Abstract
The “glycan shield” exposed on the surface of the HIV-1 gp120 env glycoprotein has been previously proposed as a novel target for anti-HIV treatments. While such targeting of these glycans provides an exciting prospect for HIV treatment, little is known about the conservation and variability of glycosylation patterns within and between the various HIV-1 group M subtypes and circulating recombinant forms. Here, we present evidence of strong strain-specific glycosylation patterns and show that the epitope for the 2G12 neutralising antibody is poorly conserved across HIV-1 group M. The unique glycosylation patterns within the HIV-1 group M subtypes and CRFs appear to explain their varying susceptibility to neutralisation by broadly cross-neutralising (BCN) antibodies. Compensatory glycosylation at linearly distant yet three-dimensionally proximal amino acid positions appears to maintain the integrity of the glycan shield while conveying resistance to neutralisation by BCN antibodies. We find that highly conserved clusters of glycosylated residues do exist on the gp120 trimer surface and suggest that these positions may provide an exciting target for the development of BCN anticarbohydrate therapies.
1. Introduction
The envelope gene of human immunodeficiency virus type 1 (HIV-1) encodes a gp160 precursor that is cleaved to form gp120 and gp41 that exists as a trimer on the surface of a HIV virion and is responsible for host cell recognition and binding. As the envelope protein moves through the endoplasmic reticulum, N-linked glycans are added to aid correct folding and processing of the protein [1–3]. The gp120 protein is one of the most heavily known glycosylated proteins [3–5]. The carbohydrates present on gp120 are created by the host cell and, as such, are recognised as immunologically “self” by the host immune system. Studies have shown that the “glycan shield” bound to gp120 can prevent neutralisation of the virus by antibodies [6–13]. It has been suggested that lowly glycosylated viruses may be replicatively fitter and are thus selected early on in infection with glycosylated viruses only being selected for following the activation of the host humoral immune response [14–18]. This trend does not occur in all cases; however, it has been suggested that it occurs more frequently in particular viral subtypes [17, 19].
Domains on gp120 responsible for receptor binding and trimer interactions tend to exhibit low levels of glycosylation resulting in the designation of three domains within gp120: the neutralizing face, the nonneutralizing face, and the silent face [20–22]. The neutralizing face comprises the receptor-binding sites while the non-neutralizing face contains epitopes that are accessible to neutralizing antibodies in monomeric gp120 but which are hidden in the gp120-gp41 trimer. The highly glycosylated domain has been termed the silent face given that is immunologically “self” to the host immune system. It has been suggested, however, that the highly conserved glycans on the gp120 surface may, themselves, provide an ideal target for neutralizing antibodies [18, 23]. In fact, the neutralizing antibody 2G12 binds to a well-defined epitope comprising solely of N-linked glycans bound to the gp120 surface [24–26]; however 2G12 has been shown to have varying efficacy for different subtypes and is particularly ineffective against subtype C and CRF01_AE [27–29]. More recently, a number of studies have isolated BCN antibodies whose activity appears to be highly dependent on the presence of glycosylation at a number of positions on the gp120 trimer, particularly position 332 [30–33]. Work has also shown that lectins isolated from various sources exhibit antiviral activity by interacting with the carbohydrates bound to gp120 and, thus, block cell-to-cell contact between gp120 and the host cell thereby inhibiting cell binding and fusion [34–39]. Similarly, Balzarini and colleagues have shown that Pradimicin A, an antifungal antibiotic, displays properties that inhibit virus entry into host cells [40]. Due to the high degree of glycosylation of gp120, targeting these carbohydrates means that there are multiple targets available, and the emergence of resistance to such anticarbohydrate therapies will most likely involve the removal of multiple glycosylated sites thereby exposing the surface of the virus to neutralizing antibodies [40–42].
HIV-1 group M, which is responsible for the vast majority of HIV infections worldwide, exhibits incredibly high levels of genetic diversity, and phylogenetic analysis has determined a number of major clades/subtypes within the group M phylogeny (designated A-D, F-K) [43]. While subtype B predominates in North America and Europe [44], subtype C accounts for greater than 50% of worldwide infections [45]. Recombination occurs frequently between HIV-1 group M subtypes with established recombinants described as circulating recombinant forms (CRFs). These CRFs now account for as much as 18% of the worldwide infections [46, 47] with CRF01_AE and CRF02_AG representing the most prevalent forms in Southeast Asia and West/West Central Africa, respectively [48–52]. In light of numerous studies highlighting the vast potential of anti-carbohydrate therapies in HIV treatment, it is important that we fully understand the complexities and dynamics of N-linked glycosylation in the most predominant strains of HIV currently circulating within the worldwide population.
