Abstract
2-chloro-2-fluoro-deoxy-9-D-arabinofuranosyladenine (Clofarabine), a purine nucleoside analog, is used in the treatment of hematologic malignancies and as induction therapy for stem cell transplantation. The discovery of pharmacogenomic markers associated with chemotherapeutic efficacy and toxicity would greatly benefit the utility of this drug. Our objective was to identify genetic and epigenetic variants associated with clofarabine toxicity using an unbiased, whole genome approach. To this end, we employed International HapMap lymphoblastoid cell lines (190 LCLs) of European (CEU) or African (YRI) ancestry with known genetic information to evaluate cellular sensitivity to clofarabine. We measured modified cytosine levels to ascertain the contribution of genetic and epigenetic factors influencing clofarabine-mediated cytotoxicity. Association studies revealed 182 single nucleotide polymorphisms (SNPs) and 143 modified cytosines associated with cytotoxicity in both populations at the threshold P≤ 0.0001. Correlation between cytotoxicity and baseline gene expression revealed 234 genes at P ≤ 3.98 × 10−6. Six genes were implicated as: (i) their expression was directly correlated to cytotoxicity, (ii) they had a targeting SNP associated with cytotoxicity, and (iii) they had local modified cytosines associated with gene expression and cytotoxicity. We identified a set of three SNPs and three CpG sites targeting these six genes explaining 43.1% of the observed variation in phenotype. siRNA knockdown of the top three genes (SETBP1, BAG3, KLHL6) in LCLs revealed altered susceptibility to clofarabine, confirming relevance. As clofarabine's toxicity profile includes acute kidney injury, we examined the effect of siRNA knockdown in HEK293 cells. siSETBP1 led to a significant change in HEK293 cell susceptibility to clofarabine.
INTRODUCTION
2-chloro-2-fluoro-deoxy-9-D-arabinofuranosyladenine (Clofarabine) is a purine nucleoside analogue used in the treatment of relapsed or refractory lymphocytic leukemia (1,2). Recently, phase II trials have explored expanding the indications of clofarabine for additional hematologic malignancies (3,4), as well as induction of hematologic stem cell transplantation (5). To exert its antitumor effect, clofarabine requires metabolic conversion to clofarabine-triphosphate through intracellular phosphorylation by deoxycytidine kinase (6,7) and the triphosphate inhibits both DNA polymerase and ribonucleotide reductase. Of additional mechanistic interest is clofarabine's hypomethylating activity, either alone or in conjunction with decitabine (4,8–10).
The toxicity profile of clofarabine includes its intended bone marrow suppression. Additionally, clofarabine administration has been associated with transient transaminitis, emesis and infectious sequelae. Of particular interest is the oft under-recognized severe acute kidney injury associated with the drug. Clofarabine is subject to renal filtration, secretion and reabsorption with over 60% of administered drug excreted unchanged in the urine (11). Case reports and several phase II trials have noted rates of grade 3–4 renal toxicity in up to 21% of patients (2,3,5,12).
Genetic factors predicting drug efficacy and toxicity have frequently been extrapolated from other well-studied nucleoside analogues, including cytarabine (6). However, in a panel of acute lymphoblastic leukemia cell lines of the B-lineage, cells were found to be over 7-fold more sensitive to clofarabine compared with cytarabine, while cells of T-lineage were not differentially affected (13). Further, nucleoside analogue transport studies revealed differences in hENT1 transport efficiency and intracellular accumulation between clofarabine and other analogues (14). Based on these studies, there is reason to believe important differences exist between nucleoside analogues and an independent genomic analysis is warranted.
Human cell-based models have been successfully utilized for pharmacogenetic discovery and validation (15–19). Although these models have limitations, the lymphoblastoid cell lines (LCLs) of the International HapMap Consortium (www.hapmap.org) are a renewable resource with a remarkable amount of publicly available genetic information. Several identified genetic variants in LCLs have been replicated in patient samples, with particular success in hematologic malignancies (20–23). For example, variants found in LCLs contributing to cytarabine sensitivity also affected sensitivity in leukemic blasts of patients with acute myeloid leukemia (24). Our laboratory has shown that single nucleotide polymorphisms (SNPs) categorized as expression quantitative trait loci (eQTLs, SNPs that are associated with expression of a target gene, whether cis or trans acting) are enriched in genome wide association study (GWAS) results of pharmacologic and complex traits (25,26). Therefore, regulatory SNPs are important contributors to explaining cellular sensitivity to chemotherapeutics.
This study represents an unbiased, comprehensive approach that identifies the contribution of modified cytosine sites and SNPs associated with clofarabine cytotoxicity through their effect on baseline gene expression. We included cytosine modification data in our analysis to provide a more complete understanding of the contributing factors of gene expression and understand whether the addition of epigenetic variants would improve the predictive quality of our models. In the evaluation of baseline gene expression, as well as genetic and epigenetic variants, we identified six genes most relevant to this drug, further validating the importance of three with siRNA knockdown in LCLs. A subset of genetic and epigenetic markers targeting the six genes explains 43.1% of phenotypic variation in sensitivity to clofarabine. We considered whether some of these genes also mediated susceptibility to clofarabine-mediated kidney injury and interrogated their relevance in human embryonic kidney cells (HEK293) with siRNA knockdown. The novel prioritization, and incorporation of both genetic and epigenetic data, undertaken in this study can be applied to other drugs and future pharmacogenomic discovery. Furthermore, pharmacogenomic markers identified through these studies can be evaluated in a clinical setting.
RESULTS
Clofarabine-induced cytotoxicity
Populations of LCLs with African ancestry (YRI, n = 102) and European ancestry (CEU, n = 88) were phenotyped for sensitivity to clofarabine using a short-term growth inhibition assay. Areas under the curve (AUCs) obtained by fitting percent survival against drug concentration were normally distributed, passing the Kolmogorov–Smirnov normality test (P > 0.2, Supplementary Material, Fig. S1A). Two example cytotoxicity curves are provided in the inset of Figure 1, one resistant LCL (high AUC) and one susceptible LCL (low AUC). The mean (SEM) AUC for all 190 LCLs was 69 828.2 [±1146.0 (%·nM)] with no significant population difference between the CEU and YRI populations (P = 0.204, Supplementary Material, Fig. S1B).
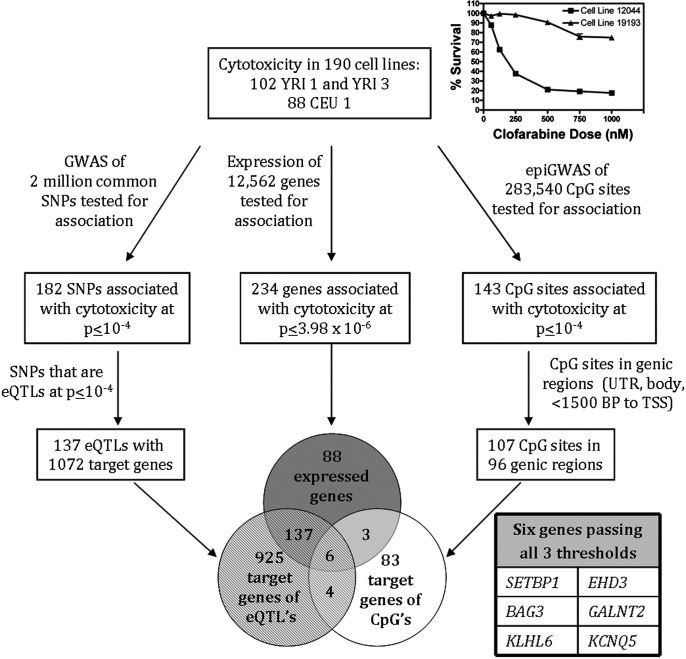
Figure 1.
Schematic diagram of prioritization for target gene validation. Three simultaneous thresholds were applied and six genes passed all three thresholds. Prioritized genes were directly associated with cytotoxicity (after Bonferroni multiple corrections testing). Genes also possessed at least 1 targeting SNP and 1 CpG site in its genic region associated with cytotoxicity at P < 10−4. Inset: Example cytotoxicity curves of a cell line sensitive to clofarabine (LCL GM12044) and resistant to clofarabine (LCL GM19193).
Association studies of clofarabine cytotoxicity
As there was no significant population difference in cytotoxicity, all 190 LCLs were analyzed in combined analyses. Simultaneously, we performed three unbiased genome-wide analyses to evaluate clofarabine AUC in 190 LCLs as phenotype against: (i) 2 million common SNPs; (ii) baseline expression of 12 562 genes as measured by Affymetrix exon array; and (iii) cytosine modification at 283 540 CpG sites as measured by an Illumina Infinium Human Methylation 450 beadchip array. The workflow, data analysis and thresholds used are summarized in Figure 1.
