Abstract
Background
Redox-cofactor balancing constrains product yields in anaerobic fermentation processes. This challenge is exemplified by the formation of glycerol as major by-product in yeast-based bioethanol production, which is a direct consequence of the need to reoxidize excess NADH and causes a loss of conversion efficiency. Enabling the use of CO2 as electron acceptor for NADH oxidation in heterotrophic microorganisms would increase product yields in industrial biotechnology.
Results
A hitherto unexplored strategy to address this redox challenge is the functional expression in yeast of enzymes from autotrophs, thereby enabling the use of CO2 as electron acceptor for NADH reoxidation. Functional expression of the Calvin cycle enzymes phosphoribulokinase (PRK) and ribulose-1,5-bisphosphate carboxylase (Rubisco) in Saccharomyces cerevisiae led to a 90% reduction of the by-product glycerol and a 10% increase in ethanol production in sugar-limited chemostat cultures on a mixture of glucose and galactose. Co-expression of the Escherichia coli chaperones GroEL and GroES was key to successful expression of CbbM, a form-II Rubisco from the chemolithoautotrophic bacterium Thiobacillus denitrificans in yeast.
Conclusions
Our results demonstrate functional expression of Rubisco in a heterotrophic eukaryote and demonstrate how incorporation of CO2 as a co-substrate in metabolic engineering of heterotrophic industrial microorganisms can be used to improve product yields. Rapid advances in molecular biology should allow for rapid insertion of this 4-gene expression cassette in industrial yeast strains to improve production, not only of 1st and 2nd generation ethanol production, but also of other renewable fuels or chemicals.
Keywords: Metabolic engineering; Synthetic biology; Rubisco; Ribulose-1,5-bisphosphate carboxylase; Phosphoribulokinase; NADH re-oxidation; Carbon dioxide fixation; Saccharomyces cerevisiae; Glycerol; Bioethanol
Background
The yeast Saccharomyces cerevisiae is not only used for the large-scale production of fuel ethanol [1], but also for industrial production of a broad and rapidly expanding range of other chemical compounds from renewable carbohydrate feedstocks [2,3]. In anaerobic, ethanol-producing cultures of S. cerevisiae, excess NADH generated from biosynthetic reactions, such as NAD+-dependent oxidative decarboxylations involved in synthesis of the precursors acetyl-CoA and 2-oxoglutarate, is reoxidized by reducing part of the sugar substrate to glycerol [4]. In growing anaerobic yeast cultures, glycerol production typically accounts for 4-10% of the total sugar consumption and therefore has a significant impact on ethanol yields and process economy in both 1st and 2nd generation large-scale bioethanol production [5,6].
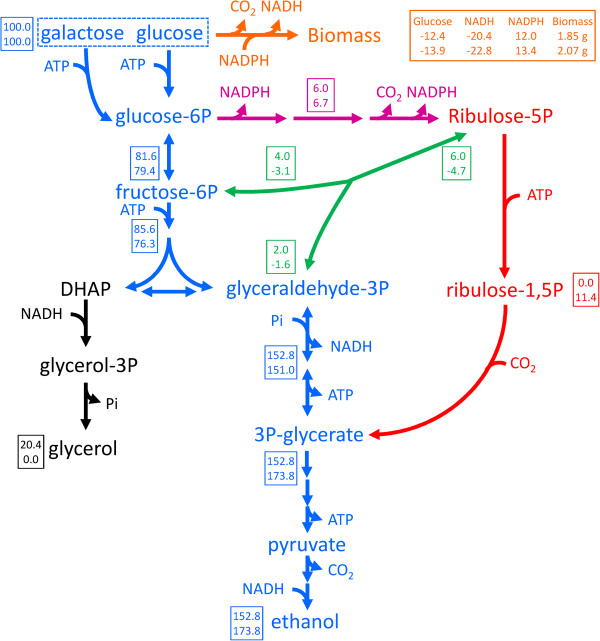
Using CO2 as electron acceptor for the reoxidation of NADH would be a highly attractive metabolic engineering strategy, in particular when CO2 reduction can be coupled to the formation of the product of interest. Functional expression of the Calvin cycle enzymes phosphoribulokinase (PRK) and ribulose-1,5-bisphosphate carboxylase (Rubisco) in S. cerevisiae should enable the coupling of CO2, a major product of alcoholic fermentation, to ribulose-5-phosphate, a normal intermediate of the S. cerevisiae pentose-phosphate pathway (Figure 1). The resulting two molecules of 3-phosphoglycerate can subsequently be converted to 2 molecules each of ethanol and CO2, with the concomitant net oxidation of 2 molecules of NADH to NAD+ (Figure 1). When ribulose-5-phosphate is formed via the oxidative pentose-phosphate pathway (Figure 1), this route results in a transhydrogenase-type conversion of redox cofactors (NADP+ + NADH → NADPH + NAD+). Since the total amount of NADPH required in biosynthesis is smaller than the amount of NADH generated [7], such a transhydrogenase-like activity cannot fully replace glycerol formation as a mechanism for reoxidizing biosynthetic NADH. However, no such constraint exists when ribulose-5-phosphate is formed from intermediates of glycolysis via the rearrangement reactions of the non-oxidative pentose-phosphate pathway (Figure 1). A theoretical analysis shows that complete replacement of glycerol production with CO2 incorporation through PRK and Rubisco can increase the ethanol yield of sugar by as much as 14% (Figure 1).
Figure 1.
Schematic representation of central carbon metabolism and the introduced Calvin-cycle enzymes in Saccharomyces cerevisiae. Orange: Formation of biomass and NADH from glucose and NADPH. Stoichiometries are according to Verduyn et al.[7]; Blue: Redox-neutral, ATP-yielding alcoholic fermentation of glucose and galactose via the Embden-Meyerhof-Parnas glycolysis and Leloir pathways, respectively; Magenta: NADPH generation via the oxidative part of the pentose-phosphate pathway; Green: rearrangement of sugar-phosphate carbon skeletons via the non-oxidative pentose-phosphate pathway; Black: NADH oxidation by formation of glycerol through glycerol-3-phosphate dehydrogenase and glycerol-3-phosphatase; Red: heterologously expressed Calvin-cycle enzymes phosphoribulokinase and Rubisco. Numbers in boxes represents the distribution of carbon along the different pathways (in mmol) normalized for a combined glucose and galactose uptake of 100 mmol for a wild-type, glycerol-producing reference strain (top) and for a scenario in which the alternative pathways via the Calvin cycle enzymes completely replace glycerol formation as the mechanism for reoxidizing NADH formed in biosynthetic reactions (bottom). In the scenario with the Calvin cycle enzymes, ribulose-5-phosphate was assumed to be preferentially derived from the oxidative reactions of the pentose phosphate pathway. Once the generation of NADPH from these reactions matched the requirement for NADPH in biosynthesis, further ribulose-5-phosphate was derived from glycolytic intermediates via the non-oxidative pentose-phosphate pathway rearrangement reactions. The biomass yield on ATP was assumed to be identical for both scenarios.