Previous work has examined glycosylation patterns within a selection of sequences representing the HIV-1 group M subtypes and CRFs and concluded that despite the extreme genetic variability between the HIV-1 group M subtypes and CRFs, the patterns of glycosylation are essentially conserved between them [53]. Poon and colleagues studied the evolutionary interactions between N-linked glycosylation proposing that mutual exclusion of glycosylation occurs between colocalized glycans while mutual dependency occurs between structurally distant glycans [54]. Here, we have expanded upon these previous studies by looking at the conservation and strain-specific patterns of N-linked glycosylation across all available sequences from full-length genomes representing the HIV-1 group M subtypes and CRFs. We examine the prevalence of the 2G12 epitope and identify mutations that cause loss of sensitivity to 2G12 neutralisation yet still maintain the protective glycan shield on the gp120 trimer. Further, we identify clusters of glycosylated positions that are highly conserved across the entire spectrum of the HIV-1 group M subtypes and CRFs that show potential as targets for BCN carbohydrate-binding therapies.
2. Materials and Methods
2.1. Data Selection
All available full-length genomes representing the HIV-1 group M subtypes and circulating recombinant forms (CRFs) were retrieved from the Los Alamos National Laboratory (LANL) HIV Sequence Database (http://www.hiv.lanl.gov/). Full genomes were used in order to avoid the possibility of including intersubtype recombinants in the analysis. Using the patient and isolate annotation, where available, multiple sequences representing single individuals were removed to ensure that the dataset representing each subtype/CRF was nonredundant. Subtypes and CRFs with significant numbers of representative sequences were selected for subsequent analysis (subtypes A (n = 73), B (n = 161), C (n = 385), D (n = 48), CRF01_AE (n = 90), and CRF02_AG (n = 38), see Supplementary Table 1 Supplementary Material available online at doi: 10.5402/2012/823605). For all selected subtypes/CRFs, the fragment of the env gene encoding gp120 was excised from the full genome, and all sequences were manually aligned using MacClade [55]. Regions of the variable loops that proved difficult to align were excluded in order to avoid false positives due to alignment ambiguities. The final dataset comprised 820 taxa and was 2394 nucleotides (798 amino acids) in length.
2.2. Glycosylation Prediction
Potential glycosylated positions (sequins) were predicted by scanning the amino acid sequences to identify the presences of NX[ST] motifs where X is any amino acid. Studies have shown that NXT motifs where X is P and NXS motifs, where X is P, W, D, E, or L are poor oligosaccharide acceptors and are less likely to be glycosylated [56], and, thus, we discounted these motifs as potential glycosylated positions. Any sequon utilizing the NNTT motif or NNSS motif was categorised as a single glycosylation event as steric occlusion means that glycosylation at both adjacent positions is highly unlikely. To enable direct comparisons between subtypes within this study and between other studies all results presented here correspond to HXB2 gp120 amino acid numbering. Positions predicted as glycosylated in more than 75% of sequences from at least one of the subtypes/CRFs were identified and, as such, represent highly conserved glycosylated positions.
2.3. Glycosylation within Variable Loops
Due to their high sequence diversity, the variable loops could not be aligned directly to each other. For each gp120 sequence, each of the variable loops was extracted and analysed independently of all other sequences without prior alignment. Therefore, while direct comparison between specific glycosylated positions was not possible, identification of the number of predicted glycosylated positions in each sequence as well as correlation of these numbers with variable loop length was performed. The variable loops were defined according to the information contained on the LANL HIV Sequence Database with V1/V2 ranging from 131C to 195S, V3 from 296C to 330H, V4 from 385C to 418P and V5 from 461S to 470P (HXB2 numbering).
2.4. Gp120 Structure Analysis
3D structural analysis was performed using the gp120 V3 loop-containing structure covering residues 84V-492E [57]. An in silico approach was used to add sugars to the gp120 structure using GlyProt [58] with the type of carbohydrate added, where available, having been previously determined [59]. It must be noted, however, that these carbohydrate types are only used for representation purposes and there is a strong likelihood that carbohydrate type may differ between subtypes/CRFs. The gp120 structure corresponds to a subtype B virus [57] and, where necessary, the amino acid sequence of the structure was mutated using MacPyMol, the Macintosh version of PyMol (http://pymol.sourceforge.net/).
3. Results
In general, the average numbers of predicted glycosylated positions are relatively conserved across the HIV-1 group M subtypes/CRFs showing an average of 15-17 glycosylated positions in gp120. In order to identify sequons that are highly conserved within and/or between subtypes, all positions where sequons were identified in greater than 75% of sequences for at least one subtype/CRF were plotted (Figure 1). Positions 88N, 156N, 160N, 197N, 234N, 241N, 262N, 276N, 301N, 356N, 386N, 392N, and 448N all showed extremely high levels of conservation of glycosylation across all subtypes/CRFs studied. A number of subtype-specific patterns were identified, however, with 230N and 442Q showing high levels of predicted glycosylation within subtype C (81% and 76% of subtype C sequences, resp.) when compared to all other subtypes/CRFs (Figure 1). Also, position 295N is predicted as being glycosylated in a smaller number of subtype A and C sequences (47% and 16% resp.), when compared with the other subtypes/CRFs (Figure 1). Position 334S is predicted as glycosylated in 95% of CRF01_AE sequences with it only being observed as glycosylated in between 6% and 22% of sequences representing the other subtypes/CRFs (Figure 1). CRF01_AE also shows very low predicted levels of glycosylation at position 332N while between 74% and 85% of sequences representing the other subtypes/CRFs are predicted as glycosylated at 332N.