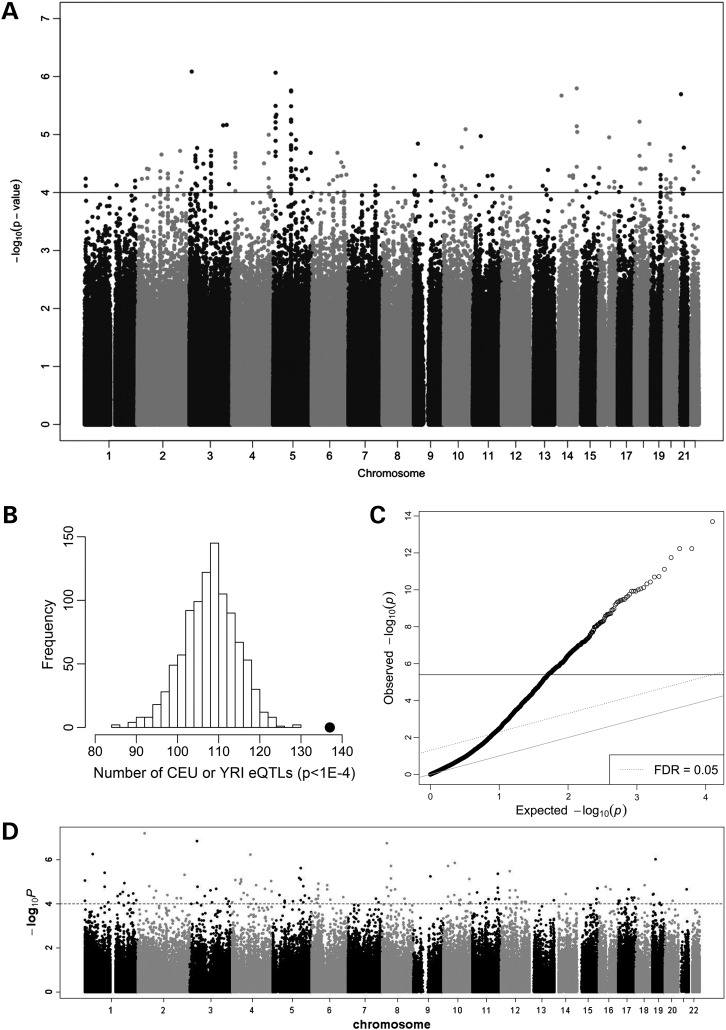
In the first analysis, we identified 182 SNPs associated with clofarabine-induced cytotoxicity at a P ≤ 0.0001 threshold (Fig. 2A). To discern whether identified SNPs were eQTLs, we evaluated the effect of each of the 182 SNPs on global baseline gene expression. A full index of the 182 SNPs and their annotated target genes are listed in Supplementary Material, Table S1. Of the 182 SNPS, 137 were associated with baseline expression of one or more genes at a P ≤ 0.0001 threshold using SCAN database, a results database of eQTLs in LCLs (27). Permutation analysis revealed that these 137 eQTLs represent a significantly enriched functional class of SNPs when compared with the number of eQTLs expected by chance (Fig. 2B, empirical P < 0.001). The 137 eQTLs and their 1072 target genes were used in further analysis.
Figure 2.
Genetic, expression and epigenetic associations. (A) Manhattan plot of association between SNPs and clofarabine AUC. At P < 10−4, 182 SNPs were associated with cytotoxicity. (B) Analysis of distribution of eQTL count in 1000 simulations, each matching the MAF distribution of all clofarabine-associated SNPs at P < 10−4. The black dot is the observed eQTL count (n = 137) in the clofarabine susceptibility-associated SNPs. (C) Q–Q plot representing the observed associations between baseline gene expression and clofarabine AUC. At P < 3.98 × 10−6, 234 genes were significantly associated. Solid line indicates FDR < 0.05. (D) Manhattan plot of association between modified CpG sites and clofarabine AUC. At P < 10−4, 143 CpG sites were associated with cytotoxicity.
In the second analysis, the direct correlation between baseline expression of 12 562 genes and clofarabine AUC was queried. The strength of association for all genes is provided in a Q–Q plot (Fig. 2C) and indexed in Supplementary Material, Table S2. Of all genes interrogated, 234 genes passed this multiple testing correction threshold (P ≤ 3.98 × 10−6) and were used in further analysis.
In the third simultaneous analysis, we identified 143 CpG sites associated with clofarabine cytotoxicity at a P < 0.0001 and the Manhattan plot of modified cytosines is provided (Fig. 2D). A CpG site was considered to be in a genic region if it was found in the gene body, 5′ UTR, 3′ UTR or within 1500 base pairs of a gene's transcriptional start site. A complete index of the 143 CpG sites is provided in Supplementary Material, Table S3, including the gene where these CpG sites were located near and the location relative to the gene coding sequence. Of the 143 CpG sites identified, 107 were in genic regions. The 96 genes containing these 107 modified cytosines were used in further analysis.
Prioritization of genes for further investigation
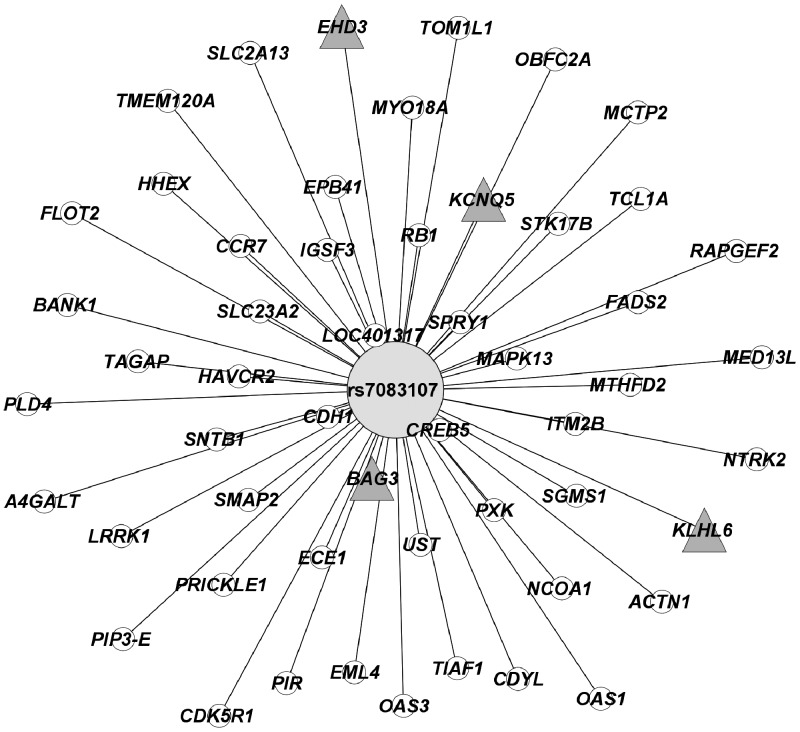
The principal goal of this study is to identify the genes most highly associated with clofarabine cytotoxicity and whose expression is significantly regulated by genetic and epigenetic factors because these markers may be used to identify patients at risk for toxicity or non-response prior to therapy. We determined the overlap among the 1072 target genes of the eQTLs associated with cytotoxicity, the 234 genes whose expression was associated with cytotoxicity, and the 96 genes containing modified cytosines that were associated with cytotoxicity. Six genes were common among the three analyses (SETBP1, BAG3, KLHL6, EHD3, GALNT2 and KCNQ5). Therefore, each of the six genes had one or more of the eQTLs targeting it and had one or more associated modified cytosines in its genic region (Table 1). Both the eQTLs and modified cytosines were associated with cellular sensitivity to clofarabine. Of note, a single SNP, rs7083107, was associated with expression of 53 target genes (Fig. 3) and was associated with gene expression of four of the top six genes. This SNP was associated with clofarabine cytotoxicity at P = 8.08 × 10−6.
Table 1.
Genetic and epigenetic variants associated with the identified target genes for susceptibility to clofarabine from the whole genome analysis for AUC in both CEU and YRI populations
| Target gene | P, gene expression–cytotoxicity | Beta, gene expression–cytotoxicity | SNPs targeting gene at P < 10−4 | Host gene (chr) | P, SNP–cytotoxicity | Beta, SNP–cytotoxicity | P, SNP–gene expression | CpG sites associated | CpG location | P, CpG–cytotoxicity | Coefficient, CpG–cytotoxicity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SETBP1 | 6.32E-11 | −11 570 | rs2137664 | DIAPH3 (13) | 7.66E−5 | 6771 | 5.00E−5 | cg25162588 | Body | 1.43E−5 | −8624 |
| BAG3 | 6.36E-10 | −20 216 | rs7083107 | ENTPD7 (10) | 8.08E−6 | −14 220 | 3.00E−6 | cg18908677 | Body | 7.21E−6 | −9950 |
| cg08756008 | Body | 3.51E−5 | −6904 | ||||||||
| KLHL6 | 6.63E-10 | −8071 | rs6452564 | EDIL3 (5) | 1.74E−6 | −8000 | 1.00E−4 | cg18262615 | Body | 7.35E−5 | 13 602 |
| rs4145660 | EDIL3 (5) | 1.81E−6 | −8626 | 1.00E−4 | |||||||
| rs7083107 | ENTPD7 (10) | 8.08E−6 | −14 220 | 9.00E−5 | |||||||
| rs1445747 | EDIL3 (5) | 3.81E−5 | −6902 | 1.00E−4 | |||||||
| rs10415181 | Intergenic (19) | 4.97E−5 | −7712 | 2.00E−5 | |||||||
| rs8112998 | Intergenic (19) | 5.87E−5 | −8656 | 3.00E−5 | |||||||
| rs10408356 | Intergenic (19) | 7.97E−5 | −7703 | 8.00E−5 | |||||||
| EHD3 | 1.02E-9 | −12 721 | rs7083107 | ENTPD7 (10) | 8.08E−6 | −14 220 | 1.00E−4 | cg20203365 | Body | 6.11E−8 | −12 932 |
| rs10415181 | Intergenic (19) | 4.97E−5 | −7712 | 1.00E−6 | |||||||
| rs7535740 | Intergenic (1) | 5.75E−5 | 6561 | 1.00E−4 | |||||||
| rs8112998 | Intergenic (19) | 5.87E−5 | −8656 | 2.00E−6 | |||||||
| rs10408356 | Intergenic (19) | 7.97E−5 | −7703 | 3.00E−6 | |||||||
| GALNT2 | 1.68E-8 | 19 614 | rs7124929 | Intergenic (11) | 5.06E−5 | −8523 | 6.00E−5 | cg13359998 | Body | 9.35E−5 | −6311 |
| KCNQ5 | 4.59E-7 | 9706 | rs7083107 | ENTPD7 (10) | 8.08E−6 | −14 220 | 4.00E−5 | cg22394948 | 3′ UTR | 2.14E−5 | 8538 |
| rs7124929 | Intergenic (11) | 5.06E−5 | −8523 | 4.00E−5 | cg23221791 | Body | 1.42E−5 | 11 176 |
Gene names: SETBP1, SET binding protein 1; BAG3, BCL2-binding protein; KLHL6, Kelch-like protein 6; EHD3, EH domain containing protein 3; GALNT2, UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2; KCNQ5, K+ voltage gated channel.
Figure 3.
One SNP targeted four of the top six prioritized target genes. This SNP (rs7083107) is a master regulator and targets 53 genes at P < 10−4. The distance between nodes reflects the strength of the association between the SNP genotype and gene expression levels: the closer the gene is to the SNP, the higher the strength of association between the SNP genotype and gene expression. Prioritized genes indicated by Δ node.
