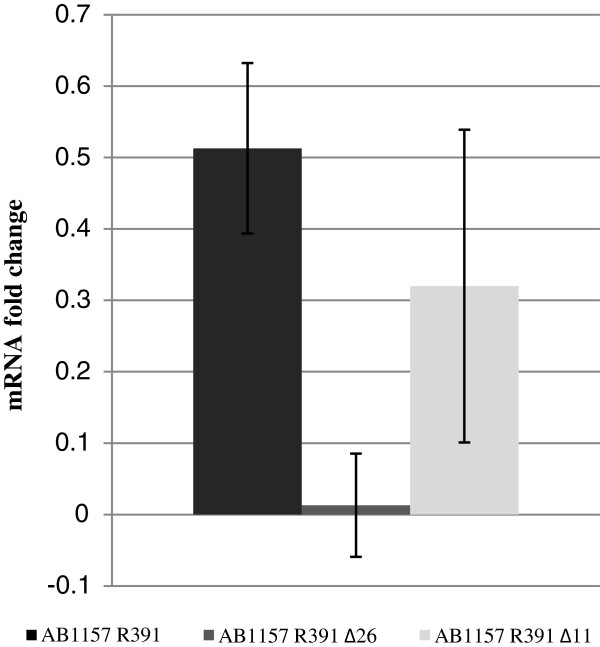
qRT-PCR analysis of expression levels pre and post exposure to 40 J.m-2 UV-irradiation indicated that orfs90/91, orf43 and the previously documented UV-inducible orf4 (jef, Figure 1) [14] were up-regulated after exposure to UV irradiation. Analysis indicated that orf4 (jef) specific mRNA levels were up-regulated 0.78 fold, orf43, 0.513 fold and orfs9091, 0.339 fold. In contrast other ICE R391 genes not involved in cell sensitisation [8] were not up-regulated post exposure: aph (encoding Kanamycin resistance) was down-regulated 0.23 fold post-exposure while orf31 (encoding a putative Lon protease) was also down-regulated 0.19 fold post-exposure. Analysis of the up-regulated genes in mutant backgrounds indicated that in a Δorfs90/91 (∆26) background, orf43 up-regulation was abolished (Figure 2) while analysis of orfs90/91 transcription in a Δorf43 (∆14) background did not prevent orfs90/91 specific mRNA up-regulation following UV irradiation (orfs90/91 up-regulated 0.61 fold in AB1157 R391 ∆14). This indicated a dependency on orfs90/91 for orf43 up-regulation but not vice versa. Further analysis of orf43 transcription in a Δorfs40/41 mutant (Δ11) [8] demonstrated that deletion of these genes, upstream of orf43, did not prevent the UV-induced up-regulation of orf43 mRNA, suggesting that inducible orf43 transcription was stimulated through a region directly in front of the orf43 gene (Figure 2) and that this region should be investigated further. This observation is supported by previous deletion analysis where orfs40/41 (Δ11) and ∆orf42 (∆13) were deleted but retained the UV-inducible sensitising phenotype [8]. Analysis of the up-regulated orfs90/91 and orf43 mRNA decay rate post-exposure (Figure 3) revealed that orfs90/91 mRNA levels were maximally up-regulated directly after exposure and decayed rapidly with a return to basal levels within 5 minutes post-exposure. However orf43 mRNA levels were maximally up-regulated 7 minutes post-exposure and up-regulated levels were sustained for a longer period of time, minimally over 30 minutes (Figure 3). The observation of the rapid increase and decay of orfs90/91 specific mRNA levels followed by a slower and longer sustained increase in orf43 specific mRNA levels supports the hypothesis that UV irradiation acts as an inducing agent for orfs90/91, which subsequently up-regulates the transcription of orf43 possibly from a site preceding the gene.
Figure 2.
Increase in orf43 mRNA levels after exposure to 40 J.m-2 UV irradiation. Backgrounds analysed were AB1157 R391, AB1157 R391 ∆26 (∆orfs90/91) and AB1157 R391 ∆11 (∆orfs40/41). All results were normalised using the endogenous constitutively expressed proC gene. Average values were calculated from a minimum of 9 replicates for each strain analysed.
Figure 3.