Figure 1.
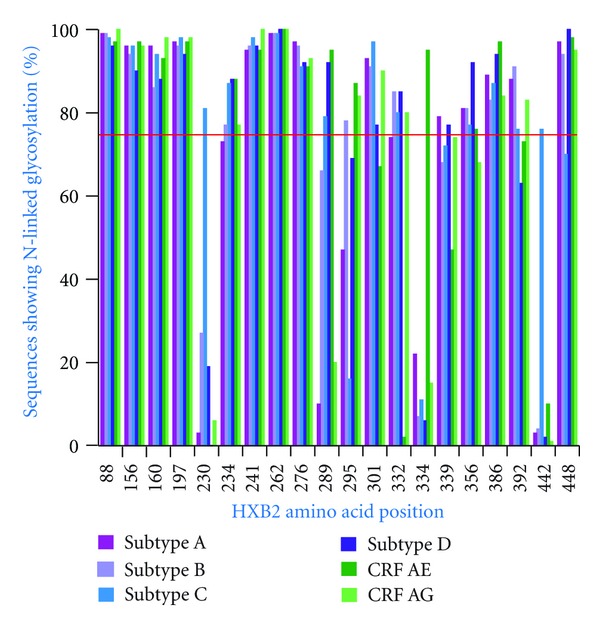
Prevalence of glycosylation at selected positions. The relative frequency of glycosylation between subtypes/CRFs at positions where at least one subtype/CRFs shows high levels of glycosylation. Positions where greater than 75% of sequences were predicted as glycosylated for at least one subtype/CRF are shown.
3.1. Variable Loop Glycosylation
As a number of domains within the variable loops were impossible to align with confidence, these had been excluded from the analysis of glycosylation in gp120 in all subtypes and CRFs. For each sequence representing the subtypes/CRFs, glycosylation within the variable loops was studied without prior alignment. While glycosylation at specific positions cannot be directly compared within or between subtypes/CRFs, these results do give us an overview to the levels of glycosylation within the variable loops. The average length of the V1/V2 loop for each of the subtypes/CRFs ranged between 65 and 69 (Table 1) amino acids suggesting that V1/V2 length is relatively conserved between the majority of virus strains in all subtypes/CRFs. Within the V1/V2 domain there was a large spread of predicted glycosylated positions ranging from one to eleven positions with a mean of six glycosylated positions for all of the subtypes/CRFs (Table 1). Interestingly, regression analysis suggests that as the length of the V1/V2 variable loop increases so does the number of glycosylated positions suggesting that the two are dependent (data not shown). For all subtypes/CRFs, the V3 loop averaged 34 amino acids in length and had a highly conserved glycosylation pattern with the vast majority only predicted as glycosylated at one position (301N) and a small number of sequences from B (2%), C (2%), AE (1%), and AG (2%) being predicted as glycosylated at two positions within V3 (Table 1). Both V4 and V5 show conserved patterns of glycosylation between the subtypes/CRFs with an average of 4-5 glycosylated positions for V4 and 1-2 for V5 (Table 1).
Table 1.
gp120 variable loop glycosylation. The average, median, range of lengths (number of amino acids), and number of predicted N-linked glycosylated positions for the gp120 variable loops of each of the HIV-1 group M subtypes and CRFs studied.
| A | B | C | D | AE | AG | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AV | MED | RA | AV | MED | RA | AV | MED | RA | AV | MED | RA | AV | MED | RA | AV | MED | RA | |
| Length | ||||||||||||||||||
|
| ||||||||||||||||||
| V1/V2 | 68 | 68 | 57–92 | 68 | 68 | 46–85 | 68 | 67 | 51–89 | 65 | 67 | 57–82 | 69 | 69 | 55–85 | 66 | 65 | 53–89 |
| V3 | 34 | 34 | 25–35 | 34 | 34 | 31–38 | 34 | 34 | 24–36 | 33 | 33 | 33–35 | 34 | 34 | 34–35 | 34 | 34 | 33–34 |
| V4 | 31 | 30 | 22–40 | 31 | 31 | 15–41 | 27 | 27 | 17–39 | 30 | 30 | 21–37 | 27 | 27 | 18–39 | 31 | 31 | 24–38 |
| V5 | 10 | 10 | 9–13 | 11 | 11 | 9–19 | 11 | 11 | 7–19 | 11 | 10.5 | 8–16 | 10 | 10 | 8–13 | 10 | 10 | 9–15 |
|
| ||||||||||||||||||
| Number of glycosylated positions | ||||||||||||||||||
|
| ||||||||||||||||||
| V1/V2 | 6 | 6 | 3–10 | 6 | 6 | 3–9 | 6 | 6 | 1–10 | 6 | 6 | 3–8 | 6 | 6 | 3–11 | 6 | 6 | 3–10 |
| V3 | 1 | 1 | NA | 1 | 1 | 1-2 | 1 | 1 | 1-2 | 1 | 1 | NA | 1 | 1 | 1-2 | 1 | 1 | 1-2 |
| V4 | 5 | 5 | 2–7 | 5 | 5 | 2–7 | 4 | 4 | 1–7 | 5 | 5 | 2–7 | 4 | 4 | 2–6 | 5 | 5 | 3–7 |
| V5 | 2 | 2 | 1–3 | 1 | 1 | 1–3 | 1 | 1 | 1–4 | 2 | 2 | 1–3 | 2 | 2 | 1–3 | 2 | 2 | 1–3 |
AV: average, MED: median, RA: range.