Linear modeling to quantify the impact of the genetics and epigenetics on phenotype
To determine the impact of genetics and epigenetics on phenotypic variance, variables in a linear regression model were selected using backward elimination (28). In the genetic-only model, three (rs4145660, rs2137664, rs8112998) of the ten unique SNPs (Table 1) analyzed were included in the final optimal model. In the epigenetic-only model, four (cg23221791, cg20203365, cg25162588, cg13359998) of the eight unique CpG sites (Table 1) analyzed were included in the final model. The optimal combined model included three SNPs (rs4145660, rs2137664, rs8112998) and three CpG sites (cg23221791, cg13359998, cg25162588). These six variants target expression of five of the six top genes (SETBP1, KLHL6, EHD3, GALNT2 and KCNQ5). The proportion of phenotypic variance attributed to the reduced SNP (genetic) and modified cytosine (epigenetic) models are provided in Table 2. The contribution of genetics and epigenetics were 29.8 and 34.8%, respectively, and the proportion of phenotypic variance explained by the optimal combined model was 43.1%. The combined model explained variance significantly better than the null model (P = 1.65 × 10−12). It is worth noting that none of the 137 eQTLs was directly associated with the 143 modified CpG sites (i.e. methyl-QTLs) as the vast majority of SNPs were trans eQTLs and the modified CpG sites were local, and presumably cis-acting.
Table 2.
Proportion of AUC variance explained by linear models
| Independent variables | Predictors included in model | % Phenotypic variance explained |
|---|---|---|
| Optimal genetic model (3 SNPs) | rs4145660 | 29.8 |
| rs2137664 | ||
| rs8112998 | ||
| Optimal epigenetic model (4 CpG sites) | cg23221791 | 34.8 |
| cg20203365 | ||
| cg25162588 | ||
| cg13359998 | ||
| Optimal combined model (3 SNPs + 3 CpG sites) | rs4145660 | 43.1 |
| rs2137664 | ||
| rs8112998 | ||
| cg23221791 | ||
| cg25162588 | ||
| cg13359998 |
No cis-acting eQTLs for the six prioritized genes were associated with cytotoxicity below a threshold of P < 10−4. The eQTL database (eqtl.uchicago.edu) is a searchable web browser summarizing eQTLs (mostly cis-acting) from 17 studies. From the eQTL database, one cis-acting eQTL, rs11199066, was found to target BAG3 (29) and was also associated with clofarabine cytotoxicity (P = 0.0031, β = −9176). This cis-eQTL was not a methyl-QTL of either of the CpG sites in the gene body of BAG3.
In an analogous exercise to our optimal genetic and epigenetic model above, a correlation matrix and backward-elimination linear regression model were created for gene expression associations. All gene expression associations exhibited some degree of correlation with Pearson's R coefficients ranging 0.37–0.64 (Supplementary Material, Table S4); however, these were not so high as to infer complete dependence. The optimal gene expression association model included only SETBP1, KLHL6 and GALNT2. The proportion of phenotypic variance explained by these three gene associations was 28.8% (P = 8.61 × 10−9). Thus, our optimal combined SNP/CpG model outperformed the gene expression model.
As our cytotoxicity to gene expression associations were strong, we sought to clarify whether the SNPs and CpG sites in our optimal model retained significance after adjustment for the contribution of gene expression. As expected, the SNP/CpG P-value is attenuated when the correlated gene expression term is added to each model. However, in each case, some SNP/CpG signal remains (P < 0.05), indicating that some of the SNP/CpG association with AUC cannot be explained by the respective gene expression. These data are included in Supplementary Material, Table S5.
Gene expression of SETBP1, BAG3 and KLHL6 correlate with phenotype in both populations
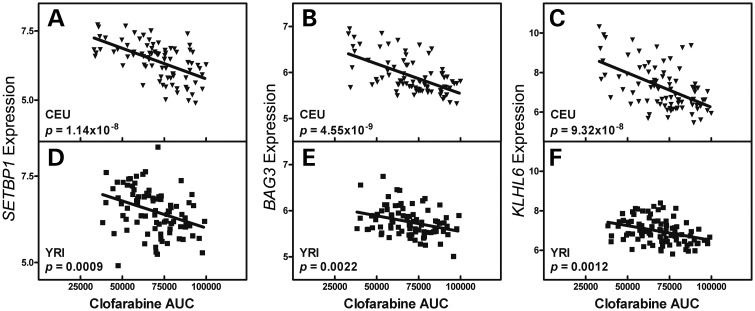
The top three (SETBP1, BAG3 and KLHL6) of the six genes identified were prioritized for functional validation with siRNA knockdown based on strength of correlation between gene expression and sensitivity to drug. Linear regression was used to correlate baseline log2-transformed gene expression with clofarabine AUC. Higher gene expression of SETBP1, BAG3 and KLHL6 is associated with susceptibility to clofarabine (lower AUC); thus lower expression confers resistance to clofarabine (Fig. 4). In each population, SETBP1 expression was negatively associated with clofarabine AUC (CEU r2 = 0.323, P = 1.14 × 10−8; YRI r2 = 0.122, P = 0.0009), BAG3 expression was negatively associated with clofarabine AUC (CEU r2 = 0.337, P = 4.55 × 10−9; YRI r2 = 0.105, P = 0.0022) and KLHL6 expression was negatively associated with clofarabine AUC (CEU r2 = 0.289, P = 9.32 × 10−8; YRI r2 = 0.116, P = 0.0012). Population interaction was significant for SETBP1 (P = 0.036), suggesting a stronger relationship between SETBP1 expression and clofarabine AUC in the CEU population over the YRI population. Population interaction with gene expression approached significance for BAG3 (P = 0.088) and was non-significant for KLHL6 (P = 0.85). For the next three genes, expression was positively correlated with AUC in two of the three (GALNT2 and KCNQ5), and negatively correlated in EHD3 (Supplementary Material, Fig. S2).
Figure 4.
Gene expression correlation with phenotype by population. SETBP1, BAG3 and KLHL6 expression levels in LCLs were significantly associated with clofarabine-induced cytotoxicity (AUC) in both CEU and YRI populations. Log2-transformed SETBP1 expression and clofarabine AUC correlation in CEU population (A) and YRI population (D). Log2-transformed BAG3 expression and clofarabine AUC correlation in CEU population (B) and YRI population (E). Log2-transformed KLHL6 expression and clofarabine AUC correlation in CEU population (C) and YRI population (F).
Gene expression associations with clofarabine AUC are consistent across expression array platforms
The correlation of inter-individual mRNA expression measurements across laboratories and platforms is low (median rho = 0.12) (30). In this study, the cytotoxicity to gene expression associations were initially performed using data from an Affymetrix exon array (28). To account for technical variation across platforms, we assessed whether these associations were still present when using expression data from an Illumina H6 v2 bead chip (31). All six of the prioritized genes had the same direction of association in linear regression as present in the Affymetrix exon array analysis (Supplementary Material, Fig. S3). Four of the six genes were in the top 500 of the 47K transcripts tested with P-values less than 0.001 (SETBP1, BAG3, KLHL6, EHD3). GALNT2 had a P-value below 0.005 and the association for KCNQ5 was non-significant (P = 0.12). These results indicate that our cytotoxicity to gene expression associations hold significance on multiple gene expression platforms.
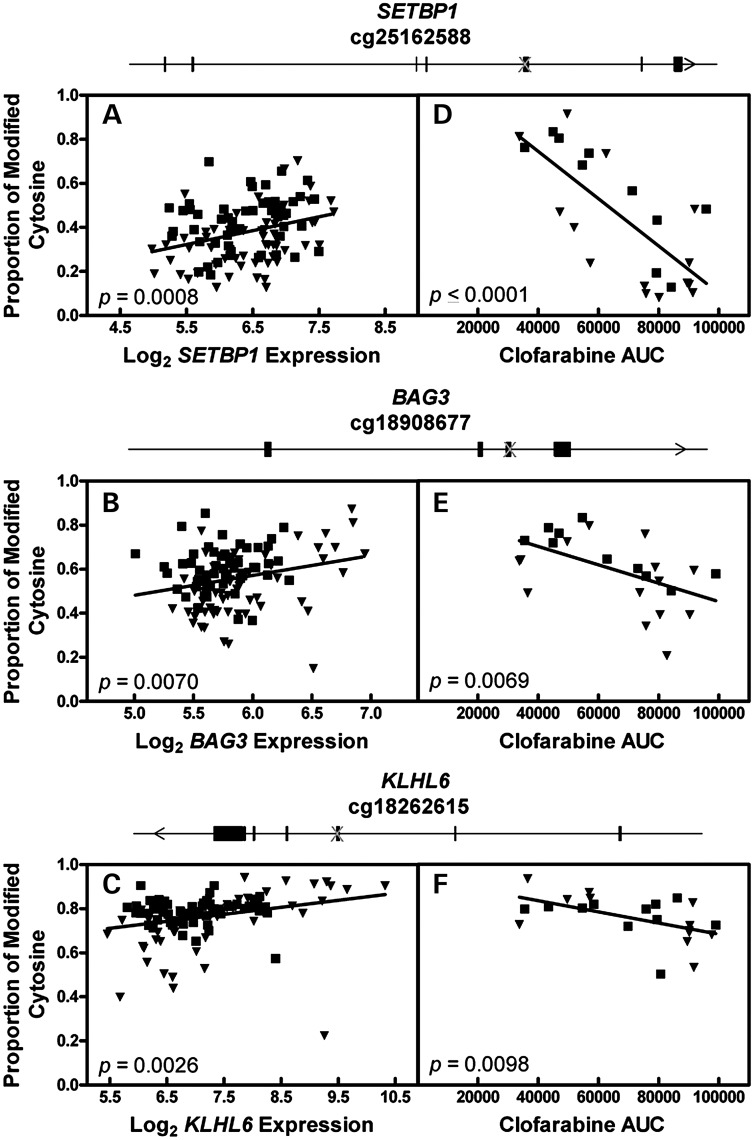
Validation of cytosine modification in SETBP1, BAG3, and KLHL6
The proportion of cytosine modification for CpG sites within SETBP1, BAG3 and KLHL6 was positively associated with expression of its respective gene (Fig. 5A–C; SETBP1 r2 = 0.096, P = 0.0008; BAG3 r2 = 0.063, P = 0.0070; KLHL6 r2 = 0.077, P = 0.0026). All unrelated LCLs from the methylation and gene expression arrays were used in this expression analysis. For all three genes, increased cytosine modification is associated with both greater gene expression and increased susceptibility to clofarabine. One modified cytosine found in each gene body of SETBP1, BAG3 and KLHL6 was selected for validation with bisulfite sequencing. For each gene, DNA was isolated from 24 unrelated LCLs, bisulfite treated and sequenced. Twenty-four LCLs were selected to represent samples from each population (YRI and CEU) as well as represent the most sensitive and resistant samples to clofarabine. The correlation between the degree of cytosine modification and clofarabine AUC was significant (negative association) for all three CpG sites interrogated (Fig. 5D–F). For cg25162588 in SETBP1, cytosine modification and phenotype were strongly correlated (r2 = 0.540, P < 0.0001). For cg18908677 in BAG3 and cg18262615 in KLHL6, cytosine modification and phenotype were correlated (r2 = 0.288, P = 0.0069 and r2 = 0.278, P = 0.0098, respectively). The cytosine modification associations of the additional three top genes (EHD3, GALNT2 and KCNQ5) are provided in Supplementary Material, Figure S4. The proportion of cytosine modification in EHD3 and GALNT2 was negatively correlated with expression and cytosine modification in KCNQ5 was positively correlated with expression. Nonetheless, the directionality of associations between expression, phenotype and cytosine modification of all six genes were internally consistent (Figs 4 and 5, Supplementary Material, Figs S2 and S4).