The PRK gene from Spinacia oleracea[8] has previously been expressed in the yeast Pichia pastoris[9] and is therefore an interesting candidate for heterologous expression in S. cerevisiae. For Rubisco, a key enzyme in the Calvin cycle for autotrophic carbon fixation, three catalytically active forms have been described [10,11]. Prokaryotic form-II Rubisco’s are encoded by single structural genes and several have been heterologously expressed in E. coli[12,13]. Functional expression of form-II Rubisco’s in E. coli was shown to be strongly stimulated by the E. coli protein-folding chaperones GroEL and GroES [14] and expression of Hydrogenovibrio marinus Rubisco in E. coli was further stimulated by co-expression of the CbbO and CbbQ chaperones of the donor organism [15]. Very recently the structure of GroEL/GroES encapsulating Rubisco was visualized by cryo-electron microscopy [16]. Eukaryotes such as S. cerevisiae harbour a chaperone couple (Hsp60/Hsp10) that structurally and functionally resemble GroEL/GroES. However, these proteins are located in the mitochondria, whereas a role in Rubisco expression would require their activity in the cytosol.
In this study we investigated how to achieve functional expression of PRK and Rubisco in yeast. In view of the envisioned benefit of being encoded by single structural genes, a prokarytic form-II Rubisco gene was expressed in S. cerevisiae in combination with the PRK gene from Spinacia oleracea. Both the promoters and coding regions for genes required for glycerol formation were left unchanged compared to the reference strain. Subsequently, the impact of the resulting CO2 incorporation on product formation was studied, with special emphasis on the yields of ethanol and the undesired by-product glycerol.
Results and discussion
Chaperone-mediated functional expression of Rubisco in Saccharomyces cerevisiae
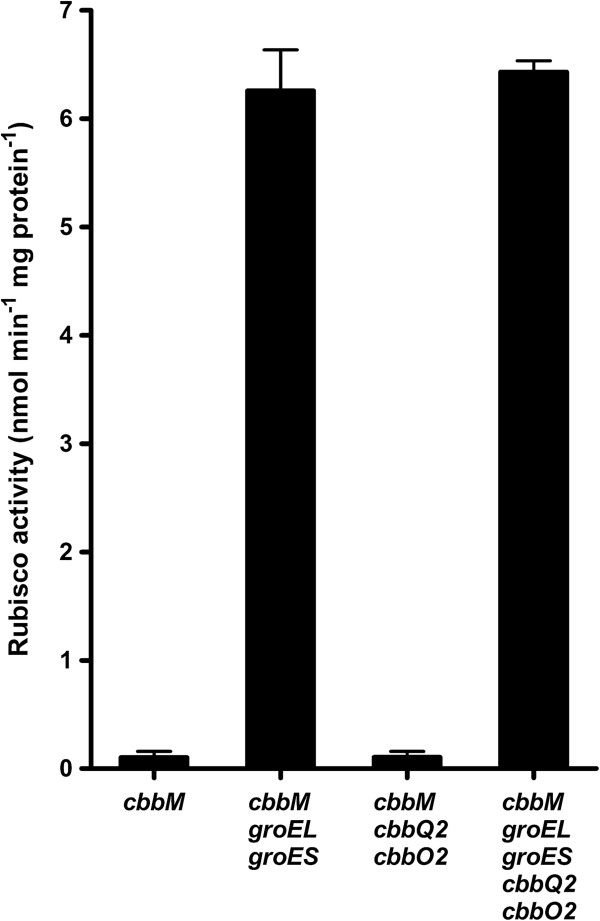
To study a possible requirement of heterologous chaperones for expression of Rubisco in S. cerevisiae, the form-II Rubisco-encoding cbbM gene from T. denitrificans[17] was codon-optimised for expression in S. cerevisiae and expressed from a centromeric vector, both alone and in combination with expression cassettes for the codon-optimised E. coli groEL/groES[18] and/or T. denitrificans cbbO2/cbbQ2 genes [19,20]. Functional expression of T. denitrificans Rubisco in S. cerevisiae, as indicated by ribulose-1,5-bisphosphate-dependent 14CO2 fixation by yeast cell extracts, was only observed upon co-expression of E. coli GroEL/GroES (Figure 2). Co-expression of CbbO2/CbbQ2 did not result in a further increase of Rubisco activity (Figure 2). Co-expression of bacterial chaperones has previously been shown to improve heterologous protein expression in Pichia pastoris and insect cells [21,22]. The positive effect of GroEL/GroES on Rubisco expression in S. cerevisiae demonstrates the potential value of co-expression of heterologous chaperones for metabolic pathway engineering that requires expression of prokaryotic enzymes in the cytosol of eukaryotes.
Figure 2.
Rubisco enzymatic activity in S. cerevisiae strains expressing different synthetic constructs. Specific ribulose-1,5-bisphosphate carboxylase (Rubisco) activity in cell extracts of S. cerevisiae expressing Rubisco form II CbbM from T. denitrificans, either alone (IMC033) or in combination with the E. coli chaperones GroEL/GroES [18] (IMC035), the T. denitrificans chaperones CbbO2/CbbQ2 [20] (IMC034) or all four chaperones (IMC014). Heterologously expressed genes were codon optimised for expression in yeast and expressed from a single centromeric vector. Biomass samples were taken from anaerobic batch cultures on synthetic media (pH 5.0, 30°C), sparged with nitrogen and containing 20 g l-1 glucose as carbon source. Rubisco activities, measured as 14CO2-fixation in cell extracts, in a wild-type reference strain and in S. cerevisiae strains expressing cbbM and cbbM-cbbQ2-cbbO2 were below the detection limit of the enzyme assay (0.2 nmol CO2 min-1 mg protein-1).
Functional expression of phosphoribulokinase in Saccharomyces cerevisiae
The Spinacia oleracea phosphoribulokinase (PRK) gene [8], which has previously been expressed in the yeast Pichia pastoris[9], was integrated together with E. coli groEL/groES and T. denitrificans cbbO2/cbbQ2 into the S. cerevisiae genome at the CAN1 locus, under control of the galactose-inducible GAL1 promotor. This resulted in high PRK activities (approximately 15 μmol mg protein-1 min-1) in cell extracts of S. cerevisiae strain IMU033 taken from carbon-limited chemostat cultures on a mixture of glucose and galactose (Table 1). Although relatively high background activities were measured in the reference strain without PRK (IMU032), this activity does not contribute to pathway activity (see below). We therefore assumed that the background activity observed in the reference strain was caused by an impurity in one of the chemicals used in the assay and did not reflect formation of ribulose-1,5-bisphosphate. The engineered strain IMU033, which additionally carried the centromeric expression cassette for T. denitrificans Rubisco, was used to quantitatively analyse the physiological impacts of the expression of Rubisco and PRK.