3.2. 2G12 Conservation
The 2G12 neutralizing antibody targets carbohydrates bound to the gp120 surface [24], yet it has been shown to have low efficacy against subtype C and CRF01_AE [27, 28]. It has been identified that the 2G12 epitope is centred around positions 295N, 332N, and 392N while peripheral glycosylated positions 339N, 386N, and 448N are not critical for (but play a role in) 2G12 binding [25, 26]. Previous work has suggested that glycosylation at the core sites of the 2G12 epitope is well conserved, with the exception of 295N in subtype C [53]. In this study, however, we find that glycosylation of all of the core sites of the 2G12 epitope (295N, 332N, and 392N) is not particularly well conserved ranging from 1% of CRF01_AE sequences to 61% of subtype B sequences (Table 2). Subtype C and CRF01_AE, in particular, show very low conservation of glycosylation at the 2G12 epitope core, 11% and 1%, respectively (Table 2). In subtype C the reason glycosylation at the core positions of the 2G12 epitope is so poorly conserved is because position 295N is only predicted as glycosylated in 16% of subtype C sequences, while for CRF01_AE position, 332N is only glycosylated in 2% of sequences (Figure 1). When the proposed peripheral positions (339N, 386N, and 448N) are included, the conservation of glycosylation of the extended 2G12 epitope drops to 1% for CRF01_AE up to 36% for subtype B (Table 2).
Table 2.
2G12 conservation percentage of sequences representing each of the subtypes/CRFs exhibiting glycosylation at all positions in the conserved 2G12 core epitope (295N, 332N, and 392N) and the core epitope with peripheral glycans that have been suggested to be involved in 2G12 binding (339N, 386N, and 448N).
| Subtype/CRF | Core epitope | Core + peripheral glycans |
|---|---|---|
| A | 33% | 27% |
| B | 61% | 36% |
| C | 11% | 5% |
| D | 35% | 23% |
| CRF01_AE | 1% | 1% |
| CRF02_AG | 60% | 35% |
3.3. Conservation of Glycosylation at Multiple Positions
In order to examine the levels of conservation of glycosylation within the subtypes/CRFs studied, we identified any glycosylated pairs where both positions were predicted to be glycosylated together in greater than 90% of sequences representing a particular subtype/CRF. The number of highly conserved glycosylated pairs ranged from 16 (subtype D) to 44 pairs (CRF01_AE; Supplementary Table 2) with only five pairs (88 and 241, 88 and 262, 197 and 262, 241 and 262, and 262 and 276) observed as conserved over all the subtypes/CRFs. Interestingly, all subtypes/CRFs showed networks where glycosylation of all residues in the network is highly conserved with all other members in the network (Figure 2). CRF01_AE shows the strongest network of dependency with 11 positions all showing high conservation of glycosylation among them while subtype D only shows a conserved network containing four residues (Figure 2). Similarly, each subtype/CRF shows a number of highly conserved pairs which do not comprise the core conserved network (Supplementary Table 2).
Figure 2.
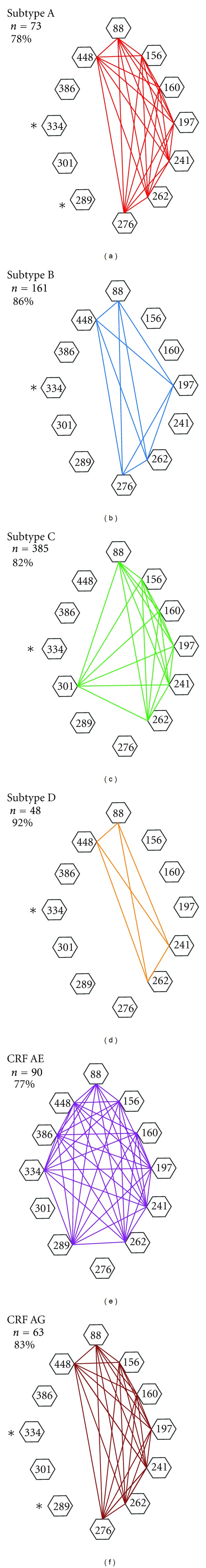
Networks of positions with highly conserved glycosylation. Networks of highly conserved glycosylated pairs identified in each HIV-1 group M subtype/CRF. Pairs where greater than 90% of sequences exhibit glycosylation at both positions are joined by a line. In all cases, the networks are mutually dependent such that each position in the network is seen as highly dependent with every other position in the network. The number of sequence in each dataset is represented as n while the percentage of sequences glycosylated at all of the positions in the network for the respective subtype/CRF is also shown. Positions that are predicted as N-linked glycosylated in less than 25% of sequences for a particular subtype/CRF are marked with a ∗.