Figure 5.
Correlation of cytosine modification with gene expression and phenotype. (A–C) The proportion of cytosine modification in 131 cell lines as measured by DNA methylation array was correlated with gene expression as measured by exon gene expression array. (A) cg25162588 modification is associated with gene expression of SETBP1. (B) cg18908677 modification is associated with gene expression of BAG3. (C) cg18262615 modification is associated with gene expression of KLHL6. (D–F) DNA harvested from 24-cell lines were bisulfite treated and sequenced for 3 CpG sites. The proportion of cytosine modification was correlated with clofarabine AUC. (D) cg25162588 is found in the gene body of SETBP1, modification in 24 LCLs is associated with clofarabine AUC. (E) cg18908677 is found in the gene body of BAG3, modification in 24 LCLs is associated with clofarabine AUC. (F) cg18262615 is found in the gene body of KLHL6, modification in 24 LCLs is associated with clofarabine AUC. Inverted triangles, CEU cell line; squares, YRI cell line; Crosses, location of CpG within gene.
Knockdown of SETBP1, BAG3 and KLHL6 gene expression increases cellular resistance to clofarabine
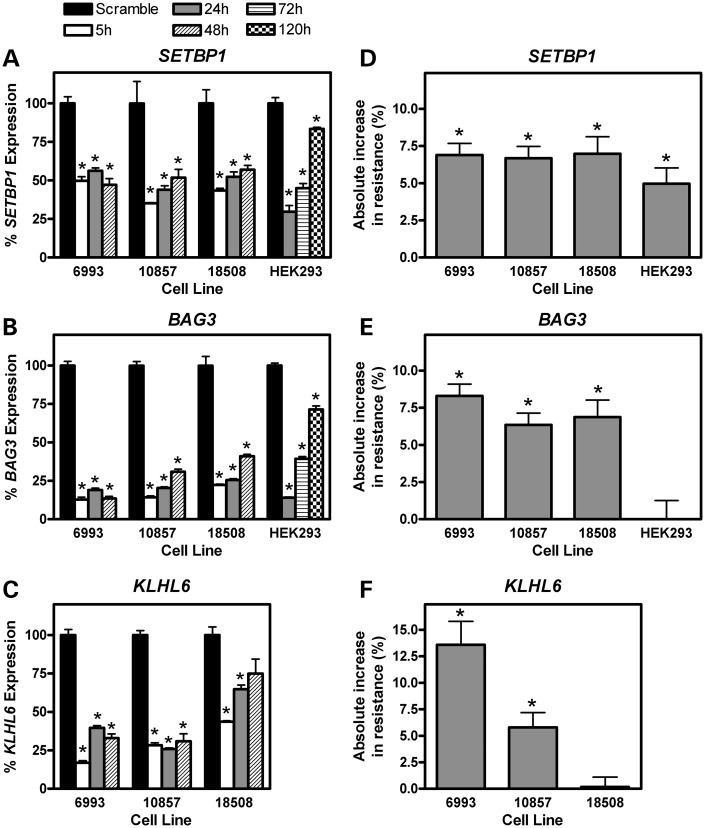
To validate the role of SETBP1, BAG3 and KLHL6, mRNA levels of each gene were decreased in LCLs with siRNA nucleofection. The relevance of these genes was assessed in three cell lines (6993, 10857, and 18508) by transfection with either a scramble siRNA control or siRNA targeting one of the three genes. By real-time polymerase chain reaction (RT-PCR), the degree of mRNA knockdown for each cell line and each gene relative to its time matched scrambled siRNA control was determined (Fig. 6A–C). For all LCL–gene combinations, there was a significant decrease in mRNA levels 5 h after exposure to siRNA relative to the scrambled control (P < 0.05). Gene knockdown was often most pronounced 5 h post-nucleofection with gradual restitution of expression over 48 h.
Figure 6.
Effect of gene knockdown on mRNA gene expression and cellular resistance to clofarabine. Following transfection with a scramble siRNA or siRNA targeting SETBP1, BAG3 or KLHL6, the effect on gene expression (A–C) and clofarabine-induced cytotoxicity (D–F) were determined. Knockdown was performed in three LCLs (6993, 10857 and 18508) and HEK293 cells with at least two biologic replicates (six or more technical replicates) for each cell line. mRNA levels were measured by Taq-man RT-PCR 5, 24 and 48 h post-siRNA transfection in LCLs and 24, 72 and 120 h in HEK293 cells. Gene expression at each time point is given as a percentage of expression relative to the scramble control. The scramble siRNA gene expression was measured at each time point; however, as the scramble expression was normalized to 100% at each time point, only one scramble expression bar is given for each gene–cell line combination. Cytotoxicity was measured 3 days after initial clofarabine exposure at the 125 nm concentration. Cell survival at the 125 nm concentration was calculated relative to control (no drug). The absolute increase in % cell survival after gene knockdown for each cell line is relative to the siRNA scramble for each cell line. *P < 0.05 by t-test between the scramble siRNA and the siRNA targeting each gene. Baseline KLHL6 gene expression in HEK293 cells was at the limits of detection by RT-PCR; thus, siRNA knockdown was not attempted in this gene–cell line combination.
Concomitant with mRNA knockdown of SETBP1, BAG3 and KLHL6 was an increase in cellular resistance to clofarabine cytotoxicity in 2 or more cell lines for each gene when compared with a scrambled siRNA control (Fig. 6D–F). The difference in cell survival (resistance to 125 nm clofarabine) between knockdown and scrambled control is significant when the absolute increase in resistance (%) is statistically different than 0. siSETBP1 knockdown conferred increased resistance to 125 nm clofarabine in LCL 6993 (6.88 ± 0.79%, P < 0.001), LCL 10857 (6.68 ± 0.79%, P < 0.001) and LCL 18508 (6.98 ± 1.15%, P < 0.001). BAG3 knockdown conferred increased resistance to 125 nm clofarabine in LCL 6993 (8.30 ± 0.79%, P < 0.001), LCL 10857 (6.35 ± 0.79%, P < 0.001) and LCL 18508 (6.86 ± 1.15%, P < 0.001). siKLHL6 knockdown conferred increased resistance to 125 nm clofarabine in LCL 6993 (13.60 ± 2.20%, P = 0.004), LCL 10857 (5.79 ± 1.39%, P = 0.005), but not in LCL 18508. Using a linear mixed effects model including all three cell lines and experimental replicates, the increase in cellular resistance to 125 nm clofarabine was significant for all three genes (siSETBP1 P = 0.00625, siBAG3 P = 0.00218, siKLHL6 P = 0.00752). These results are mechanistically consistent with our gene expression data given in Figure 4, where lower gene expression was associated with greater cellular resistance to clofarabine.
As clofarabine's toxicity profile includes acute kidney injury, we sought to determine whether the genes validated in LCLs would also impact susceptibility to clofarabine in HEK293 cells. HEK293 cells are human in origin, renally derived and easily nucleofected. siSETBP1 knockdown yielded a small yet statistically significant change in resistance to clofarabine cytotoxicity (5.0 ± 1.0%, P = 0.0089, Fig. 6D). The mRNA level 24 h following siSETBP1 nucleofection was 29.7% when compared with its scrambled control (Fig. 6A). The effects of siSETBP1 knockdown were small, but consistent with those observed in LCLs. Despite effective mRNA expression reduction, siBAG3 knockdown did not cause any measurable increase in resistance (Fig. 6E). Baseline KLHL6 gene expression was at the limits of RT-PCR detection in HEK293 cells (data not shown) compared with LCLs and siRNA knockdown was not warranted.
DISCUSSION
This study reveals the results of comprehensive association studies between clofarabine susceptibility in LCLs with two million common SNPs, expression of 12 562 genes and modification of 283 540 CpG sites. Given clofarabine's expanding set of clinical indications, we were interested in identifying both genetic and epigenetic susceptibility factors that may be readily translatable to improving clofarabine's efficacy as a chemotherapeutic agent in the treatment of hematologic malignancies. To prioritize the 234 genes correlated to sensitivity to clofarabine, we refined the list to only those genes that had both eQTLs and modified cytosines associated with gene expression and clofarabine sensitivity. Six genes (SETBP1, BAG3, KLHL6, EHD3, GALNT2 and KCNQ5) were implicated by both a targeting eQTL and local modified cytosine. The top three genes (SETBP1, BAG3 and KLHL6) were subject to siRNA knockdown resulting in a significant change in cellular resistance to clofarabine in LCLs.