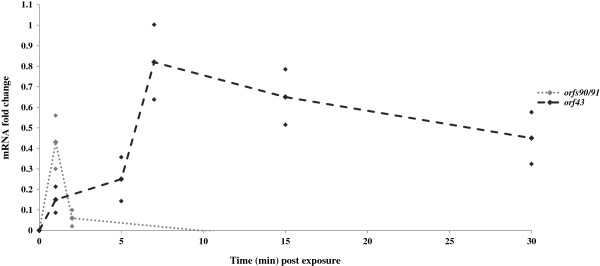
Decay of AB1157 R391 orfs90/91 and orf43 mRNA levels after exposure to 40 J.m-2 UV irradiation. All results were normalised using the endogenous constitutively expressed proC gene. Standard deviation is denoted by markers above and below all data points. orfs90/91 specific mRNA levels were up-regulated immediately post exposure to UV irradiation but quickly decayed. orf43 specific mRNA levels were maximally up-regulated 7 minutes post exposure and elevated levels were sustained for over 30 minutes post exposure.
Cytotoxic orf43 transcription is regulated through a region directly upstream of orf43
Based on previous observations with the Δ11 and ∆13 ICE R391 deletions, which deleted orfs40 to most of orf42 inclusive [8], the most likely location for an orf43 control site would be the last 36 bp specific to orf42 directly in front of orf43. Comparative bioinformatic analysis of this region and the previously documented orfs90/91 regulated orf4 (jef) [14] uncovered a short 7 bp homologous DNA sequence (5’-AGAAGAT-3’) present in front of both genes. This conserved sequence was located 77 bp upstream of orf4 (jef) but directly in front of orf43 where the last 2 base pairs of the sequence overlapped the first two base pairs of the predicted start codon of orf43. As no other recognisable promoter or operator region was predicted upstream of orf43, this 7 bp sequence may possibly represent a binding motif for the putative transcriptional enhancer (orfs90/91). However it is well known that transcriptional enhancer control sites can be difficult to predict [18] as they tend to be short DNA sequences lacking high sequence conservation even between enhancer types. To examine if the last 36 bp specific to orf42 and preceding orf43 did in fact contain a control site for orf43 transcription, orf43 specific mRNA expression was analysed in a number of specific deletion backgrounds spanning this putative control region [Table 1, Figure 4C]. Three directed ICE R391 deletion mutants were generated [Figure 4C] in an E. coli (AB1157 R391) background; the KOA deletion removed the genes orf32 to orf42 and placed the inserted ampicillin cassette on the reverse complement to ensure removal of all possible promoters of orf43 transcription except for the 36 bp directly in front of orf43, the KOB deletion removed the genes orf32 to orf42 similar to KOA but additionally removed the 36 bp directly in front of orf43 while the KOC deletion was identical to the KOA mutation, preserving the putative 36 bp control site but also contained an additional secondary zeocin resistant deletion which removed orfs90/91. These three deletion mutations were screened by both qualitative and quantitative UV survival assays to determine their effect on the cell-sensitising function [Figure 4A] and additionally were examined by RT-PCR to determine if orf43 specific mRNA transcription still occurred [Figure 4B]. The KOA mutant retained the UV-inducible sensitising function [Figure 4A] and orf43 mRNA transcription [Figure 4B], while the KOB and KOC mutations abolished the sensitising function as well as orf43 mRNA transcription. As the KOB and KOC mutations deleted either the proposed orfs90/91 control site or orfs90/91 genes respectively and both of these deletions prevented cytotoxic orf43 transcription, this analysis supported the hypothesis that orfs90/91 encode a transcriptional enhancer complex that activates cytotoxic orf43 transcription through the 36 bp sequence directly in front of orf43. In addition, cloning of orf43 with the predicted control site in front of the gene showed that the cytotoxic function could be repressed only in cells not containing orfs90/91 (data not shown), again supporting the hypothesis.
Table 1.