4. Discussion
It has been previously suggested that the glycan shield covering the HIV gp120 trimer exposed on the surface of a virion may provide a novel target for anti-HIV treatments [18, 23, 40]. While the targeting of these glycans provides an exciting prospect in HIV treatments, very little is known about the conservation and dependencies of glycosylation within and between the various HIV-1 group M subtypes and circulating recombinant forms. Here, we have performed a comprehensive study examining the glycosylation patterns present in all major circulating forms of HIV-1.
Previous work examining glycosylation patterns in a subset of HIV-1 group M sequences suggested that despite the extreme genetic variability between the HIV-1 group M subtypes and CRFs, the patterns of glycosylation are essentially conserved between them [53]. We have observed such a pattern at some position, with 13 amino acid positions in particular exhibiting high levels of predicted glycosylation across all subtypes/CRFs studied (Figure 1). However, we do find that a number of amino acid positions show strong subtype/CRF-specific glycosylation patterns (Figure 1) and that these positions form an essential part of the epitopes for many BCN antibodies [25, 26, 30, 31, 33].
4.1. Conservation of the Epitopes of BCN Antibodies
Of all of the neutralising antibodies that target the glycan shield, the epitope for 2G12 is the most well defined and is centred around glycosylation of positions 295N, 332N, and 392N in gp120 while peripheral glycosylated positions 339N, 386N, and 448N are not critical for (but play a role in) 2G12 binding [25, 26]. Previous work has suggested that glycosylation at the core sites of the 2G12 epitope is well conserved, with the exception of 295N in subtype C [53]. By studying all available env sequences, we find, however, that glycosylation of the core 2G12 epitope (295N, 332N, and 392N) is, in fact, poorly conserved across all HIV-1 group M subtypes and recombinants ranging from 1% of CRF01_AE sequences to 61% of subtype B. Including the proposed peripheral positions (339N, 386N, and 448N), the conservation of glycosylation of the 2G12 epitope drops to 1% for CRF01_AE up to 36% for subtype B. For subtype C and CRF01_AE, this low level of 2G12 conservation is not surprising as poor reactivity of 2G12 against them is well documented [27, 28, 60] due to the low levels of glycosylation at positions 295 and 332 in subtype C and CRF01_AE, respectively.
Binley and colleagues have clearly demonstrated the poor reactivity of 2G12 against both subtype C and CRF01_AE [27]. Examining the CRF01_AE nucleotide sequences from their study shows that two of the 2G12 core positions (295 and 392) are glycosylated in 100% of sequences (n = 9), and that the third core position (332) is not glycosylated in any of the sequences. However, 100% of the CRF01_AE sequences are glycosylated at 334, a position that has been suggested to compensate for loss of glycosylation at 332 in the 2G12 epitope [61]. Despite this, none of the CRF01_AE viruses were neutralized by 2G12 [27] suggesting that glycosylation at position 334 cannot replace 332 as part of the 2G12 epitope. Similarly, we have shown here that 96% of CRF01_AE sequences are glycosylated at position 334 instead of 332, and if 334 was truly compensating for 332 as part of the 2G12 epitope, one would expect CRF01_AE to be more sensitive to neutralisation than is observed [27, 28, 60].
The vast majority of subtype C viruses (84%, Figure 1) are not glycosylated at position 295, the position suggested to be responsible for the lack of subtype C neutralisation by 2G12 [28]. Gray and colleagues suggest that reintroduction of glycosylation at position 295 is not sufficient to restore 2G12 sensitivity, and that positions 442 and 448 are also responsible for conferring sensitivity to 2G12 [28]. This concurs with our examination of the sequence data from the study by Binley and colleagues where the only subtype C virus glycosylated at all positions of the 2G12 core epitope (295, 332, and 392) was not neutralized by 2G12 (most likely because it is not glycosylated at position 442). The remaining subtype C sequences (n = 11) show no glycosylation at position 295 with 10 of these being glycosylated at position 442. Consistent with previous work, none of these subtype C viruses were neutralized by 2G12 [27]. Only 4% of subtype C viruses show glycosylation at all of the core 2G12 epitope positions (295, 332, and 392) plus positions 442 and 448 suggesting that perhaps a very low number of wild-type subtype C viruses (containing the appropriate glycosylation pattern) may be neutralized by 2G12. While it has been previously suggested that 295 and 442 are mutually exclusive in subtype C [54], we see that 7% of subtype C sequences are glycosylated at both positions 295 and 442. The vast majority (77%) of subtype C sequences, however, show glycosylation at either position 295 or 442.
We observe that carbohydrates bound to positions 332 and 334 occupy overlapping three-dimensional space on the gp120 structure, as do positions 295 and 442. Therefore, it appears that while subtype C and CRF01_AE can evade neutralisation by 2G12 through the absence of glycosylation at essential positions, unique glycosylation at structurally proximal positions means that the protective qualities of the gp120 glycan shield are fully preserved.