The analysis approach undertaken here is unique as it combines genetics and epigenetics to identify genes relevant to drug sensitivity. Past approaches have utilized gene expression to prioritize genetic variants associated with a phenotype (32–34), used linkage and pedigree analyses (35,36) or used LCL findings to confirm variants in a clinical GWAS (37). In this study, we employed a novel approach to identify a set of SNPs and CpG sites that explain ∼43% of the observed variation in phenotype. Incorporating modified cytosines in this study improved the predictive quality of these markers over SNPs alone, despite utilizing the same six genes.
Each of the six prioritized genes had one or more associated modified cytosines in its gene body or 3′ UTR. It is important to note that LCLs are immortalized cell lines and that EBV immortalization could affect methylation status (38,39). Further, these changes in methylation status might not be stable across multiple passages (40,41). However, a comparison between our 450K data and Fraser et al. (42,43) suggested that cytosine modification levels in these LCLs appeared to be stable. Previous microarray platforms have allowed genome-wide investigation of the contribution of cytosine modification to gene expression; however, these platforms have focused on 27 000 CpG sites in the promoter regions of genes, largely ignoring the gene body or 3′ UTR (44). Promoter region or miRNA cytosine modification has been correlated with expression of SETBP1, BAG3, KLHL6 and EHD3 (45–48). Little is known regarding the mechanistic interaction between methylation and gene expression of either GALNT2 or KCNQ5. In our association of 283 540 CpG sites, 10–20 CpG probes cover each gene, with several found in CpG shores, shelves and gene bodies, outside of known CpG islands. Here we identify six genes whose expression are correlated with cytosine modification of their gene body or 3′ UTR, and are associated with a pharmacological phenotype. In contrast to promoter region methylation, it is presently unknown why gene body methylation is often associated with increased gene expression (49). However, evidence suggests that cytosine modification along the gene body can affect nucleosome positioning, splicing and histone modifications, all of which are involved in the transcriptional process (50–52). Therefore, associations between gene body cytosine modifications, gene expression and pharmacologic phenotypes provide a more complete foundation for studying the functional consequences of transcriptional regulation by cytosine modification.
The identified genetic and epigenetic variants associated with clofarabine-mediated cytotoxicity have clinical application. As the advent of large-scale patient population-based genotyping has begun in certain regions of the USA (53), there is increasing need to understand and effectively utilize this information. Recent phase II clinical trials for clofarabine are expanding its indications for a variety of hematologic malignancies (3–5). A straightforward predictive model such as this can be validated in clinical trials and employed to predict response to the drug. Our optimal combined genetic and epigenetic model consisting of three SNPs and three CpG sites explained 43.1% of clofarabine-mediated cytotoxicity variance. Based on existing clinical data, each of the top genes identified provides a plausible mechanism for its importance.
SETBP1 (SET binding protein 1) is a transcription factor that binds the nuclear oncogene SET (54). SETBP1 is thought to bind SET and suppress tumorigenesis, allowing apoptosis to proceed normally (55). SETBP1 mutations have been implicated in the pathogenesis of both pediatric acute T-cell lymphoblastic leukemia (56) as well as atypical chronic myeloid leukemia (57). In our study, SETBP1 gene knockdown increased resistance to clofarabine cytotoxicity, presumably reducing apoptosis. In cell culture, clofarabine has been shown to induce cell death through apoptotic pathways (58) and clofarabine activated caspase 3/7 in our own experiments (data not shown). Mechanistically, our results are consistent with the published literature as reduced SETBP1 levels increased cellular resistance to clofarabine.
In tumor cells, BAG3 (BCL2-associated anthogene) is anti-apoptotic, interacting with Bcl-2 to prevent cell death (59). BAG3 is over-expressed in a variety of solid tumors (60–62) and hematologic malignancies (63). Multiple cell culture and mouse models have shown that silencing BAG3 gene expression confers increased susceptibility to chemotherapy and cell death (60,63). Our studies revealed that BAG3 gene expression was highly associated with susceptibility to clofarabine in LCLs and BAG3 knockdown increased resistance to clofarabine-induced cytotoxicity in LCLs. Thus, high expression of BAG3 confers resistance to chemotherapy in malignant tissue, but sensitivity to clofarabine in LCLs.
KLHL6 (kelch-like protein 6) is involved in B-lymphocyte antigen receptor signaling and germinal center formation (64,65). Loss of KLHL6 expression in mice led to reduced numbers of mature-circulating B-cells (65). Using whole-genome sequencing of chronic lymphocytic leukemia patients, KLHL6 was one of four genes found with consistent somatic mutations affecting gene function (66). Little information is available regarding the role of KLHL6 in cytoprotection or cell death. Our results suggest a role for KLHL6 as a marker of cell susceptibility to clofarabine, as gene expression was negatively correlated with AUC phenotype. KLHL6 knockdown led to increased resistance to clofarabine in our mixed-effects model of LCLs.
As detailed above, prior data suggest the importance of SETBP1, BAG3 and KLHL6 in the pathogenesis of hematologic malignancy, so it is not surprising we have found associations with clofarabine-mediated cytotoxicity. As clofarabine's toxicity profile includes high rates of acute kidney injury, we considered whether some of the same set of identified genes would also mediate susceptibility in kidney cells. It is well understood that gene expression and methylation patterns are tissue specific (67,68). However, understanding the role of genes in different tissues will help uncover heretofore under-recognized pathways of kidney injury and provide an initial step to differentiate predictors of clofarabine bone marrow suppression from predictors of its other toxicities. Using siRNA knockdown, the relevance of SETBP1 and BAG3 were examined in HEK293 cells. siSETBP1, but not siBAG3, led to a small but significant change in HEK293 cell susceptibility. Of interest, germ line mutations in SETBP1 have produced a known renal phenotype in patients. SETBP1 variants have been implicated in the pathogenesis of Schinzel-Giedeon Syndrome (SGS), characterized by altered facies and mental disability (69). In a case series of 35 patients with SGS, 31 had renal manifestations including hydronephrosis (70) or renal calyceal hypertrophy (71). It is unclear whether these phenotypic changes would be associated with changes in renal cellular sensitivity to clofarabine. Our model is certainly limited in that HEK293 cells are immortalized and not an ideal model of renal epithelial cell injury. However, HEK293 cells are human in origin, renally derived and easily nucleofected. Further, a population-based approach to identifying genetic and epigenetic variants in primary renal tissue is not feasible at present as no set of renal cell lines analogous to LCLs exists. Further studies in primary renal cell lines, in vivo models, and human cohorts will be necessary to confirm the role of SETBP1 in conferring susceptibility to clofarabine-mediated renal cell death.
In conclusion, we present comprehensive genome-wide association and methylation association studies of clofarabine-mediated susceptibility. Through association studies of genetic variants, gene expression and methylation, we identified potential predictors of chemotherapeutic response and toxicity. The variants impacting expression levels of SETBP1, BAG3 and KLHL6 may serve as future biomarkers of response for patients with a variety of hematologic malignancies. Additionally, through siRNA knockdown in HEK293 cells, we have identified SETBP1 as a worthy target of future investigation in clofarabine-induced renal toxicity.
MATERIALS AND METHODS
Cell lines and drug
All LCLs were cultured in RPMI 1640 media containing 15% bovine growth serum (Hyclone, Logan, UT, USA) and 3.7 mm glutamine. Cell lines were diluted three times per week at a concentration of 300 000–350 000 cells/ml and were maintained in a 37°C, 5% CO2 humidified incubator. Medium and components were purchased from Cellgro (Herndon, VA). All HapMap LCLs were purchased from Coriell Institute for Medical Research (www.coriell.org). A total of 190 LCLs were phenotyped as follows: 88 from CEU1 (HAPMAPPT01, all except 12236 and 12716), 88 from YRI1 (HAPMAPPT03, all except 18852 and 18912) and 14 from YRI3 (HAPMAPPT04, includes LCLs 18486, 18498, 18499, 18909, 18510, 18511, 18519, 18916, 19108, 19114, 19147, 19190, 19225 and 19257).
HEK293 cells were a gift from the Benjamin S. Ko laboratory. HEK293 cells were maintained in DMEM media (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (HyClone) and 1% l-glutamine (Life Technologies). HEK293 cells were diluted to 20–30% confluency three times a week and maintained at 37°C in 95% humidified atmosphere with 5% CO2.
Clofarabine was commercially obtained (Sigma, St Louis, MO, USA). Phosphate buffered saline (PBS), pH 7.4, was purchased from Life Technologies, Inc.
Clofarabine-induced cytotoxicity
HapMap LCLs from 88 CEU Phase 1 individuals, 88 YRI Phase 1 individuals and 14 YRI Phase 3 individuals were phenotyped for cellular sensitivity to clofarabine using a short-term, colorimetric growth inhibition assay. Cell lines were maintained in exponential growth phase with >85% viability as determined using the Vi-Cell XR viability analyzer (Beckman Coulter, Fullerton, CA, USA) and plated in triplicate at a density of 1 × 105 cells/ml in 96-well round-bottom plates (Corning, Inc., NY, USA) 24 h prior to drug treatment. Clofarabine (Sigma) was prepared in the dark by dissolving powder in ice cold sterile PBS to obtain a stock solution of 500 µm with subsequent drug filtration. The drug was added to media to obtain six different concentrations of clofarabine (60, 125, 250, 375, 500 and 1000 nm) that were in turn added to the 96-well plates of each cell line. The range of concentrations was carefully chosen based on previously published in vitro cytotoxicity data (13). Alamar Blue (Life Technologies) was added 48 h after drug addition and 24 h before absorbance reading at wavelengths of 570 and 600 nm using the Synergy-HT multi-detection plate reader (BioTek, Winooski, VT, USA). Percent survival was quantified relative to a control well without drug addition. Each 5-day cytotoxicity experiment was conducted two or more separate times for each cell line (minimum of two biologic replicates and six technical replicates). The phenotype chosen for analysis was AUC calculated from the survival curve, as over 10% of cell lines were highly resistant with incalculable IC50. AUC was obtained by fitting percent survival against drug concentration, using the trapezoidal rule. The AUC phenotype has previously been shown to increase power for detection between genotypes when compared with an IC50 phenotype (72).