Table 1.
Physiological analysis of S. cerevisiae IMU033 expressing PRK and Rubisco and the isogenic reference strain IMU032 in anaerobic chemostat cultures, grown at a dilution rate of 0.05 h-1 on a synthetic medium (pH 5) supplemented with 12.5 g l-1 glucose and 12.5 g l-1 galactose as carbon sources#
| |
IMU032 |
IMU033 |
||
|---|---|---|---|---|
| (Reference strain) | (Expressing PRK and Rubisco) | |||
| CO2 in inlet gas (%) |
0 |
10 |
0 |
10 |
| CO2 in outlet gas (%) |
0.89 ± 0.03 |
10.8 ± 0.0 |
1.02 ± 0.00 |
10.8 ± 0.1 |
| Phosphoribulokinase (μmol mg protein-1 min-1) |
0.58 ± 0.09 |
0.51 ± 0.12 |
14.4 ± 1.5 |
15.2 ± 1.0 |
| Rubisco (nmol mg protein-1 min-1) |
< 0.2* |
< 0.2 |
4.59 ± 0.30 |
2.67 ± 0.28 |
| Biomass yield on sugar (g g-1) |
0.083 ± 0.000a |
0.084 ± 0.000b |
0.093 ± 0.001a |
0.095 ± 0.000b |
| Ethanol yield on sugar (mol mol-1) |
1.56 ± 0.03c |
1.56 ± 0.02d |
1.73 ± 0.02c |
1.73 ± 0.01d |
| Glycerol yield on sugar (mol mol-1) | 0.14 ± 0.00e | 0.12 ± 0.00f | 0.04 ± 0.00e, g | 0.01 ± 0.00f, g |
#Results are represented as average ± mean deviations of data from independent duplicate chemostat experiments. Data pairs labelled with the same subscripts (a,a, b,b, etc.) are considered statistically different in a standard t-test (p <0.02).
*Detection limit of enzyme activity assay.
Carbon dioxide as electron acceptor in anaerobic chemostat cultures of Saccharomyces cerevisiae
Quantitative physiological analysis is facilitated by the constant and highly reproducible process conditions in steady-state chemostat cultures [23,24]. Therefore, ethanol and glycerol yields of PRK- and Rubisco-expressing S. cerevisiae were compared to those of an isogenic reference strain in anaerobic, sugar-limited chemostats on a mixture of 12.5 g l-1 glucose and 12.5 g l-1 galactose. In nitrogen-sparged cultures, the glycerol yield on sugar in the strain expressing both Calvin-cycle enzymes was 68% lower than in the reference strain, while ethanol and biomass yields on sugar were 11% and 12% higher, respectively (Table 1). To investigate whether the low affinity of T. denitrificans form-II Rubisco for CO2 (KCO2 = 0.26 mM [17]) limited its in vivo activity in the nitrogen-sparged cultures, additional chemostats were sparged with a 10%/90% blend of CO2 and N2. Indeed, this CO2 supplementation resulted in a further decrease of the glycerol yield to a value below 10% of that of the reference strain (Table 1). Co-expression of Rubisco and chaperones without co-expression of PRK (strain IMC014) did not result in decreased glycerol yield (0.13 mol mol-1) compared to the reference strain IMU032 (0.12 mol mol-1) in carbon-limited chemostat cultures supplemented with CO2. This observation confirmed that expression of a heterologous phosphoribulokinase (PRK) gene is required for in vivo carbon fixation via Rubisco in yeast.
Carbon dioxide as electron acceptor in anaerobic batch fermentations
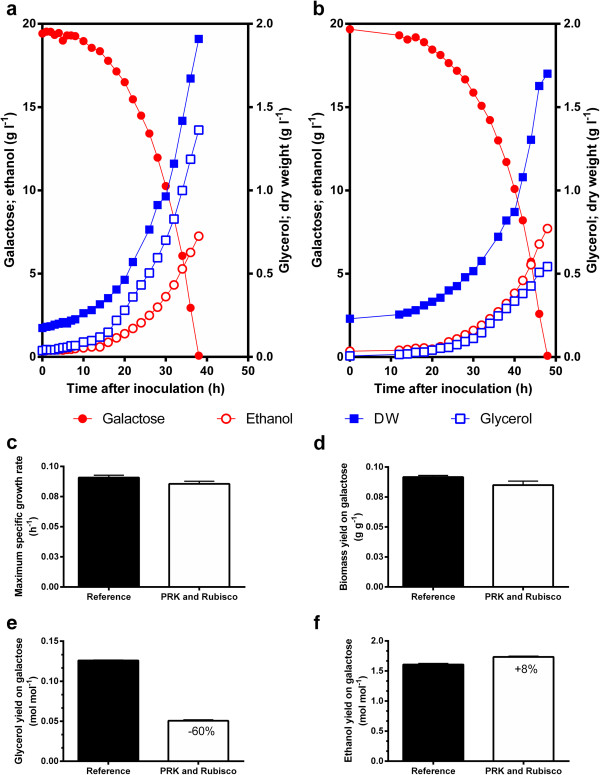
Since industrial-scale ethanol production is routinely performed in batch fermentations [25], the impact of the expression of PRK and Rubisco was also investigated in anaerobic, CO2-supplemented batch cultures (Figure 3). Galactose was used as the carbon source for these experiments to enable efficient expression of PRK from the GAL1 promoter. Despite an almost 10 h difference in the lag phase, the specific growth rates of the engineered and reference strains on galactose in these anaerobic cultures were not significantly different (Figure 3) and in good agreement with values reported elsewhere for this yeast strain family [26]. Consistent with the observations in chemostat cultures, expression of the two Calvin cycle enzymes reduced glycerol formation in the batch cultures by 60% and increased the ethanol yield on galactose by 8% (Figure 3e-f). However, the biomass yield on sugar, which in the chemostat cultures increased according to the predictions, did not increase in the anaerobic batch cultures on galactose. These observations together might indicate that under the galactose excess conditions used for these batch cultivations increased expression levels of PRK result in a small metabolic burden, but still result in an overall positive effect on the ethanol yield.
Figure 3.
Physiological impact of expression of Calvin cycle enzymes on growth, substrate consumption and product formation in galactose-grown anaerobic batch cultures of S. cerevisiae. a: growth curves of isogenic reference strain S. cerevisiae IMU032, b: growth curves of S. cerevisiae IMU033 expressing PRK and Rubisco. Growth conditions: T = 30°C, pH 5.0, 10% CO2 in inlet gas. Each graph represents values for one of two independent replicate experiments, whose growth kinetic parameters differed by less than 5%. c-f: Calculated parameters: Maximum specific growth rate (c), biomass yield (d), glycerol yield (e), and ethanol yield (f) on galactose of the isogenic S. cerevisiae reference (black bars) and strain expressing PRK and Rubisco (white bars). Results are represented as average ± mean deviations of data from independent duplicate cultures. Values inside the white bars represent statistically significant differences in a standard t-test (p value < 0.02) relative to the reference strain.