Similarly, glycosylation at position 332 has been shown to significantly affect susceptibility to other recently described BCN antibodies [30–33] with Moore and colleagues observing that viruses that are glycosylated at position 334 are not susceptible to neutralisation by BCN antibodies described in their study [33]. Walker and colleagues suggest that positions 332 and/or 301 are important for neutralisation by at least six of the BCN antibodies described in their study [30] and this observation was supported by a structural study by Pejchal and colleagues [31]. While we observe that glycosylation at position 301 is relatively highly conserved across all of the subtypes and CRFs, the low level of glycosylation at position 332 in CRF01_AE means that it is rarely susceptible to neutralisation by any of the known BCN antibodies.
Thus, if fully broadly cross-clade neutralising synthetic anti-carbohydrate therapies are to be investigated, researchers should take into account the glycosylation anomalies observed in CRF01_AE and focus on amino acid positions where glycosylation is conserved across the entire spectrum of the HIV-1 group M subtypes and circulating recombinant forms. In order to identify such positions, we employed an approach to identify pairs and networks of glycosylated residues that show high levels of conservation across all of the HIV-1 group M subtypes and recombinant forms. Only five pairs of highly dependent glycosylated residues were identified as being conserved over all of the subtypes/CRFs (Figure 2). Of these five pairs only positions 88N and 241N exhibit similar levels of three-dimensional proximity (12.5 Å) to those of the 2G12 epitope (mean of 14.6 Å) when mapped onto the V3-containing gp120 core structure [57]. Similarly, positions 262N and 448N show high levels of glycosylation dependency in all but subtype C (Figure 2) and are proximal in the 3D structure. Whilst glycosylation at positions 262N and 448N is not identified as significant (shared in greater than 90% of sequences) for subtype C, the level of conservation (69%) is still greater than that of the 2G12 epitope in subtype B (61%). Similarly, networks of highly conserved glycosylated positions were identified for all subtypes/CRFs (Figure 2). While these highly conserved pairs/networks of glycosylated residues are evident, it is impossible to know whether these perceived dependencies are mutually dependent. Despite the extreme sequence diversity observed between HIV-1 group M subtypes/CRFs, glycosylation of these positions is highly conserved suggesting that there is a functional constraint on retaining glycosylation of these positions. These glycosylated positions form potential epitopes that are highly conserved across all HIV-1 group M subtypes and recombinants, and, as such, further investigation of potential epitopes for the development of BCN synthetic anti-carbohydrate therapeutics is warranted.
5. Conclusions
Here, we have shown that while N-linked glycosylation at many positions is highly conserved across the HIV-1 group M subtypes and CRFs, strong strain-specific glycosylation patterns exist and appear to affect the susceptibility broadly cross-neutralising antibodies. We suggest that alternate glycosylation patterns in some HIV strains allow evasion from neutralisation from BCN antibodies while still maintaining the overall protective qualities of the gp120 glycan shield. Further, we have identified highly conserved, structurally proximal, clusters of glycosylated positions that should be investigated further to explore their potential as epitopes against which BCN carbohydrate-binding therapies can be developed.
Supplementary Material
For each subtype/CRF, all available full length sequences were retrieved from the Los Alamos National Laboratory (LANL) HIV Sequence Database (http://www.hiv.lanl.gov/). Using the patient and isolate annotation, where available, multiple sequences representing single individuals were removed to ensure that the dataset representing each subtype/CRF was non-redundant. The fragment of the envelope gene that encodes for gp120 was excised from each full genome and separate datasets generated for each of the subtypes/CRFs studied. This supplementary table details all of the DNA sequences used in this study together with their designated subtype/CRF, genbank accession number, the country of origin and, where available, the year of sampling.
Acknowledgments
This work was supported by the Irish Health Research Board Grant (RP/2006/141) and the South African National Research Foundation Blue Skies award (Grant no. 75899) to S. A. Travers. The authors thank Mario Fares for assistance with regression analyses and Jason Egan and Chris McCrohan for assistance in obtaining the data for the circulating recombinant forms.