HEK293 cellular sensitivity was measured in an analogous fashion with two notable exceptions. First, as cells rapidly divided and were adherent, they were plated at a density of 2 × 104 in 96-well flat-bottom plates. Second, growth inhibition was measured using the CellTiter-Glo assay (Promega, Madison, WI, USA).
Genome-wide association and gene expression association with phenotype
A GWAS was performed, using the quantitative trait disequilibrium test (QTDT) total association model (73), to identify SNPs associated with cellular sensitivity to clofarabine. SNPs were included in the association analysis if they possessed minor allele frequency >5%, had no Mendelian errors, and were in Hardy–Weinberg equilibrium (P > 0.001). A covariate for population was used in the QTDT analysis. Regression analyses, including the relationship between clofarabine AUC and gene expression levels and the relationship between SNP and gene expression, were performed in R (http://www.r-project.org/). Global gene expression of the LCLs as measured by Affymetrix exon array has been previously published (74), GSE 7851. To associate baseline gene expression with phenotype, a Toeplitz covariance model was used with two diagonal bands in the regression analysis due to the relatedness of the HapMap samples (trio structure), as previously described (75). P-values presented are after conditioning on intrinsic growth rate (76). A nominal P-value of 1 × 10−4 was used as a filtering threshold in the GWAS and in the subsequent eQTL analysis (P < 10−4 in either the YRI or CEU population).
eQTL enrichment analysis and visualization
To test for enrichment of eQTLs among the top clofarabine-associated SNPs (P < 0.0001), we generated 1000 random sets of SNPs from the set of HapMap CEU and YRI SNPs, each with matching minor allele frequency distribution as the set of the most significantly associated SNPs. For each such random set, the number of eQTLs was determined, yielding a distribution showing the expected eQTL count from which an empirical P-value for the enrichment was calculated (77). For visualization of the relationships between a master regulator eQTL and its target genes, the open-source software Cytoscape was used (www.cytoscape.org).
Correlation analysis of cytosine modification with phenotype
Cytosine modification levels were recently measured by our group using the Illumina Infinium Human Methylation 450 bead chip array (Illumina, Inc., San Diego, CA, USA) (78), accession number GSE39672. Details of sample collection, profiling, data collection and array analysis are included (Supplementary Material) (79). The M-values, defined as log2 ratio of the intensities of modified probe versus unmodified probe, were demonstrated to provide better detection sensitivity at extreme cytosine modification levels (80). After quantile normalization and correction for batch effect, the M-values of 283540 CpG sites on autosomes were tested for association with clofarabine AUC using a linear model with population identity and gender as covariates in the unrelated CEU (n = 58) and YRI (n = 57) samples. A nominal P-value of 1 × 10−4 was used as the filtering threshold for associations.
Bisulfite sequencing of selected differentially modified probes
To confirm the cytosine modification levels measured by the 450K array, we used bisulfite sequencing to test CpG modification variation within SETBP1, BAG3 and KLHL6. Twenty-four samples from the original DNA collection used in modification profiling were used for bisulfite sequencing. Bisulfite primers were designed to amplify ∼200 bp fragments that included the CpG sites interrogated by the array as well as adjacent CpGs. ZymoTaq polymerase was used for PCR amplification and DNA sequencing was performed at the University of Chicago Sequencing Core Facility. The proportion of cytosine modification was determined from a chromatogram read in Chromas lite 2.1 software (chromas@technelysium.com.au). Primer sequences (PCR Tm °C): SETBP1 (56.0) F: AGTTATTTTAGGAGGTGTGTTTAAGT, R: ATCAAAAATAACCATAACTACCTCTC; BAG3 (61.3) F: TATAAGATTTAGGGGGATGATTGG, R: AAACATACCACCATAACCAATCTACTAA; KLHL6 (57.3) F: TTTGAATTTTGAGGAGTTTTTTGAT, R: CCAAAAAAATAATACATCCTAACTTCCTA.
Proportion of phenotypic variance estimation
To estimate the proportion of variance in clofarabine AUC explained by genetics and/or epigenetics, linear regression models were fit to the AUC data from unrelated individuals with terms for each SNP and/or each CpG site listed in Table 1 as well as a covariate for population (CEU or YRI) using R version 2.15.2. A backward-elimination approach was applied for model reduction. The term with the highest P-value was removed at each iteration until only terms with P < 0.05 remained. Adjusted R2 values represent the proportion AUC variance explained by each reduced model (SNPs only, CpG sites only, SNPs and CpG sites combined). Analogously, a backward-elimination approach was also employed for the genes in Table 1 whose baseline expression was associated with AUC. Multiple linear regression was further used to determine population:gene expression interaction P-values and SNP/CpG site associations corrected for gene expression (Supplementary Material, Table S5). A gene expression Pearson correlation matrix was generated using the corr.test function in the R psych package (Supplementary Material, Table S4).
siRNA nucleofection
LCLs were diluted to 500 000 cells/ml 1 day prior to nucleofection. The cells were nucleofected using the SF Cell Line 96-well Nucleofector Kit (Lonza, Inc., Basel, Switzerland). Cells were centrifuged at 90g for 10 min at room temperature and re-suspended at a concentration of 1 000 000 cells/20 µl in SF/supplement solution (included in SF Kit Lonza Catalog V4SC2096) and 2000 nm final concentration of All Stars Negative Control siRNA (Qiagen, Inc., Valencia, CA, USA) or a pool of four siRNA constructs from each of the following genes: SETBP1 (SI04954474, SI04954453, SI04954460, SI04954467), BAG3 (SI02632819, SI03066378, SI00062132, SI02632812) and KLHL6 (SI04328562, SI04374524, SI04194792, SI04264547) (Qiagen). The cells were nucleofected using the DN-100 program on Lonza's Nucleofector 96-well shuttle system. Cells were allowed to rest for 10 min prior to the addition of pre-warmed (37°C water bath) RPMI media and then for another 5 min in the warm RPMI media. Cells were then plated for mRNA harvest or drug treatment. LCLs were harvested at 5, 24 and 48 h post-nucleofection for gene expression measurement. HEK293 cells were nucleofected in an analogous manner using DMEM media and harvested at 24, 72 and 120 h for gene expression measurement.
Quantitative real-time PCR
Quantitative RT-PCR (qRT-PCR) was performed to measure the level of expression of SETBP1, BAG3 and KLHL6 in LCLs. A total of 3 million cells were pelleted at 5 (corresponds to time 0 h for clofarabine cytotoxicity assay), 24 and 48 h after nucleofection, washed in ice-cold PBS and centrifuged to remove PBS. In HEK293 cells, 3 million cells were pelleted at 24, 72 and 120 h after nucleofection given the relative stability of nucleofection in this cell line. All pellets were flash-frozen and stored at −80°C until RNA isolation. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) following the manufacturer's protocol. RNA quality assessment and quantification were conducted using the optical spectrometry 260/280 nm ratio. Subsequently, mRNA was reverse transcribed to cDNA using Applied Biosystems High Capacity Reverse Transcription Kit (Life Technologies). The final concentration of cDNA was 25 ng/ml. qRT-PCR was performed for the three genes above and huB2M (beta-2-microglobulin) as an endogenous control using TaqMan Gene Expression Assays (Life Technologies) on the Applied Biosystems Viia 7 RT-PCR system. Total reaction was carried out in 12 µl volume, which consisted of 6 µl of TaqMan Fast Advanced Master Mix, 0.6 ml of each primer/probe mix (final of 900 nm forward and reverse primers and 250 nm of probe) and 0.4 ml of water, along with 5 µl of 1.25 ng/ml cDNA. All Taqman primer/probe mixtures were labeled with a FAM or VIC reporter dye and the MGB quencher dye. The fast thermocycler parameters were 95°C for 20 s, 40 cycles of 95°C for 1 s and then 60°C for 20 s, with ramping speeds of 1.6–1.98 C/s. Viia 7 automatically set the CT threshold and baseline for each experiment.
A relative standard curve method was used to obtain the relative expression of each gene for samples treated with their associated pool of siRNA or scrambled siRNA. Each sample's gene expression of SETBP1, BAG3 or KLHL6 was first divided by huB2M expression to create a normalized ratio of gene expression relative to the housekeeping gene. Then, the normalized ratio of the siRNA experimental sample was compared with a time-matched scrambled siRNA control. For Figure 6, gene expression at each time point is given as a percentage of expression relative to the scramble control. While the scramble siRNA gene expression was measured at each time point, only one scramble expression bar is given for each gene–cell line combination because the scrambled siRNA expression was set to 100% for each time point. Each knockdown experiment was conducted two or more separate times, with the RT-PCR samples run in triplicate. Within each cell line, statistical significance was assessed based on an ANOVA between time points.
Drug treatment after nucleofection
LCLs were plated in triplicate at a density of 1 × 105 cells per milliliter in 96-well round-bottom plates (Corning, Inc.) after a 15 min resting period following nucleofection. Five hours after plating, either PBS vehicle or clofarabine was added to the cells at a concentration of 125 nm. HEK293 cells were plated in triplicate at a density of 5 × 104 cells/ml in 96-well flat-bottom plates and clofarabine 125 nm was added 24 h later. Each knockdown experiment was conducted two or more separate times (at least two biologic replicates and six technical replicates). Cell survival was measured at 72 h post-drug treatment using the Cell-Titer Glo assay (Promega). Absolute increase in resistance to clofarabine was calculated as follows: (SurvivalsiGene,125nM/SurvivalsiGene,PBS) – (SurvivalSCR,125nM/SurvivalSCR,PBS), where 125 nm = clofarabine treated cells, PBS is the vehicle treated cells, siRNA experimental knockdown group and SCR the scrambled siRNA control group.
siRNA statistical analysis
To assess the size and significance of the effect of siRNA on LCL survival after clofarabine treatment, the following linear mixed model was fit for each gene: survival ∼ siRNA + (1|id) + (1|experiment), where siRNA is 1 for the gene knockdown and 0 for scrambled control. Cell line id and experiment were used as random effects to properly account for the correlation between responses as previously described (25). For HEK293 cells, the linear mixed model used was: survival ∼ siRNA + (1|experiment). The mixed-effects model was fit using the lmer function from the lme4 package in R version 2.15.2. Significance of the siRNA term in the model was assessed using a likelihood ratio test.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a Specialized Center of Research Grant from the Leukemia and Lymphoma Society, Pharmacogenetics of Anticancer Agents Research Group (U01GM61393), and R21HG006367 (W.Z. and M.E.D.). M.T.E. was supported by the National Institutes of Health (T32 GM007019). H.E.W. was supported by the NIH/NCI National Research Service Award (F32CA165823).