Genotype of bacterial strains, plasmids and ICE R391 mutants used
| Strain | Genotype | Source |
|---|---|---|
| AB1157 |
F-, thr-1, araC14, leuB6, ∆(gpt-proA)62, lacY1, tsx-33, qsr’-0, glnV44, galK2, λ-, Rac-0, hisG4, rfbC1, mgl-51, rpoS396, rpsL31 (StrR), kdgK51, xylA5, mtl-1, argE3, thi-1 |
E. coli genetic stock centre (CGSC), Yale University, New Haven, Connecticut, USA |
| TOP10 |
F-, mcrA0, ∆(mrr-hsdRMS-mcrBC), φ80dlacZ58(M15), ∆lacX74, recA1, araD139, ∆(araA-leu)7697, galU-, galK0, rpsL- (StrR), endA1, nupG- |
Bio-Sciences, Dun Laoghaire, Dublin, Ireland |
| P125109 |
S. Enteritidis PT4 wild type (NCTC 13349), NalR |
National Collection of Type Cultures (NCTC), Salisbury, UK |
|
Plasmid |
Genotype |
Source |
| pBAD33-orf43 |
CmR, p15A ori, PBAD L-arabinose inducible, orf43 |
Armshaw and Pembroke, 2013 [8] |
| pBAD33-orf43[SM12] |
CmR, p15A ori, PBAD L-arabinose inducible, orf43 containing mutation converting two leucines to prolines at a.a. position 47 and 48. |
This study |
| pBAD33-orf43[SM56] |
CmR, p15A ori, PBAD L-arabinose inducible, orf43 containing mutation converting glutamine at position 115 to asparagine. |
This study |
| pKOBEG |
Ts, PBAD-gam-bet-exo cat (CmR) |
Dr. P. Latour-Lambert, Institut Pasteur, 25 rue du Dr Roux, Paris, France |
| pUC18 |
AmR template for deletion mutant construction |
Sigma-Aldrich, Arklow, Wicklow, Ireland |
| pcDNA3.1(+) |
ZeR template for deletion mutant construction |
Invitrogen, Bio-Sciences, Dun Laoghaire, Dublin, Ireland |
|
ICE |
Genotype |
Source |
| R391 |
KmR, HgR |
Dr R.W. Hedges, Royal Postgraduate Medical School, London, UK |
|
R391 Mutant |
Genotype |
Source |
| AB1157 R391 ∆14 (∆orf43) |
ICE R391 orf43 deletion strain, AmR, UV-, tra- |
Armshaw and Pembroke, 2013 [8] |
| AB1157 R391 ∆26 (∆orfs90/91) |
ICE R391 orfs90/91 deletion strain, AmR, UV-, tra- |
Armshaw and Pembroke, 2013 [8] |
| AB1157 R391 ∆11 (∆orfs40/41) |
ICE R391 orfs40/41 deletion strain, AmR, tra- |
Armshaw and Pembroke, 2013 [8] |
| AB1157 R391 ∆25AmR∆14ZeR |
ICE R391 orf90 – orf94 and orf43 deletion strain, AmR, ZeR, UV-, tra- |
This study |
| AB1157 R391 KOA |
ICE R391 orf32 - orf42 (29575 bp – 41491 bp) deletion strain, AmR, tra- |
This study |
| AB1157 R391 KOB |
ICE R391 orf32 - orf42 (29575 bp – 41527 bp) deletion strain, AmR, UV-, tra- |
This study |
| AB1157 R391 KOC | ICE R391 orf32 - orf42 (29575 bp – 41491 bp) and orfs90/91 deletion strain, AmR, ZeR, UV-, tra- | This study |
StrR is streptomycin resistant; CmR is chloramphenicol resistant; KmR is kanamycin resistant; HgR is mercury resistant; ZeR is zeocin resistant; Ts is temperature sensitive; NalR is nalidixic acid resistant and AmR is ampicillin resistant. UV- is UV-inducible cell-sensitising function abolished; tra- is conjugative transfer deficient mutant.
Figure 4.