Conclusions
This study provides a compelling proof of principle for the replacement of glycerol formation as the predominant redox sink in anaerobic yeast metabolism by PRK- and Rubisco-mediated incorporation of CO2 into yeast central carbon metabolism. The loss of sugar feedstock due to glycerol production in industrial bioethanol processes has been estimated at 4% of the consumed sugar [5]. If expression of PRK and Rubisco in industrial yeast strains were to completely eliminate this loss, this could enable an additional production of 5 billion liters of ethanol from the amount of sugar used for the 2011 global ethanol production of 110 billion liters [1]. Use of CO2 as an external electron acceptor offers important advantages over previously proposed strategies for reducing glycerol production in yeast-based bioethanol production. Optimizing the redox cofactor specificity of nitrogen assimilation in S. cerevisiae[5] only enables a partial reduction of glycerol production and its impact further depends on the nitrogen sources present in industrial feedstocks. Similarly, a metabolic engineering strategy that enables NADH-dependent reduction of acetic acid [6] to ethanol is dependent on the presence of acetic acid in industrial feedstocks. Further optimization of PRK and Rubisco gene expression and regulation in S. cerevisiae should enable the design and construction of DNA cassettes that can be easily introduced in the genomes of industrial yeast strains. This should include the replacement of the GAL1-promotor, that was used for the expression of PRK in this study, by a promotor that is compatible with fast growth at high glucose concentrations and that further balances the expression of PRK and Rubisco. Since ribulose-5-phosphate is also an intermediate in pentose metabolism by engineered S. cerevisiae strains [27], this approach should also be readily applicable to the yeast-based conversion of lignocellulosic hydrolysates. The observed stimulatory effect of CO2 on the engineered strains will not hinder application of this concept in industrial bioethanol production, since large-scale processes for bioethanol production are characteristically CO2 saturated.
Our results illustrate how metabolic engineering strategies based on the functional integration of extensively studied reactions in the central carbon metabolism of distantly related organisms enables the optimization of product yields in industrial biotechnology. Although the present study focuses on ethanol production by yeast, functional integration of autotrophic carbon-fixing enzymes in the metabolic networks of industrial microorganisms should also enable optimization of yields of other existing and novel products whose synthesis results in a net positive ATP yield.
Methods
Construction of expression modules
Phosphoribulokinase (PRK) cDNA from Spinacia oleracea (spinach) [9] (accession number: X07654.1) was PCR-amplified using Phusion Hot-Start Polymerase (Finnzymes, Landsmeer, the Netherlands) and the oligonucleotides XbaI_prk_FW2 and RV1_XhoI_prk (Table 2), and was ligated in pCR®-Blunt II-TOPO® (Life Technologies Europe BV, Bleiswijk, the Netherlands). After restriction by XbaI and XhoI, the PRK-containing fragment was ligated into pTEF424 [28]. The TEF1p was later replaced by GAL1p from plasmid pSH47 [29] by XbaI and SacI restriction/ligation, creating plasmid pUDE046 (Table 3).
Table 2.
Oligonucleotides used in this study
| Number | Name | Sequence (5′ to 3′) | Purpose |
|---|---|---|---|
| |
Cloning |
|
|
| 1 |
XbaI_prk_FW2 |
TGACATCTAGATGTCACAACAACAAACAATTG |
Cloning of PRK into pUDE046. |
| 2 |
RV1 XhoI prk |
TGACATCTAGATGTCACAACAACAAACAATTG |
Cloning of PRK into pUDE046. |
| |
Primers used for in vivo plasmid assembly |
|
|
| 3 |
HR-cbbM-FW-65 |
TTGTAAAACGACGGCCAGTGAGCGCGCGTAATACGACTCACTATAGGGCGAATTGGGTACAGCTGGAGCTCAGTTTATCATTATC |
Rubisco cbbM cassette for plasmids pUDC075, pUDC099, and pUDC100. |
| 4 |
HR-cbbM-RV-65 |
GGAATCTGTGTAGTATGCCTGGAATGTCTGCCGTGCCATAGCCATGTATGCTGATATGTCGGTACCGGCCGCAAATTAAAG |
Rubisco cbbM cassette for plasmids pUDC075, pUDC099, and pUDC100 |
| 5 |
linker-cbbO2-pRS416 |
ATCACTCTTACCAGGCTAGGACGACCCTACTCATGTATTGAGATCGACGAGATTTCTAGGCCAGCTTTTGTTCCCTTTAGTGAGGGTTAATTGCGCGCTTGGCGTAATCATGGTCATAGC |
Linker fragment for assembly of plasmid pUDC099. |
| 6 |
linker-cbbM-GroEL |
GACATATCAGCATACATGGCTATGGCACGGCAGACATTCCAGGCATACTACACAGATTCCATCACTCTTACCAGGCTAGGACGACCCTACTCATGTATTGAGATCGACGAGATTTCTAGG |
Linker fragment for assembly of plasmid pUDC100. |
| |
Primers used for in vivo integration assembly |
|
|
| 7 |
FW pTDH3- HR-CAN1up |
GTTGGATCCAGTTTTTAATCTGTCGTCAATCGAAAGTTTATTTCAGAGTTCTTCAGACTTCTTAACTCCTGTAAAAACAAAAAAAAAAAAAGGCATAGCAAGCTGGAGCTCAGTTTATC |
1st cloning expression cassette linker fragment between CAN1 upstream and PRK expression cassette (IMI229), and CAN1up-linker and KlLEU2 expression cassette (IMI232). |
| 8 |
RV linker-iHR2B |
AGATATACTGCAAAGTCCGGAGCAACAGTCGTATAACTCGAGCAGCCCTCTACTTTGTTGTTGCGCTAAGAGAATGGACC |
1st cloning fragment: linker fragment between CAN1up-linker and PRK expression cassette (IMI229). |