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the work of the sequencing core at the University of Chicago and the help of the PAAR cell core run by Dr Stephanie Huang. The technical advice and expertise of Dr Yujia Wen, Nirav Antao, Claudia Wing, and Shan Wong is appreciated.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kantarjian H., Gandhi V., Cortes J., Verstovsek S., Du M., Garcia-Manero G., Giles F., Faderl S., O'Brien S., Jeha S., et al. Phase 2 clinical and pharmacologic study of clofarabine in patients with refractory or relapsed acute leukemia. Blood. 2003;102:2379–2386. doi: 10.1182/blood-2003-03-0925. [DOI] [PubMed] [Google Scholar]
- 2.Advani A.S., Gundacker H.M., Sala-Torra O., Radich J.P., Lai R., Slovak M.L., Lancet J.E., Coutre S.E., Stuart R.K., Mims M.P., et al. Southwest Oncology Group Study S0530: a phase 2 trial of clofarabine and cytarabine for relapsed or refractory acute lymphocytic leukaemia. Br. J. Haematol. 2010;151:430–434. doi: 10.1111/j.1365-2141.2010.08387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnett A.K., Russell N.H., Kell J., Dennis M., Milligan D., Paolini S., Yin J., Culligan D., Johnston P., Murphy J., et al. European development of clofarabine as treatment for older patients with acute myeloid leukemia considered unsuitable for intensive chemotherapy. J. Clin. Oncol. 2010;28:2389–2395. doi: 10.1200/JCO.2009.26.4242. [DOI] [PubMed] [Google Scholar]
- 4.Faderl S., Ravandi F., Huang X., Wang X., Jabbour E., Garcia-Manero G., Kadia T., Ferrajoli A., Konopleva M., Borthakur G., et al. Clofarabine plus low-dose cytarabine followed by clofarabine plus low-dose cytarabine alternating with decitabine in acute myeloid leukemia frontline therapy for older patients. Cancer. 2012;118:4471–4477. doi: 10.1002/cncr.27429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Besien K., Stock W., Rich E., Odenike O., Godley L.A., O'Donnell P.H., Kline J., Nguyen V., Del Cerro P., Larson R.A., et al. Phase I-II study of clofarabine-melphalan-alemtuzumab conditioning for allogeneic hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 2012;18:913–921. doi: 10.1016/j.bbmt.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamba J.K. Genetic factors influencing cytarabine therapy. Pharmacogenomics. 2009;10:1657–1674. doi: 10.2217/pgs.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamba J.K., Crews K., Pounds S., Schuetz E.G., Gresham J., Gandhi V., Plunkett W., Rubnitz J., Ribeiro R. Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants. J. Pharmacol. Exp. Ther. 2007;323:935–945. doi: 10.1124/jpet.107.128595. [DOI] [PubMed] [Google Scholar]
- 8.Thudium K.E., Ghoshal S., Fetterly G.J., Haese J.P., Karpf A.R., Wetzler M. Synergism between clofarabine and decitabine through p53R2: a pharmacodynamic drug-drug interaction modeling. Leuk. Res. 2012;36:1410–1416. doi: 10.1016/j.leukres.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadia T.M., Jabbour E., Kantarjian H. Failure of hypomethylating agent-based therapy in myelodysplastic syndromes. Semin. Oncol. 2011;38:682–692. doi: 10.1053/j.seminoncol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Shahriar M., Zhang J., Ahmed S.U., Lim S.H. Clofarabine induces hypomethylation of DNA and expression of Cancer-Testis antigens. Leuk. Res. 2009;33:1678–1683. doi: 10.1016/j.leukres.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Ajavon A.D., Bonate P.L., Taft D.R. Renal excretion of clofarabine: assessment of dose-linearity and role of renal transport systems on drug excretion. Eur. J. Pharm. Sci. 2010;40:209–216. doi: 10.1016/j.ejps.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Kintzel P.E., Visser J.A., Campbell A.D. Clofarabine-associated acute kidney injury and proteinuria. Pharmacotherapy. 2011;31:923. doi: 10.1592/phco.31.9.923. [DOI] [PubMed] [Google Scholar]
- 13.Beesley A.H., Palmer M.L., Ford J., Weller R.E., Cummings A.J., Freitas J.R., Firth M.J., Perera K.U., de Klerk N.H., Kees U.R. In vitro cytotoxicity of nelarabine, clofarabine and flavopiridol in paediatric acute lymphoblastic leukaemia. Br. J. Haematol. 2007;137:109–116. doi: 10.1111/j.1365-2141.2007.06527.x. [DOI] [PubMed] [Google Scholar]
- 14.King K.M., Damaraju V.L., Vickers M.F., Yao S.Y., Lang T., Tackaberry T.E., Mowles D.A., Ng A.M., Young J.D., Cass C.E. A comparison of the transportability, and its role in cytotoxicity, of clofarabine, cladribine, and fludarabine by recombinant human nucleoside transporters produced in three model expression systems. Mol. Pharmacol. 2006;69:346–353. doi: 10.1124/mol.105.015768. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler H.E., Dolan M.E. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics. 2012;13:55–70. doi: 10.2217/pgs.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox N.J., Gamazon E.R., Wheeler H.E., Dolan M.E. Clinical translation of cell-based pharmacogenomic discovery. Clin. Pharmacol. Ther. 2012;92:425–427. doi: 10.1038/clpt.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters E.J., Kraja A.T., Lin S.J., Yen-Revollo J.L., Marsh S., Province M.A., McLeod H.L. Association of thymidylate synthase variants with 5-fluorouracil cytotoxicity. Pharmacogenet. Genomics. 2009;19:399–401. doi: 10.1097/FPC.0b013e328329fdec. [DOI] [PubMed] [Google Scholar]
- 18.Niu N., Schaid D.J., Abo R.P., Kalari K., Fridley B.L., Feng Q., Jenkins G., Batzler A., Brisbin A.G., Cunningham J.M., et al. Genetic association with overall survival of taxane-treated lung cancer patients - a genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study. BMC Cancer. 2012;12:422. doi: 10.1186/1471-2407-12-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown C.C., Havener T.M., Medina M.W., Auman J.T., Mangravite L.M., Krauss R.M., McLeod H.L., Motsinger-Reif A.A. A genome-wide association analysis of temozolomide response using lymphoblastoid cell lines shows a clinically relevant association with MGMT. Pharmacogenet. Genomics. 2012;22:796–802. doi: 10.1097/FPC.0b013e3283589c50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra A.K., Crews K., Pounds S., Cao X., Downing J.R., Raimondi S., Campana D., Ribeiro R.C., Rubnitz J.E., Lamba J.K. Impact of genetic variation in FKBP5 on clinical response in pediatric acute myeloid leukemia patients: a pilot study. Leukemia. 2011;25:1354–1356. doi: 10.1038/leu.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziliak D., O'Donnell P.H., Im H.K., Gamazon E.R., Chen P., Delaney S., Shukla S., Das S., Cox N.J., Vokes E.E., et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Transl. Res. 2011;157:265–272. doi: 10.1016/j.trsl.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S.H., Yang W., Fan Y., Stocco G., Crews K.R., Yang J.J., Paugh S.W., Pui C.H., Evans W.E., Relling M.V. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia. 2011;25:66–74. doi: 10.1038/leu.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan X.L., Moyer A.M., Fridley B.L., Schaid D.J., Niu N., Batzler A.J., Jenkins G.D., Abo R.P., Li L., Cunningham J.M., et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin. Cancer Res. 2011;17:5801–5811. doi: 10.1158/1078-0432.CCR-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra A.K., Crews K.R., Pounds S., Cao X., Feldberg T., Ghodke Y., Gandhi V., Plunkett W., Dolan M.E., Hartford C., et al. Genetic variants in cytosolic 5’-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J. Pharmacol. Exp. Ther. 2011;339:9–23. doi: 10.1124/jpet.111.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark A.L., Delaney S.M., Wheeler H.E., Im H.K., Dolan M.E. Functional consequences of PRPF39 on distant genes and cisplatin sensitivity. Hum. Mol. Genet. 2012;21:4348–4355. doi: 10.1093/hmg/dds266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen Y., Gamazon E.R., Bleibel W.K., Wing C., Mi S., McIlwee B.E., Delaney S.M., Duan S., Im H.K., Dolan M.E. An eQTL-based method identifies CTTN and ZMAT3 as pemetrexed susceptibility markers. Hum. Mol. Genet. 2012;21:1470–1480. doi: 10.1093/hmg/ddr583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamazon E.R., Zhang W., Konkashbaev A., Duan S., Kistner E.O., Nicolae D.L., Dolan M.E., Cox N.J. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R.S., Duan S., Bleibel W.K., Kistner E.O., Zhang W., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang R.S., Duan S., Kistner E.O., Bleibel W.K., Delaney S.M., Fackenthal D.L., Das S., Dolan M.E. Genetic variants contributing to daunorubicin-induced cytotoxicity. Cancer Res. 2008;68:3161–3168. doi: 10.1158/0008-5472.CAN-07-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang R.S., Duan S., Shukla S.J., Kistner E.O., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Dolan M.E. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J. Hum. Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartford C.M., Duan S., Delaney S.M., Mi S., Kistner E.O., Lamba J.K., Huang R.S., Dolan M.E. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113:2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welsh M., Mangravite L., Medina M.W., Tantisira K., Zhang W., Huang R.S., McLeod H., Dolan M.E. Pharmacogenomic discovery using cell-based models. Pharmacol. Rev. 2009;61:413–429. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson V.G., Motsinger-Reif A., Hardison N.E., Peters E.J., Havener T.M., Everitt L., Auman J.T., Comins D.L., McLeod H.L. Identification and replication of loci involved in camptothecin-induced cytotoxicity using CEPH pedigrees. PLoS ONE. 2011;6:e17561. doi: 10.1371/journal.pone.0017561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler H.E., Gamazon E.R., Wing C., Njiaju U.O., Njoku C., Baldwin R.M., Owzar K., Jiang C., Watson D., Shterev I., et al. Integration of cell line and clinical trial genome-wide analyses supports a polygenic architecture of paclitaxel-induced sensory peripheral neuropathy. Clin. Cancer Res. 2012;19:491–499. doi: 10.1158/1078-0432.CCR-12-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caliskan M., Cusanovich D.A., Ober C., Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum. Mol. Genet. 2011;20:1643–1652. doi: 10.1093/hmg/ddr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernando H., Shannon-Lowe C., Islam A.B., Al-Shahrour F., Rodriguez-Ubreva J., Rodriguez-Cortez V.C., Javierre B.M., Mangas C., Fernandez A.F., Parra M., et al. The B cell transcription program mediates hypomethylation and overexpression of key genes in Epstein-Barr virus-associated proliferative conversion. Genome Biol. 2013;14:R3. doi: 10.1186/gb-2013-14-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grafodatskaya D., Choufani S., Ferreira J.C., Butcher D.T., Lou Y., Zhao C., Scherer S.W., Weksberg R. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 2010;95:73–83. doi: 10.1016/j.ygeno.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Noer A., Lindeman L.C., Collas P. Histone H3 modifications associated with differentiation and long-term culture of mesenchymal adipose stem cells. Stem Cells Dev. 2009;18:725–736. doi: 10.1089/scd.2008.0189. [DOI] [PubMed] [Google Scholar]