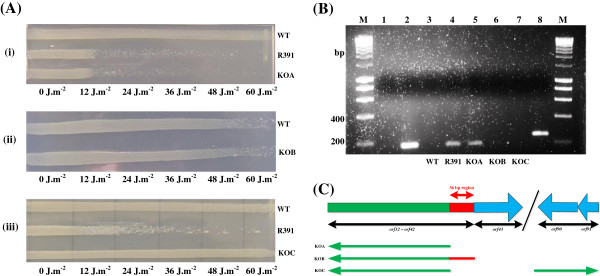
Qualitative UV assay and mRNA analysis of E. coli R391 mutants KOA, KOB and KOC. (A) AB1157 R391 mutants KOA, KOB and KOC. UV254nm exposure increasing (12 J.m-2) from left to right. (i) From top to bottom, AB1157, AB1157 R391, AB1157 R391 KOA. (ii) AB1157, AB1157 R391 KOB. (iii) AB1157, AB1157 R391, AB1157 R391 KOC. (B) SYBR® Safe stained 1% (w/v) agarose gel confirming orf43 mRNA transcription in AB1157 R391 KOA. M, Bioline Hyperladder I DNA marker; 1, AB1157 R391 RNA negative control; 2, AB1157 R391 genomic DNA positive control; 3, AB1157 orf43 cDNA; 4, AB1157 R391 orf43 cDNA; 5, KOA orf43 cDNA; 6, KOB orf43 cDNA; 7, KOC orf43 cDNA; 8, KOB orf20 cDNA. Primers used specific to orf43 generated a 188 bp PCR product. Primers used for lane 8 only were specific for the kanamycin resistance gene of ICE R391, orf20, which generated a PCR product of 223 bp. Amplification of orf20 specific cDNA was carried out to show KOB and KOC RNA was not degraded. Lane 1 negative control was DNase treated RNA that was not converted to cDNA. (C) Map of exact locations of KOA, KOB and KOC deletions on ICE R391 genome. The KOA, KOB and KOC ampicillin resistance cassettes and associated promoter were inserted into the ICE R391 genome in the reverse complement to prevent the ampicillin resistance cassette promoter inducing the transcription of orf43 mRNA. The KOA deletion removed all possible promoters of orf43 in front of the gene and left the last 36 bp specific to the preceding orf42 gene. The KOB deletion removed the same region as KOA and the 36 bp region. The KOC deletion was a duplicate of KOA with an additional zeocin resistant orfs90/91 deletion.
Site-directed mutagenesis of Orf43
Bioinformatic analysis of orf43 indicated that it belongs to a highly conserved TraV-like family of transfer proteins involved in type IV secretion systems required for conjugation [8]. Site-directed mutagenesis of pBAD33-orf43 was carried out to convert two leucines at a.a. positions 47 and 48 to prolines in the predicted Orf43 protein (GenBank: AAM08037). Insertion of two prolines was expected to disrupt the α-helical transmembrane spanning region of Orf43 by creating a 30° bend [19]. This mutation was found to cause loss of the cytotoxic function of pBAD33-orf43[8] as there was no observable decline in host cell growth rates after induction of the mutant clone compared to the wild type clone [Figure 5A,B]. Since introduction of membrane disruptive mutations abolish the effect, this is suggestive that membrane association is required in addition to over-expression of the Orf43 protein for sensitisation and cytotoxicity associated with this ICE product. Other directed mutations such as conversion of glutamine at position 115 (predicted in the periplasmic region of Orf43) to asparagine (Q115 to N115) was shown to have no effect on UV sensitisation or orf43 transcription [Figure 5C,D].
Figure 5.
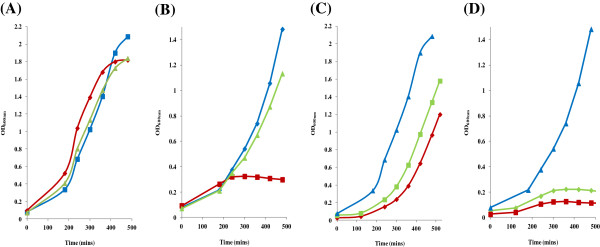
Effect on growth rates of the pBAD33-orf43 SM12 and SM56 mutations in E. coli TOP10. (A) Un-induced growth rates for pBAD33 (blue curve), pBAD33-orf43 (red curve) and pBAD33-orf43 SM12 (green curve). (B) Induced growth rates for pBAD33 (blue curve), pBAD33-orf43 (red curve) and pBAD33-orf43[SM12] (green curve). (C) Un-induced growth rates for pBAD33 (blue curve), pBAD33-orf43 (red curve) and pBAD33-orf43 SM56 (green curve). (D) Induced growth rates for pBAD33 (blue curve), pBAD33-orf43 (red curve) and pBAD33-orf43[SM56] (green curve). Note that the SM12 mutation in pBAD33-orf43 caused a return to exponential growth behaviour expected with E. coli cells.