References
- 1.Land A, Braakman I. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie. 2001;83(8):783–790. doi: 10.1016/s0300-9084(01)01314-1. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Luo L, Rasool N, Kang CY. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. Journal of Virology. 1993;67(1):584–588. doi: 10.1128/jvi.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. Journal of Biological Chemistry. 1990;265(18):10373–10382. [PubMed] [Google Scholar]
- 4.Mizuochi T, Spellman MW, Larkin M, Solomon J, Basa LJ, Feizi T. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochemical Journal. 1988;254(2):599–603. doi: 10.1042/bj2540599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myers G, MacInnes K, Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Research and Human Retroviruses. 1992;8(3):373–386. doi: 10.1089/aid.1992.8.373. [DOI] [PubMed] [Google Scholar]
- 6.Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 7.Ohgimoto S, Shioda T, Mori K, Nakayama EE, Hu H, Nagai Y. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. Journal of Virology. 1998;72(10):8365–8370. doi: 10.1128/jvi.72.10.8365-8370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schønning K, Jansson B, Olofsson S, Hansen JES. Rapid selection for an N-linked oligosaccharide by monoclonal antibodies directed against the V3 loop of human immunodeficiency virus type 1. Journal of General Virology. 1996;77(4):753–758. doi: 10.1099/0022-1317-77-4-753. [DOI] [PubMed] [Google Scholar]
- 9.Bolmstedt A, Hinkula J, Rowcliffe E, Biller M, Wahren B, Olofsson S. Enhanced immunogenicity of a human immunodeficiency virus type 1 env DNA vaccine by manipulating N-glycosylation signals: effects of elimination of the V3 N306 glycan. Vaccine. 2001;20(3-4):397–405. doi: 10.1016/s0264-410x(01)00358-9. [DOI] [PubMed] [Google Scholar]
- 10.Chackerian B, Rudensey LM, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. Journal of Virology. 1997;71(10):7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Brown A, Harouse J, Luciw PA, Mayer AJ. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIV(SF33A) variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. Journal of Virology. 1999;73(7):5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang SM, Shi Quan F, Huang C, et al. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology. 2005;331(1):20–32. doi: 10.1016/j.virol.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 13.McCaffrey RA, Saunders C, Hensel M, Stamatatos L. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. Journal of Virology. 2004;78(7):3279–3295. doi: 10.1128/JVI.78.7.3279-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. Journal of Virology. 2006;80(19):9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost SDW, Wrin T, Smith DM, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(51):18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derdeyn CA, Decker JM, Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303(5666):2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Salazar-Gonzalez JF, Derdeyn CA, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. Journal of Virology. 2006;80(23):11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446(7139):1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Hoffman F, He J, et al. Characterization of HIV-1 subtype C envelope glycoproteins from perinatally infected children with different courses of disease. Retrovirology. 2006;3, article 73 doi: 10.1186/1742-4690-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp 120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyatt R, Kwong PD, Desjardins E, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 22.Pantophlet R, Ollmann Saphire E, Poignard P, Parren PWHI, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. Journal of Virology. 2003;77(1):642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balzarini J. Targeting the glycans of gp120: a novel approach aimed at the Achilles heel of HIV. Lancet Infectious Diseases. 2005;5(11):726–731. doi: 10.1016/S1473-3099(05)70271-1. [DOI] [PubMed] [Google Scholar]
- 24.Trkola A, Purtscher M, Muster T, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. Journal of Virology. 1996;70(2):1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanlan CN, Pantophlet R, Wormald MR, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1—>2 mannose residues on the outer face of gp120. Journal of Virology. 2002;76(14):7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders RW, Venturi M, Schiffner L, et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. Journal of Virology. 2002;76(14):7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binley JM, Wrin T, Korber B, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. Journal of Virology. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray ES, Moore PL, Pantophlet RA, Morris L. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. Journal of Virology. 2007;81(19):10769–10776. doi: 10.1128/JVI.01106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bures R, Morris L, Williamson C, et al. Regional clustering of shared neutralization determinants on primary isolates of Clade C human immunodeficiency virus type 1 from South Africa. Journal of Virology. 2002;76(5):2233–2244. doi: 10.1128/jvi.76.5.2233-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LM, Huber M, Doores KJ, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pejchal R, Doores KJ, Walker LM, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334(6059):1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nandi A, Lavine CL, Wang P, et al. Epitopes for broad and potent neutralizing antibody responses during chronic infection with human immunodeficiency virus type 1. Virology. 2010;396(2):339–348. doi: 10.1016/j.virol.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore PL, Gray ES, Wibmer CK, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nature Medicine. 2012;18(11):1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balzarini J, Neyts J, Schols D, et al. The mannose-specific plant lectins from Cymbidium hybrid and Epipactis helleborine and the (N-actylglucosamine)(n)-specific plant lectin from Urtica dioica are potent and selective inhibitors of human immunodeficiency virus and cytomegalovirus replication in vitro. Antiviral Research. 1992;18(2):191–207. doi: 10.1016/0166-3542(92)90038-7. [DOI] [PubMed] [Google Scholar]