| 9 |
RV linker-iHR6 |
GCTATGACCATGATTACGCCAAGCGCGCAATTAACCCTCACTAAAGGGAACAAAAGCTGGTTGCGCTAAGAGAATGGACC |
1st cloning fragment: linker fragment between CAN1up-linker and KlLEU2 expression cassette (IMI232). |
| 10 |
FW pGAL1-prk HR2B |
CAACAAAGTAGAGGGCTGCTCGAGTTATACGACTGTTGCTCCGGACTTTGCAGTATATCTGCTGGAGCTCTAGTACGGATT |
2nd cloning fragment: GAL1p-PRK-CYC1t expression cassette (IMI229) from pUDE046. |
| 11 |
RV CYC1t-prk HR2 |
GGAATCTGTGTAGTATGCCTGGAATGTCTGCCGTGCCATAGCCATGTATGCTGATATGTCGTACCGGCCGCAAATTAAAG |
2nd cloning fragment: GAL1p-PRK-CYC1t expression cassette (IMI229) from pUDE046. |
| 12 |
FW HR2-cbbQ2-HR3 |
GACATATCAGCATACATGGCTATGG |
3rd cloning fragment: PGI1p-cbbQ2-TEF2t cassette (IMI229). |
| 13 |
RV HR2-cbbQ2-HR3 |
GGACACGCTTGACAGAATGTCAAAGG |
3rd cloning fragment: PGI1p-cbbQ2-TEF2t cassette (IMI229). |
| 14 |
FW HR3-cbbO2-HR4 |
CGTCCGATATGATCTGATTGG |
4th cloning fragment: PGK1p-cbbO2-ADH1t cassette (IMI229). |
| 15 |
RV HR3-cbbO2-HR4 |
CCTAGAAATCTCGTCGATCTC |
4th cloning fragment: PGK1p-cbbO2-ADH1t cassette (IMI229). |
| 16 |
FW HR4-GroEL-HR5 |
ATCACTCTTACCAGGCTAGG |
5th cloning fragment: TEF1p-groEL-ACT1t cassette (IMI229). |
| 17 |
RV HR4-GroEL-HR5 |
CTGGACCTTAATCGTGTGCGCATCCTC |
5th cloning fragment: TEF1p-groEL-ACT1t cassette (IMI229). |
| 18 |
FW HR5-GroES-HR6 |
CCGTATAGCTTAATAGCCAGCTTTATC |
6th cloning fragment: TPI1p-groES-PGI1t cassette (IMI229). |
| 19 |
RV HR5-GroES-HR6 |
GCTATGACCATGATTACGCCAAGC |
6th cloning fragment: TPI1p-groES-PGI1t cassette (IMI229). |
| 20 |
FW HR6-LEU2-CAN1dwn |
CCAGCTTTTGTTCCCTTTAGTGAGGGTTAATTGCGCGCTTGGCGTAATCATGGTCATAGCCTGTGAAGATCCCAGCAAAG |
7th (IMI229) or 2nd (IMI232) cloning fragment: KlLEU2 cassette from pUG73. |
| 21 |
RV LEU2 HR-CAN1 |
AGCTCATTGATCCCTTAAACTTTCTTTTCGGTGTATGACTTATGAGGGTGAGAATGCGAAATGGCGTGGAAATGTGATCAAAGGTAATAAAACGTCATATATCCGCAGGCTAACCGGAAC |
7th (IMI229) or 2nd (IMI232) cloning fragment: KlLEU2 cassette from pUG73. |
| |
Primers used for verification of the in vivo assembled constructs |
|
|
| 22 |
m-PCR-HR1-FW |
GGCGATTAAGTTGGGTAACG |
Diagnostic for assembly of plasmids pUDC075, pUDC099, and pUDC100, and integration in strain IMI229. |
| 23 |
m-PCR-HR1-RV |
AACTGAGCTCCAGCTGTACC |
Diagnostic for assembly of plasmids pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 24 |
m-PCR-HR2-FW |
ACGCGTGTACGCATGTAAC |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229 |
| 25 |
m-PCR-HR2-RV |
GCGCGTGGCTTCCTATAATC |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229 |
| 26 |
m-PCR-HR3-FW |
GTGAATGCTGGTCGCTATAC |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 27 |
m-PCR-HR3-RV |
GTAAGCAGCAACACCTTCAG |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 28 |
m-PCR-HR4-FW |
ACCTGACCTACAGGAAAGAG |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 29 |
m-PCR-HR4-RV |
TGAAGTGGTACGGCGATGC |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 30 |
m-PCR-HR5-FW |
ATAGCCACCCAAGGCATTTC |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 31 |
m-PCR-HR5-RV |
CCGCACTTTCTCCATGAGG |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 32 |
m-PCR-HR6-FW |
CGACGGTTACGGTGTTAAG |
Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
| 33 | m-PCR-HR6-RV | CTTCCGGCTCCTATGTTGTG | Diagnostic for assembly of pUDC075, pUDC099, pUDC100, and integration in strain IMI229. |
Table 3.
Plasmids used in this study
| Name | Relevant genotype | Source/reference |
|---|---|---|
| pFL451 |
AOX1p-prk (Spinach)-AOX1t (pHIL2-D2 HIS4 Amp centromeric) |
Brandes et al.[9]. |
| pCR®-Blunt II-TOPO |
Bla |
Life Technologies Europe BV |
| pTEF424_TEF |
TRP1 2μ bla |
Mumberg et al.[28]. |
| pSH47 |
URA3 CEN6 ARS4 GAL1p-cre-CYC1tbla |
Güldener et al. [29]. |
| pUD0E46 |
TRP1 2μ GAL1p-prk-CYC1tbla |
This study. |
| pPCR-Script |
Bla |
Life Technologies Europe BV. |
| pGPD_426 |
URA3 2μ bla |
Mumberg et al.[28]. |
| pRS416 |
URA3 CEN6 ARS4 bla |
Mumberg et al.[28]. |
| pBTWW002 |
URA3 2μ TDH3p-cbbM-CYC1tbla |
This study. |
| pUDC098 |
URA3 CEN6 ARS4 TDH3p-cbbM-CYC1tbla |
This study. |
| pMK-RQ |
nptII |
Life Technologies Europe BV. |
| pUD230 |
PGI1p-cbbQ2-TEF2tnptII |
Life Technologies Europe BV. |
| pUD231 |
PGK1p-cbbO2-ADH1tnptII |
Life Technologies Europe BV. |
| pUD232 |
TEF1p-groEL-ACT1tnptII |
Life Technologies Europe BV. |
| pUD233 |
TPI1p-groES-PGI1tnptII |
Life Technologies Europe BV. |
| pUDC075 |
URA3 CEN6 ARS4 TDH3p-cbbM-CYC1t;PGI1p-cbbQ2-TEF2t;PGK1p-cbbO2-ADH1t;TEF1p-groEL-ACT1t;TPI1p-groES-PGI1tbla |
This study. |
| pUDC099 |
URA3 CEN6 ARS4 TDH3p-cbbM-CYC1t;PGI1p-cbbQ2-TEF2t;PGK1p-cbbO2-ADH1tbla |
This study. |
| pUDC100 | URA3 CEN6 ARS4 TDH3p-cbbM-CYC1t;TEF1p-groEL-ACT1t;TPI1p-groES-PGI1tbla | This study. |
Rubisco form II gene cbbM from T. denitrificans[17] flanked by KpnI and SacI sites was codon optimized [30] (accession number: KC699554), synthesized at GeneArt (Life Technologies Europe BV), and ligated into pPCR-Script. The cbbM-containing fragment was ligated into the BamHI and SacI restricted vector pGPD_426 [28] creating plasmid pBTWW002. The cbbM expression cassette was transferred into pRS416 using KpnI and SacI, yielding pUDC098.