Conclusions
Hierarchical control of the ICE R391 UV-inducible sensitising effect
Many SXT/R391-like ICEs reduce post UV survival rates of E. coli host cells through the action of a recA-dependent process [6,20]. Mutational analysis of the ICE R391 determined that the core genes orfs90/91 and orf43 were required for expression of the cell-sensitising function [8] while bioinformatic analysis indicated that orf96 likely encodes a λ cI-like repressor similar to RecA substrates in other phage systems that are cleaved following SOS induction [9]. Initial attempts to delete orf96 proved fruitless and no deletion could be isolated. However a Δorf96 (Δ28) deletion [8] could be isolated in an ∆orfs90/91 mutant background suggesting that orf96 may control expression of orfs90/91 which we have shown here directly control expression of orf43, the ultimate instigator of the cytotoxicity associated with ICE R391. The data presented here and in Armshaw and Pembroke (2013) [8] have led to the development of a model to explain the control of UV-inducible sensitisation (Figure 1). We hypothesise that UV irradiation of E. coli induces the host RecA protein which results in cleavage of the ICE R391 encoded product of orf96, the phage λ434 cI-like ICE repressor. We propose that cleavage of Orf96 in turn leads to expression of orfs90/91 which in turn leads to up-regulation of orf43 and other ICE R391 genes such as orf4 (jef) [14]. We have previously demonstrated that up-regulation of orf4 (jef) leads to increased ICE R391 transfer [14]. In the related ICE SXT, Beaber et al., (2004) [17] demonstrated that SetR, the SXT homolog of Orf96, acted as a repressor of ICE SXT transfer and that it is bound to ICE operators that controlled setC/D, SXT homologs of orfs90/91, in a similar way to our proposal for ICE R391. They also proposed that repression was lifted by induced RecA protein cleaving the SetR repressor in a similar manner to our proposal for orfs90/91. The recA dependence for the ICE R391 UV-sensitising effect [6], the similarity to the SXT system [17], the deletion data and qRT-PCR data presented here support the model presented.
It would thus appear that UV irradiation is the instigator of the control loop leading to over expression of orf43 which leads to cytotoxicity. Since normal levels of Orf43 are required for ICE transfer and play a key part in formation of the conjugative transfer system of ICE R391, it appears that the associated cytotoxicity is related to the induced overexpression. Evidence in support of this comes from data showing that overexpression of orf43 from the arabinose inducible clone pBAD33-orf43 leads directly to cytotoxicity [8]. The UV-inducible sensitising effect is conserved amongst many SXT/R391 ICE family members [6,20]. A sophisticated control system is in place to control this effect yet the exact nature and reason for conservation of such an unusual apparently ‘evolutionary negative’ effect remains to be elucidated. We are currently examining the nature of the cytotoxicity and developing theories for its function and retention.
Methods
Bacterial strains, elements and media
The bacterial strains, plasmids and ICE R391 deletion mutants utilised as part of this study are listed in Table 1. Strains were stored at −80°C in either Luria-Bertani (LB) broth or M9 minimal media containing 50% (v/v) glycerol. Media was supplemented with appropriate antimicrobial agents: nalidixic acid, 30 μg ml-1; ampicillin, 100 μg ml-1; chloramphenicol, 25 μg ml-1, kanamycin, 30 μg ml-1, streptomycin, 100 μg ml-1; mercuric chloride, 20 μg ml-1; zeocin, 25 μg ml-1 as required. For growth and analysis of strains containing pBAD33-orf43, M9 minimal media containing 0.4% (v/v) glycerol was used with either 0.4% (w/v) glucose or 0.02%-0.2% (w/v) L-arabinose to repress or induce gene expression respectively as previously described [8].