- 35.Boyd MR, Gustafson KR, McMahon JB, et al. Discovery of cyanovirin-N, a novel human immunodeficiency virus- inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrobial Agents and Chemotherapy. 1997;41(7):1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolmstedt AJ, O’Keefe BR, Shenoy SR, Mcmahon JB, Boyd MR. Cyanovirin-N defines a new class of antiviral agent targeting N-linked, high-mannose glycans in an oligosaccharide-specific manner. Molecular Pharmacology. 2001;59(5):949–954. doi: 10.1124/mol.59.5.949. [DOI] [PubMed] [Google Scholar]
- 37.Bokesch HR, O’Keefe BR, McKee TC, et al. A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Biochemistry. 2003;42(9):2578–2584. doi: 10.1021/bi0205698. [DOI] [PubMed] [Google Scholar]
- 38.Balzarini J, Hatse S, Vermeire K, et al. Mannose-specific plant lectins from the Amaryllidaceae family qualify as efficient microbicides for prevention of human immunodeficiency virus infection. Antimicrobial Agents and Chemotherapy. 2004;48(10):3858–3870. doi: 10.1128/AAC.48.10.3858-3870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai CC, Emau P, Jiang Y, et al. Cyanovirin-N gel as a topical microbicide prevents rectal transmission of SHIV89.6P in macaques. AIDS Research and Human Retroviruses. 2003;19(7):535–541. doi: 10.1089/088922203322230897. [DOI] [PubMed] [Google Scholar]
- 40.Balzarini J, Van Laethem K, Daelemans D, et al. Pradimicin A, a carbohydrate-binding nonpeptidic lead compound for treatment of infections with viruses with highly glycosylated envelopes, such as human immunodeficiency virus. Journal of Virology. 2007;81(1):362–373. doi: 10.1128/JVI.01404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witvrouw M, Fikkert V, Hantson A, et al. Resistance of human immunodeficiency virus type 1 to the high-mannose binding agents cyanovirin N and concanavalin A. Journal of Virology. 2005;79(12):7777–7784. doi: 10.1128/JVI.79.12.7777-7784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balzarini J, Van Laethem K, Peumans WJ, et al. Mutational pathways, resistance profile, and side effects of cyanovirin relative to human immunodeficiency virus type 1 strains with N-glycan deletions in their gp120 envelopes. Journal of Virology. 2006;80(17):8411–8421. doi: 10.1128/JVI.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–57. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 44.Peeters M, Sharp PM. Genetic diversity of HIV-1: the moving target. AIDS. 2000;14(3):S129–S140. [PubMed] [Google Scholar]
- 45.Buonaguro L, Tornesello ML, Buonaguro FM. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. Journal of Virology. 2007;81(19):10209–10219. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 47.Osmanov S, Pattou C, Schwardländer B, et al. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. Journal of Acquired Immune Deficiency Syndromes. 2002;29(2):184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- 48.Menu E, Lien TTX, Lafon ME, et al. HIV type 1 Thai subtype E is predominant in South Vietnam. AIDS Research and Human Retroviruses. 1996;12(7):629–633. doi: 10.1089/aid.1996.12.629. [DOI] [PubMed] [Google Scholar]
- 49.Motomura K, Kusagawa S, Kato K, et al. Emergence of new forms of human immunodeficiency virus type 1 intersubtype recombinants in central Myanmar. AIDS Research and Human Retroviruses. 2000;16(17):1831–1843. doi: 10.1089/08892220050195793. [DOI] [PubMed] [Google Scholar]
- 50.Piyasirisilp S, McCutchan FE, Carr JK, et al. A recent outbreak of human immunodeficiency virus type 1 infection in Southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. Journal of Virology. 2000;74(23):11286–11295. doi: 10.1128/jvi.74.23.11286-11295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCutchan FE, Carr JK, Bajani M, et al. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology. 1999;254(2):226–234. doi: 10.1006/viro.1998.9505. [DOI] [PubMed] [Google Scholar]
- 52.Montavon C, Toure-Kane C, Liegeois F, et al. Most env and gag subtype A HIV-1 viruses circulating in west and West Central Africa are similar to the’ prototype AG recombinant virus IBNG. Journal of Acquired Immune Deficiency Syndromes. 2000;23(5):363–374. doi: 10.1097/00126334-200004150-00001. [DOI] [PubMed] [Google Scholar]
- 53.Zhang M, Gaschen B, Blay W, et al. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14(12):1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 54.Poon AFY, Lewis FI, Kosakovsky Pond SL, Frost SDW. Evolutionary interactions between N-Linked glycosylation sites in the HIV-1 envelope. PLoS Computational Biology. 2007;3(1, article e11):0110–0119. doi: 10.1371/journal.pcbi.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maddison WP, Maddison DR. MacClade. 4.5 edition. Sunderland, Mass, USA: Sinauer; 1992. [Google Scholar]
- 56.Kasturi L, Chen H, Shakin-Eshleman SH. Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochemical Journal. 1997;323(2):415–419. doi: 10.1042/bj3230415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang CC, Tang M, Zhang MY, et al. Structural biology: structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohne-Lang A, Von der Lieth CW. GlyProt: in silico glycosylation of proteins. Nucleic Acids Research. 2005;33(2):W214–W219. doi: 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;39(37):11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 60.Trkola A, Pomales AB, Yuan H, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. Journal of Virology. 1995;69(11):6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manrique A, Rusert P, Joos B, et al. In vivo and in vitro escape from neutralizing antibodies 2G12, 2F5, and 4E10. Journal of Virology. 2007;81(16):8793–8808. doi: 10.1128/JVI.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For each subtype/CRF, all available full length sequences were retrieved from the Los Alamos National Laboratory (LANL) HIV Sequence Database (http://www.hiv.lanl.gov/). Using the patient and isolate annotation, where available, multiple sequences representing single individuals were removed to ensure that the dataset representing each subtype/CRF was non-redundant. The fragment of the envelope gene that encodes for gp120 was excised from each full genome and separate datasets generated for each of the subtypes/CRFs studied. This supplementary table details all of the DNA sequences used in this study together with their designated subtype/CRF, genbank accession number, the country of origin and, where available, the year of sampling.