Expression cassette of the specific Rubisco form II chaperones from T. denitrificans cbbQ2 and cbbO2[20], and chaperones groEL and groES[18] from E. coli were codon optimized [30] (accession numbers: KC699555 and KC699556, respectively). The expression cassettes contained a yeast constitutive promoters and terminator, flanking the codon optimized gene. The cassette was flanked by unique 60-bp regions obtained by randomly combining bar-code sequences used in the Saccharomyces Genome Deletion Project [31] and an EcoRV site (GeneArt). The expression cassettes were inserted in plasmid pMK-RQ (GeneArt) using the SfiI cloning sites yielding pUD230 (PGI1p-cbbQ2-TEF2t), pUD231 (PGK1p-cbbO2-ADH1t), pUD232 (TEF1p-groEL-ACT1t), and pUDE233 (TPI1p-groES-PGI1t) (Table 3). The expression cassette TDH3p-cbbM-CYC1t was PCR-amplified from plasmid pBTWW002 using Phusion Hot-Start Polymerase (Finnzymes) and primers HR-cbbM-FW-65 and HR-cbbM-RV-65 in order to incorporate the 60-bp region for recombination cloning.
Strain construction, isolation and maintenance
All Saccharomyces cerevisiae strains used (Table 4) belong to the CEN.PK family [32,33]. All strains were grown in 2% w/v glucose synthetic media [7] supplemented with 150 mg l-1 uracil when required [34] until they reached end exponential phase, then sterile glycerol was added up to ca. 30% v/v and aliquot of 1 ml were stocked −80°C.
Table 4.
Saccharomyces cerevisiae strains used in this study
| Relevant genotype | Source/reference | |
|---|---|---|
| CEN.PK113-5D |
MATa ura3-52 |
Euroscarf. |
| CEN.PK102-3A |
MATa ura3-52 leu2-3, 112 |
Euroscarf. |
| IMC014 |
MATa ura3-52 pUDC075 (CEN6 ARS4 URA3 TDH3p-cbbM-CYC1tPGI1p-cbbQ2-TEF2tPGK1p-cbbO2-ADH1tTEF1p-groEL-ACT1tTPI1p-groES-PGI1t) |
This study. |
| IMC033 |
MATa ura3-52 pUDC098 (CEN6 ARS4 URA3 TDH3p-cbbM-CYC1t) |
This study. |
| IMC034 |
MATa ura3-52 pUDC099 (CEN6 ARS4 URA3 TDH3p-cbbM-CYC1tPGI1p-cbbQ2-TEF2tPGK1p-cbbO2-ADH1tcbbO2-pRS416 linker) |
This study. |
| IMC035 |
MATa ura3-52 pUDC100 (CEN6 ARS4 URA3 TEF1p-groEL-ACT1tTPI1p-groES-PGI1tcbbM-GroEL linker) |
This study. |
| IMI229 |
MATa ura3-52 leu2-3, 112 can1Δ::GAL1p-prk-CYC1tPGI1p-cbbQ2-TEF2t,PGK1p-cbbO2-ADH1t,TEF1p-groEL-ACT1t,TPI1p-groES-PGI1tKlLEU2 |
This study. |
| IMI232 |
MATa ura3-52 leu2-3, 112 can1::KlLEU2 |
This study. |
| IMU032 |
IMI232 p426_GPD (2μ URA3) |
This study. |
| IMU033 | IMI229 pUDC100 (CEN6 ARS4 URA3 TEF1p-groEL-ACT1tTPI1p-groES-PGI1tcbbM-GroEL linker) | This study. |
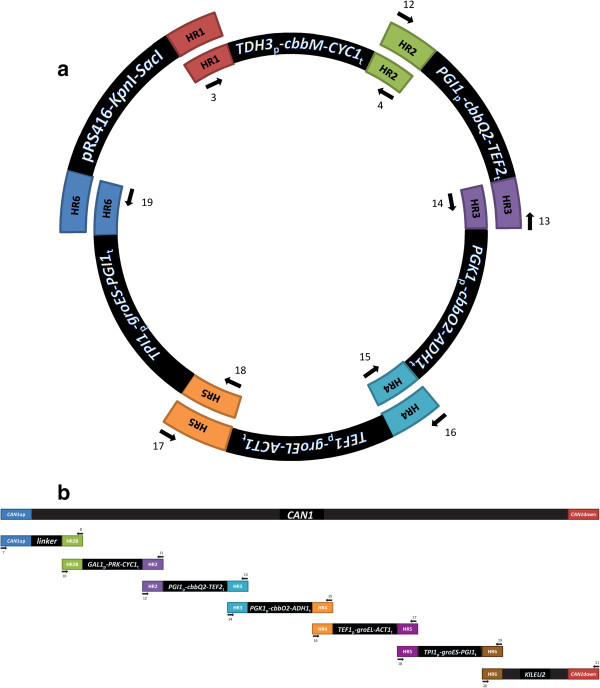
The strain IMC014 that co-expressed the Rubisco form II cbbM and the four chaperones cbbQ2, cbbO2, groEL, and groES was constructed using a previously published in vivo transformation associated recombination [35]. 200 fmol of each expression cassette were pooled with 100 fmol of the KpnI/SacI linearized pRS416 backbone in a final volume of 50 μl and transformed in CEN.PK 113-5D using the lithium acetate protocol [36] (Figure 4a). Cells were selected on synthetic medium. Correct assembly of the fragment of pUDC075 was performed by multiplex PCR on transformant colonies using primers enabling amplification over the regions used for homologous recombination (Table 2) and by restriction analysis after retransformation of the isolated plasmid in E. coli DH5α. pUDC075 was sequenced by Next Gen Seq Illumina (100-bp reads paired-end, 50 Mb) and assembled with Velvet [37]. The assembled sequence did not contain mutations in any of the assembled expression cassettes. The strains IMC034 and IMC035 that expressed cbbM/cbbQ2/cbbO2 and cbbM/groEL/groES respectively were constructed using the same in vivo assembly method with the following modification. To construct plasmids pUDC099 and pUDC100, 120 bp cbbO2-pRS416 linker and cbbM-GroEL linker were used to close the assembly respectively (Table 2), 100 fmol of each of complementary 120 bp oligonucleotides were added to the transformation. The strain IMC033 that only expressed the cbbM gene was constructed by transforming CEN.PK113-5D with pUDC098.