Directed deletions of ICE R391 and subsequent deletion mutant screening
ICE R391 specific deletions were generated as previously described [8]. Screening of resulting ICE R391 deletion mutants for loss of cell-sensitising function by qualitative and quantitative UV survival assays were carried out as described [8]. Screening of ICE R391 deletion mutants’ conjugative transfer ability to recipient Salmonella enterica serotype Enteritidis strain P125109 was performed as described [21].
Qualitative reverse transcriptase PCR
Cells were collected by centrifugation, washed twice with diethyl pyrocarbonate-treated distilled water and resuspended in 10 mM Tris, [pH8.0]. Total RNA was isolated using the Absolutely RNA Miniprep kit (Agilent Technologies) according to the manufacturer’s protocol. Absence of contaminating DNA was verified by PCR. Qualitative reverse transcriptase PCR was performed using the AccuScript High Fidelity 1st Strand cDNA Synthesis Kit (Agilent Technologies) according to the manufacturer’s protocol. Resulting cDNA was analysed immediately by PCR using gene-specific primers or stored at −20°C.
Quantitative reverse transcriptase PCR (qRT-PCR)
Quantitative UV assays were carried out as described [8]. Unirradiated and irradiated cells were collected by centrifugation and total RNA isolated as described. Absence of contaminating DNA was verified by PCR. qRT-PCR was performed using the Brilliant III Ultra-Fast SYBR Green qRT-PCR Master Mix (Agilent Technologies) according to the manufacturer’s protocol using the Stratagene Mx3000P Real Time PCR System and appropriate gene-specific PCR primers with the following temperature profile: 1 cycle at 42°C for 30 minutes to convert RNA to cDNA, 1 denaturation cycle at 95°C for 3 minutes followed by 40 cycles at 95°C for 30 seconds, 54°C for 60 seconds, 72°C for 30 seconds followed by melting curve analysis from 65°C to 95°C to determine specificity of the PCR reaction. Specificity of the PCR reaction was verified by SYBR safe staining on a 2% (w/v) agarose gel. The internal standard curve using the unirradiated RNA sample to estimate the change in target RNA quantity consisted of: undiluted RNA, a 1 in 2 dilution, a 1 in 4 dilution and a 1 in 10 dilution of unirradiated RNA. A no template negative control was also included. In addition, qRT-PCR was also carried out on the known endogenous housekeeping gene proC as an internal control to quantify the relative change in transcription of the gene of interest [22].
Site-directed mutagenesis of pBAD33-orf43
Site-directed mutagenesis of pBAD33-orf43[8] was performed using specifically designed complementary mutagenic primers to linearly amplify pBAD33-orf43 to generate a mutated nicked DNA product. Non-mutated methylated template DNA was eliminated by incubation with the DpnI restriction enzyme. Mutated DNA products were then transformed into TOP10 and plated on appropriate media containing chloramphenicol, 25 μg ml-1. Resulting TOP10 colonies were cultured, had plasmid content extracted using the QIAprep Spin Miniprep Plasmid extraction kit from QIAGEN (West Sussex, RH10, 9NQ, UK) according to the manufacturer’s protocol and screened for the presence of pBAD33-orf43 by restriction enzyme digestion. Mutated pBAD33-orf43 was verified by DNA sequencing to contain the desired mutation without additional mutations. Mutated pBAD33-orf43 was confirmed to still transcribe orf43 specific mRNA by RT-PCR as described. Determination of the effect of induction of mutated pBAD33-orf43 on host cell growth rate was carried out as described [8].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PA and JTP conceived and designed the study. PA did the laboratory work and analysed the data. PA and JTP wrote the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Patricia Armshaw, Email: patricia.armshaw@ul.ie.
J Tony Pembroke, Email: tony.pembroke@ul.ie.
Acknowledgements
This work was funded by the Irish Research Council for Science, Engineering and Technology (IRSCET) to PA. The authors would like to thank Dr. P. Latour-Lambert for providing the pKOBEG plasmids and Drs. John O’Halloran and Michael P. Ryan for helpful discussion.