- 42.Fraser H.B., Lam L.L., Neumann S.M., Kobor M.S. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Mu W., Zhang W. On the analysis of the illumina 450k array data: probes ambiguously mapped to the human genome. Front. Genet. 2012;3:73. doi: 10.3389/fgene.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell J.T., Pai A.A., Pickrell J.K., Gaffney D.J., Pique-Regi R., Degner J.F., Gilad Y., Pritchard J.K. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desmond J.C., Raynaud S., Tung E., Hofmann W.K., Haferlach T., Koeffler H.P. Discovery of epigenetically silenced genes in acute myeloid leukemias. Leukemia. 2007;21:1026–1034. doi: 10.1038/sj.leu.2404611. [DOI] [PubMed] [Google Scholar]
- 46.Poage G.M., Houseman E.A., Christensen B.C., Butler R.A., Avissar-Whiting M., McClean M.D., Waterboer T., Pawlita M., Marsit C.J., Kelsey K.T. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clin. Cancer Res. 2011;17:3579–3589. doi: 10.1158/1078-0432.CCR-11-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herbst F., Ball C.R., Tuorto F., Nowrouzi A., Wang W., Zavidij O., Dieter S.M., Fessler S., van der Hoeven F., Kloz U., et al. Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol. Ther. 2012;20:1014–1021. doi: 10.1038/mt.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang J.T., Wang J.L., Du W., Hong J., Zhao S.L., Wang Y.C., Xiong H., Chen H.M., Fang J.Y. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis. 2011;32:1207–1215. doi: 10.1093/carcin/bgr114. [DOI] [PubMed] [Google Scholar]
- 49.Jjingo D., Conley A.B., Yi S.V., Lunyak V.V., Jordan I.K. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462–474. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz S., Meshorer E., Ast G. Chromatin organization marks exon-intron structure. Nat Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 51.Anastasiadou C., Malousi A., Maglaveras N., Kouidou S. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell. Biol. 2011;30:267–275. doi: 10.1089/dna.2010.1094. [DOI] [PubMed] [Google Scholar]
- 52.Hahn M.A., Wu X., Li A.X., Hahn T., Pfeifer G.P. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS ONE. 2011;6:e18844. doi: 10.1371/journal.pone.0018844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffmann T.J., Kvale M.N., Hesselson S.E., Zhan Y., Aquino C., Cao Y., Cawley S., Chung E., Connell S., Eshragh J., et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minakuchi M., Kakazu N., Gorrin-Rivas M.J., Abe T., Copeland T.D., Ueda K., Adachi Y. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur. J. Biochem. 2001;268:1340–1351. doi: 10.1046/j.1432-1327.2001.02000.x. [DOI] [PubMed] [Google Scholar]
- 55.Cristobal I., Blanco F.J., Garcia-Orti L., Marcotegui N., Vicente C., Rifon J., Novo F.J., Bandres E., Calasanz M.J., Bernabeu C., et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115:615–625. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 56.Panagopoulos I., Kerndrup G., Carlsen N., Strombeck B., Isaksson M., Johansson B. Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12) Br. J. Haematol. 2007;136:294–296. doi: 10.1111/j.1365-2141.2006.06410.x. [DOI] [PubMed] [Google Scholar]
- 57.Piazza R., Valletta S., Winkelmann N., Redaelli S., Spinelli R., Pirola A., Antolini L., Mologni L., Donadoni C., Papaemmanuil E., et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat. Genet. 2012;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valdez B.C., Li Y., Murray D., Champlin R.E., Andersson B.S. The synergistic cytotoxicity of clofarabine, fludarabine and busulfan in AML cells involves ATM pathway activation and chromatin remodeling. Biochem. Pharmacol. 2011;81:222–232. doi: 10.1016/j.bcp.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J.H., Takahashi T., Yasuhara N., Inazawa J., Kamada S., Tsujimoto Y. Bis, a Bcl-2-binding protein that synergizes with Bcl-2 in preventing cell death. Oncogene. 1999;18:6183–6190. doi: 10.1038/sj.onc.1203043. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y., Wang J.H., Lu Q., Wang Y.J. Bag3 promotes resistance to apoptosis through Bcl-2 family members in non-small cell lung cancer. Oncol. Rep. 2012;27:109–113. doi: 10.3892/or.2011.1486. [DOI] [PubMed] [Google Scholar]
- 61.Liao Q., Ozawa F., Friess H., Zimmermann A., Takayama S., Reed J.C., Kleeff J., Buchler M.W. The anti-apoptotic protein BAG-3 is overexpressed in pancreatic cancer and induced by heat stress in pancreatic cancer cell lines. FEBS Lett. 2001;503:151–157. doi: 10.1016/s0014-5793(01)02728-4. [DOI] [PubMed] [Google Scholar]
- 62.Festa M., Del Valle L., Khalili K., Franco R., Scognamiglio G., Graziano V., De Laurenzi V., Turco M.C., Rosati A. BAG3 protein is overexpressed in human glioblastoma and is a potential target for therapy. Am. J. Pathol. 2011;178:2504–2512. doi: 10.1016/j.ajpath.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu P., Xu B., Li J., Lu H. BAG3 gene silencing sensitizes leukemic cells to Bortezomib-induced apoptosis. FEBS Lett. 2009;583:401–406. doi: 10.1016/j.febslet.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 64.Gupta-Rossi N., Storck S., Griebel P.J., Reynaud C.A., Weill J.C., Dahan A. Specific over-expression of deltex and a new Kelch-like protein in human germinal center B cells. Mol. Immunol. 2003;39:791–799. doi: 10.1016/s0161-5890(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 65.Kroll J., Shi X., Caprioli A., Liu H.H., Waskow C., Lin K.M., Miyazaki T., Rodewald H.R., Sato T.N. The BTB-kelch protein KLHL6 is involved in B-lymphocyte antigen receptor signaling and germinal center formation. Mol. Cell Biol. 2005;25:8531–8540. doi: 10.1128/MCB.25.19.8531-8540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puente X.S., Pinyol M., Quesada V., Conde L., Ordonez G.R., Villamor N., Escaramis G., Jares P., Bea S., Gonzalez-Diaz M., et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A., et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goring H.H. Tissue specificity of genetic regulation of gene expression. Nat. Genet. 2012;44:1077–1078. doi: 10.1038/ng.2420. [DOI] [PubMed] [Google Scholar]
- 69.Hoischen A., van Bon B.W., Gilissen C., Arts P., van Lier B., Steehouwer M., de Vries P., de Reuver R., Wieskamp N., Mortier G., et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat. Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 70.Albano L.M., Sakae P.P., Mataloun M.M., Leone C.R., Bertola D.R., Kim C.A. Hydronephrosis in Schinzel-Giedion syndrome: an important clue for the diagnosis. Rev. Hosp. Clin. Fac. Med. Sao Paulo. 2004;59:89–92. doi: 10.1590/s0041-87812004000200008. [DOI] [PubMed] [Google Scholar]
- 71.Herman T.E., Sweetser D.A., McAlister W.H., Dowton S.B. Schinzel-Giedion syndrome and congenital megacalyces. Pediatr. Radiol. 1993;23:111–112. doi: 10.1007/BF02012399. [DOI] [PubMed] [Google Scholar]
- 72.Brown C., Havener T.M., Everitt L., McLeod H., Motsinger-Reif A.A. A comparison of association methods for cytotoxicity mapping in pharmacogenomics. Front. Genet. 2011;2:86. doi: 10.3389/fgene.2011.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abecasis G.R., Cardon L.R., Cookson W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang W., Duan S., Kistner E.O., Bleibel W.K., Huang R.S., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am. J. Hum. Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang R.S., Kistner E.O., Bleibel W.K., Shukla S.J., Dolan M.E. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol. Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Im H.K., Gamazon E.R., Stark A.L., Huang R.S., Cox N.J., Dolan M.E. Mixed effects modeling of proliferation rates in cell-based models: consequence for pharmacogenomics and cancer. PLoS Genet. 2012;8:e1002525. doi: 10.1371/journal.pgen.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gamazon E.R., Huang R.S., Cox N.J., Dolan M.E. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl Acad. Sci. USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 79.Moen E.L., Zhang X., Mu W., Delaney S.M., Wing C., McQuade J., Myers J., Godley L.A., Dolan M.E., Zhang W. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2012. in press. [DOI] [PMC free article] [PubMed]
- 80.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.