Figure 4.
Strategy for the heterologous expression of Rubisco and PRK in S. cerevisiae. (a)In vivo assembly of Rubisco expression plasmid pUDC075, and (b)in vivo assembly and integration of PRK and chaperone proteins in CAN1 locus of Saccharomyces cerevisiae strain IMI229. Each fragment represents a different expression cassette or plasmid backbone. All fragments used in assembly experiments were flanked by 60-bp sequences used for in vivo recombination, either enabling the assembly of plasmids or the integration assembled constructs into the S. cerevisiae genome. Arrows and numbers indicate primers used in the construction of the cassette.
To construct the strain IMU033 that co-expressed PRK, cbbM, cbbQ2, cbbO2, the intermediate strain IMI229 was constructed by integrating PRK, the four chaperones and KlLEU2[38] at the CAN1 locus by in vivo homologous integration in CEN.PK102-3A (Figure 4b). The expression cassettes were PCR amplified using Phusion Hot-Start Polymerase (Finnzymes), the corresponding oligonucleotides and DNA templates (Table 2). Finally, the strain IMI229 was transformed with pUDC100 that carries the Rubisco form II cbbM and the two E. coli chaperones groEL and groES.
Strain IMI232 was constructed by transforming CEN.PK102-3A with the KlLEU2 cassette. IMI232 was finally transformed with the plasmid p426GPD to restore prototrophy resulting in the reference strain IMU032.
Experimental set-up of chemostat and batch experiments
Anaerobic chemostat cultivation was performed essentially as described [39] but with 12.5 g l-1 glucose and 12.5 g l-1 galactose as the carbon source and where indicated, a mixture of 10% CO2/90% N2 replaced pure nitrogen as the sparging gas. Residual glucose and galactose concentrations were determined after rapid quenching [40] using commercial enzymatic assays for glucose (Boehringer, Mannheim, Germany) and D-galactose (Megazyme, Bray, Ireland). Anaerobic bioreactor batch cultures were grown essentially as described [6], but with 20 g l-1 galactose and a sparging gas consisting of 10% CO2 and 90% N2. Biomass and metabolite concentrations in batch and chemostat and batch cultures were determined as described by Guadalupe et al.[6]. In calculations of ethanol fluxes and yields, ethanol evaporation was corrected for based on a first-order evaporation rate constant of 0.008 h-1 in the bioreactor setups and under the conditions used in this study [6,39].
Enzyme assays for phosphoribulokinase and Rubisco
Cell extracts for analysis of phosphoribulokinase (PRK) activity were prepared as described previously [41]. PRK activity was measured at 30°C by a coupled spectrophotometric assay [42]. Reaction rates were proportional to the amounts of cell extract added. Protein concentrations were determined by the Lowry method [43] using bovine serum albumin as a standard.
Cell extracts for Rubisco activity assays were prepared as described [41], with two modifications: Tris–HCl (1 mM, pH 8.2) containing 20 mM MgCl2•6H2O, 5 mM of DTT and 5 mM NaHCO3 was used as sonication buffer and Tris–HCl (100 mM, pH 8.2), 20 mM MgCl2•6H2O and 5 mM of DTT as freezing buffer. Rubisco activity was determined by measuring 14CO2-fixation (PerkinElmer, Groningen, The Netherlands) as described [44] and measuring radioactive counts in a TRI-CARB® 2700TR Series liquid scintillation counter (PerkinElmer, Groningen, The Netherlands), using Ultima Gold™ scintillation cocktail (PerkinElmer, Groningen, The Netherlands). Protein concentrations were determined by the Lowry method [43] using standard solutions of bovine serum albumin dissolved in 50 mM Tris–HCl (pH 8.2).
Abbreviations
Rubisco: Ribulose 1,5-bisphosphate carboxylase/oxidase; PRK: Phosphoribulokinase; NAD+: Nicotinamide adenine dinucleotide; NADP+: Nicotinamide adenine dinucleotide phosphate.
Competing interests
VGM, HWW, AJAvM, and JTP are inventors on a patent application related to the content of this article.
Authors’ contributions
VGM, JMD, JTP, AJAvM designed the experiments and wrote the manuscript. HWW selected the Rubisco and PRK genes, VGM selected the chaperone genes and constructed all yeast strains. VGM and EdH carried out and analyzed the fermentation experiments. VGM and MAHL performed the PRK and Rubisco enzymatic activity determinations. All the authors have read and approved the final manuscript.
Contributor Information
Víctor Guadalupe-Medina, Email: V.G.GuadalupeMedina@gmail.nl.
H Wouter Wisselink, Email: W.Wisselink@miccellbioservices.com.
Marijke AH Luttik, Email: M.A.H.Luttik@tudelft.nl.
Erik de Hulster, Email: A.F.deHulster@tudelft.nl.
Jean-Marc Daran, Email: J.G.Daran@tudelft.nl.
Jack T Pronk, Email: J.T.Pronk@tudelft.nl.
Antonius JA van Maris, Email: A.J.A.vanMaris@tudelft.nl.
Acknowledgements
The PhD project of VGM was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. We thank Mirjam Fuchs, Nakul Barfa, Peter de Bruijn, Astrid van der Meer and Daniel de Vries for experimental assistance and Niels Kuijpers, Tim Vos and Frank Koopman for advice on molecular biology and Gijs Kuenen for critically reading the manuscript.
References
- World Fuel Ethanol Production. http://ethanolrfa.org/pages/World-Fuel-Ethanol-Production.