References
- Taviani E, Grim CJ, Chun J, Huq A, Colwell RR. Genomic analysis of a novel integrative conjugative element in Vibrio cholerae. FEBS Lett. 2009;583(22):3630–3636. doi: 10.1016/j.febslet.2009.10.041. [DOI] [PubMed] [Google Scholar]
- Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J Antimicrob Chemoth. 2012;67(1):91–100. doi: 10.1093/jac/dkr411. [DOI] [PubMed] [Google Scholar]
- Pembroke JT, Piterina AV. A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol Lett. 2006;264(1):80–88. doi: 10.1111/j.1574-6968.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- Brochet M, Couve E, Glaser P, Guedon G, Payot S. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J Bacteriol. 2008;190(20):6913–6917. doi: 10.1128/JB.00824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Poele EM, Bolhuis H, Dijkhuizen L. Actinomycete integrative and conjugative elements. A van Leeuw J Microb. 2008;94(1):127–143. doi: 10.1007/s10482-008-9255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pembroke JT, Stevens E. The effect of plasmid R391 and other incJ plasmids on the survival of Escherichia coli after UV irradiation. J Gen Microbiol. 1984;130:1839–1844. doi: 10.1099/00221287-130-7-1839. [DOI] [PubMed] [Google Scholar]
- Wang TCV, deSaintPhalle B, Millman KL, Fowler RG. The ultraviolet-sensitizing function of plasmid R391 interferes with a late step of postreplication repair in Escherichia coli. Mutat Res-DNA Repair. 1996;362(3):219–226. doi: 10.1016/0921-8777(95)00044-5. [DOI] [PubMed] [Google Scholar]
- Armshaw PA, Pembroke JT. Generation and analysis of an ICE R391 deletion library identifies genes involved in the element encoded UV-inducible cell-sensitising function. FEMS Micro Lett. 2013;342(1):45–53. doi: 10.1111/1574-6968.12107. [DOI] [PubMed] [Google Scholar]
- Boltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol. 2002;184(18):5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL, Roberts JW. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage-lambda repressor. J Biol Chem. 1981;256(15):8039–8044. [PubMed] [Google Scholar]
- Karu AE, Belk ED. Induction of Escherichia coli RecA protein via recBC and alternate pathways - quantitation by enzyme-linked immunosorbent-assay (ELISA) Mol Gen Genet. 1982;185(2):275–282. doi: 10.1007/BF00330798. [DOI] [PubMed] [Google Scholar]
- Janion C. Inducible SOS Response System of DNA Repair and Mutagenesis in Escherichia coli. Int J Biol Sci. 2008;4(6):338–344. doi: 10.7150/ijbs.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persky NS, Lovett ST. Mechanisms of Recombination: Lessons from E. coli. Crit Rev Biochem Mol. 2008;43(6):347–370. doi: 10.1080/10409230802485358. [DOI] [PubMed] [Google Scholar]
- O’Halloran JA, McGrath BM, Pembroke JT. The orf4 gene of the enterobacterial ICE, R391, encodes a novel UV-inducible recombination directionality factor, Jef, involved in excision and transfer of the ICE. FEMS Microbiol Lett. 2007;272(1):99–105. doi: 10.1111/j.1574-6968.2007.00747.x. [DOI] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang LC, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323(5911):266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol. 2004;186(21):7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427(6969):72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- de Henestrosa AR F, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35(6):1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Nilsson I, Saaf A, Whitley P, Gafvelin G, Waller C, von Heijne G. Proline-induced disruption of a transmembrane alpha-helix in its natural environment. J Mol Biol. 1998;284(4):1165–1175. doi: 10.1006/jmbi.1998.2217. [DOI] [PubMed] [Google Scholar]
- McGrath BM, O’Halloran JA, Piterina AV, Pembroke JT. Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J Microbiol Meth. 2006;66(1):32–42. doi: 10.1016/j.mimet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- McGrath BM, O’Halloran JA, Pembroke JT. Pre-exposure to UV irradiation increases the transfer frequency of the IncJ conjugative transposon-like elements R391, R392, R705, R706 R997 and pMERPH and is recA(+) dependent. FEMS Microbiol Lett. 2005;243(2):461–465. doi: 10.1016/j.femsle.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Theis T, Skurray RA, Brown MH. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J Microbiol Meth. 2007;70(2):355–362. doi: 10.1016/j.mimet.2007.05.011. [DOI] [PubMed] [Google Scholar]