- Hong KK, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci. 2012;69:2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Larsson C, van Maris A, Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol. 2013;24:398–404. doi: 10.1016/j.copbio.2013.03.023. [DOI] [PubMed] [Google Scholar]
- van Dijken JP, Scheffers WA. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol Rev. 1986;32:199–224. [Google Scholar]
- Nissen TL, Kielland-Brandt MC, Nielsen J, Villadsen J. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab Eng. 2000;2:69–77. doi: 10.1006/mben.1999.0140. [DOI] [PubMed] [Google Scholar]
- Guadalupe Medina V, Almering MJH, van Maris AJA, Pronk JT. Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol. 2010;76:190–195. doi: 10.1128/AEM.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA, van Dijken JP. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. doi: 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- Milanez S, Mural RJ. Cloning and sequencing of cDNA encoding the mature form of phosphoribulokinase from spinach. Gene. 1988;66:55–63. doi: 10.1016/0378-1119(88)90224-7. [DOI] [PubMed] [Google Scholar]
- Brandes HK, Hartman FC, Lu TY, Larimer FW. Efficient expression of the gene for spinach phosphoribulokinase in Pichia pastoris and utilization of the recombinant enzyme to explore the role of regulatory cysteinyl residues by site-directed mutagenesis. J Biol Chem. 1996;271:6490–6496. doi: 10.1074/jbc.271.11.6490. [DOI] [PubMed] [Google Scholar]
- Shively JM, van Keulen G, Meijer WG. Something from almost nothing: Carbon Dioxide Fixation in Chemoautotrophs. Annu Rev Microbiol. 1998;52:191–230. doi: 10.1146/annurev.micro.52.1.191. [DOI] [PubMed] [Google Scholar]
- Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 2008;59:1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- Quivey J, Robert Tabita F. Cloning and expression in Escherichia coli of the form II ribulose 1,5-bisphosphate carboxylase/oxygenase gene from Rhodopseudomonas sphaeroides. Gene. 1984;31:91–101. doi: 10.1016/0378-1119(84)90198-7. [DOI] [PubMed] [Google Scholar]
- Somerville C, Somerville S. Cloning and expression of the Rhodospirillum rubrum ribulose bisphosphate carboxylase gene in Escherichia coli. Mol Gen Genet. 1984;193:214–219. doi: 10.1007/BF00330670. [DOI] [Google Scholar]
- Goloubinoff P, Gatenby AA, Lorimer GH. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- Hayashi NR, Arai H, Kodama T, Igarashi Y. The cbbQ genes, located downstream of the form I and form II RubisCO genes, affect the activity of both RubisCOs. Biochem Biophys Res Commun. 1999;265:177–183. doi: 10.1006/bbrc.1999.1103. [DOI] [PubMed] [Google Scholar]
- Chen DH, Madan D, Weaver J, Lin Z, Schröder GF, Chiu W, Rye HS. Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell. 2013;153:1354–1365. doi: 10.1016/j.cell.2013.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández JM, Baker SH, Lorbach SC, Shively JM, Tabita FR. Deduced amino acid sequence, functional expression, and unique enzymatic properties of the form I and form II ribulose bisphosphate carboxylase/oxygenase from the chemoautotrophic bacterium Thiobacillus denitrificans. J Bacteriol. 1996;178:347–356. doi: 10.1128/jb.178.2.347-356.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Nelson R, Baldwin S, Plunkett G, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M. et al. The complete genome sequence of Escherichia coli DH10B: Insights into the biology of a laboratory workhorse. J Bacteriol. 2008;190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi NR, Arai H, Kodama T, Igarashi Y. The Novel Genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem Biophys Res Commun. 1997;241:565–569. doi: 10.1006/bbrc.1997.7853. [DOI] [PubMed] [Google Scholar]
- Beller HR, Chain PSG, Letain TE, Chakicherla A, Larimer FW, Richardson PM, Coleman MA, Wood AP, Kelly DP. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J Bacteriol. 2006;188:1473–1488. doi: 10.1128/JB.188.4.1473-1488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Alonso M, Toledo-Rubio V, Noad R, Unzueta U, Ferrer-Miralles N, Roy P, Villaverde A. Rehosting of bacterial chaperones for high-quality protein production. Appl Environ Microbiol. 2009;75:7850–7854. doi: 10.1128/AEM.01532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jariyachawalid K, Laowanapiban P, Meevootisom V, Wiyakrutta S. Effective enhancement of Pseudomonas stutzeri D-phenylglycine aminotransferase functional expression in Pichia pastoris by co-expressing Escherichia coli GroEL-GroES. Microb Cell Fact. 2012;11:47. doi: 10.1186/1475-2859-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder W, Kuenen JG, Matin A. Microbial selection in continuous culture. J Appl Microbiol. 1977;43:1–24. doi: 10.1111/j.1365-2672.1977.tb00717.x. [DOI] [PubMed] [Google Scholar]
- Daran-Lapujade P, Jansen MLA, Daran JM, van Gulik W, de Winde JH, Pronk JT. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae: a chemostat culture study. J Biol Chem. 2004;279:9125–9138. doi: 10.1074/jbc.M309578200. [DOI] [PubMed] [Google Scholar]
- Bai FW, Anderson WA, Moo-Young M. Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol Adv. 2008;26:89–105. doi: 10.1016/j.biotechadv.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Garcia Sanchez R, Hahn-Hägerdal B, Gorwa-Grauslund M. PGM2 overexpression improves anaerobic galactose fermentation in Saccharomyces cerevisiae. Microb Cell Fact. 2010;9:40. doi: 10.1186/1475-2859-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund M. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müler R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nuc Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nuc Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H. et al. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Entian KD, Kötter P. In: Methods in Microbiology: Yeast Gene Analysis Second Edition. 36. Stansfield I, Stark MJR, editor. Amsterdam: Academic Press; 2007. Yeast genetic strain and plasmid collections; pp. 629–666. [Google Scholar]
- Nijkamp J, van den Broek M, Datema E, de Kok S, Bosman L, Luttik M, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne W. et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact. 2012;11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk JT. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol. 2002;68:2095–2100. doi: 10.1128/AEM.68.5.2095-2100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers N, Solis-Escalante D, Bosman L, van den Broek M, Pronk J, Daran JM, Daran-Lapujade P. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb Cell Fact. 2013;12:47. doi: 10.1186/1475-2859-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods R. In: Yeast Protocol. 313. Xiao W, editor. (Totowa, NJ, USA): Humana Press; 2006. Yeast transformation by the LiAc/SS carrier DNA/PEG method; pp. 107–120. [Google Scholar]
- Zerbino D, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nuc Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso TO, de Kok S, Dario M, do Espirito-Santo JC, Müller G, Schlölg PS, Silva CP, Tonso A, Daran JM, Gombert AK. et al. Engineering topology and kinetics of sucrose metabolism in Saccharomyces cerevisiae for improved ethanol yield. Metab Eng. 2011;13:694–703. doi: 10.1016/j.ymben.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Mashego MR, van Gulik WM, Vinke JL, Heijnen JJ. Critical evaluation of sampling techniques for residual glucose determination in carbon-limited chemostat culture of Saccharomyces cerevisiae. Biotechnol Bioeng. 2003;83:395–399. doi: 10.1002/bit.10683. [DOI] [PubMed] [Google Scholar]
- Luttik MAH, Kötter P, Salomons FA, van der Klei IJ, van Dijken JP, Pronk JT. The Saccharomyces cerevisiae ICL2 gene encodes a mitochondrial 2-methylisocitrate lyase involved in propionyl-coenzyme A metabolism. J Bacteriol. 2000;182:7007–7013. doi: 10.1128/JB.182.24.7007-7013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacElroy RD, Mack HM, Johnson EJ. Properties of phosphoribulokinase from Thiobacillus neapolitanus. J Bacteriol. 1972;112:532–538. doi: 10.1128/jb.112.1.532-538.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Beudeker RF, Cannon GC, Keunen JG, Shively JM. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch Microbiol. 1980;124:185–189. [Google Scholar]