Abstract
The cone photoreceptor cyclic nucleotide-gated (CNG) channel is essential for central and color vision and visual acuity. Mutations in the channel subunits CNGA3 and CNGB3 are associated with achromatopsia and cone dystrophy. We investigated the gene expression profiles in mouse retina with CNG channel deficiency using whole genome expression microarrays. As cones comprise only 2 to 3% of the total photoreceptor population in the wild-type mouse retina, the mouse lines with CNG channel deficiency on a cone-dominant background, i.e. Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− mice, were used in our study. Comparative data analysis revealed a total of 105 genes altered in Cnga3−/−/Nrl−/− and 92 in Cngb3−/−/Nrl−/− retinas, relative to Nrl−/− retinas, with 27 genes changed in both genotypes. The differentially expressed genes primarily encode proteins associated with cell signaling, cellular function maintenance and gene expression. Ingenuity pathway analysis (IPA) identified 26 and 9 canonical pathways in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas, respectively, with 6 pathways being shared. The shared pathways include phototransduction, cAMP/PKA-mediated signaling, endothelin signaling, and EIF2/endoplasmic reticulum (ER) stress, whereas the IL-1, CREB, and purine metabolism signaling were found to specifically associate with Cnga3 deficiency. Thus, CNG channel deficiency differentially regulates genes that affect cell processes such as phototransduction, cellular survival and gene expression, and such regulations play a crucial role(s) in the retinal adaptation to impaired cone phototransduction. Though lack of Cnga3 and Cngb3 shares many common pathways, deficiency of Cnga3 causes more significant alterations in gene expression. This work provides insights into how cones respond to impaired phototransduction at the gene expression levels.
INTRODUCTION
The cone cyclic nucleotide-gated (CNG) channel plays a pivotal role in cone phototransduction, a process essential for central and color vision and visual acuity. In darkness or dim light, the channel is opened by cGMP, maintaining an inward current. Light induces a hydrolysis of cGMP, resulting in closure of the channel and hyperpolarization of the cell (1). Structurally, the cone CNG channel belongs to the superfamily of voltage-gated ion channels. It is comprised of two structurally related subunit types, CNGA3 and CNGB3, of which the human genes are located in 2q11.2 and 8q21-q22, respectively. In a heterologous expression system, CNGA3 forms a functional channel, while CNGB3 does not form channels in the absence of CNGA3. However, co-expression of CNGA3 and CNGB3 forms heteromeric channels displaying a number of properties of typical native CNG channels (1,2). Biochemical characterization has demonstrated the interaction between CNGA3 and CNGB3 in the mouse cones and suggested a stoichiometry with three CNGA3 subunits and one CNGB3 subunit (3,4), similar to the rod CNG channel (3,5–7).
Naturally occurring mutations in CNGA3 and CNGB3 are highly associated with human (and canine) cone diseases including achromatopsia, progressive cone dystrophy and early-onset macular degeneration (8–11). Achromatopsia is a devastating hereditary visual disorder characterized by reduced cone-mediated electroretinographic responses, color blindness, visual acuity loss, pendular nystagmus, extreme light sensitivity, and daytime blindness, and it affects ∼1 in every 33 000 Americans. As the disease is primarily caused by mutations in CNG channel subunits, achromatopsia is often referred to as a ‘channelopathy’. Indeed, ∼80 and 40 mutations have been identified in CNGA3 and CNGB3, respectively, and these mutations account for over 75% of achromatopsia patients (9,10,12).
Loss of cone function and cone dystrophy has been documented in patients with CNG channel mutations by electrophysiological examinations and by optical coherence tomography studies (13–15). The defective retinal phenotype has also been well characterized in Cnga3−/− and Cngb3−/− mouse models (16–18). Cone function in Cnga3−/− mice was completely abolished (16,19), while Cngb3−/−mice retained residual cone light responses (18–20). Cnga3−/− and Cngb3−/− mice showed early-onset, progressive cone degeneration characterized with apoptotic cell death which peaked between postnatal 15 and 30 days (17,18). Cnga3−/− and Cngb3−/− mice also displayed opsin mis-trafficking/mis-localization (17,21) and remodeling of inner retinal circuits (22). Recently, by using Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− mice (CNG channel deficiency on a cone-dominant background), we demonstrated an endoplasmic reticulum (ER) stress-associated cone degeneration (19). Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− mice have a retinal phenotype similar to that in their respective single knockout mice, i.e. loss of cone light response in Cnga3−/−/Nrl−/− mice and significantly reduced light response in Cngb3−/−/Nrl−/− mice, cone apoptosis and reduced levels of cone-specific proteins (19). This work investigated the cellular responses at the gene expression levels in CNG channel-deficient retinas by microarray analysis. We tested our hypothesis that loss of CNG channel/cone function leads to alterations in pathways that are involved in modulating light response and cellular stress response in the retina. We used Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− mice, which allowed us to profile gene expression in a cone-dominant retina (cones comprise only 2 to 3% of the total photoreceptor population in the wild-type mouse retina). The cone-dominant Nrl−/− mouse line is a commonly used model for studies of cone cell biology and disease. NRL is a basic-motif leucine zipper transcription factor essential for the normal development of rods. Mice lacking the Nrl gene have no rods, but have increased numbers of S-cones, functionally manifested as a loss of rod function coupled with super-normal cone function (23). Morphologically, Nrl−/− retinas have a cone-like nucleus, short and disorganized outer segment, and a rosette-like structure (23,24). Electrophysiological studies on the isolated photoreceptors from Nrl−/− retinas demonstrate the expression of functional S- and M-opsins (25), whereas analysis of retinal gene expression in Nrl−/− relative to wild-type mice by both whole genome expression microarray and the recently developed technology next generation sequencing confirms the transformation of rods into cones (26–29). Our analyses show that CNG channel deficiency differentially regulates expression of a wide range of genes that affect phototransduction cascade and cellular stress response, and such regulations play a crucial role(s) in retinal adaptation to impaired phototransduction. Though lack of each subunit affects gene expression differently and CNGA3 deficiency causes more significant gene alterations, defects of the two subunits indeed share many common alterations. This work sheds light on the understanding of the cellular adaptation in response to impaired cone phototransduction associated with CNG channel deficiency.
RESULTS
Differentially expressed genes in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas
We used whole genome expression microarrays to investigate gene expression profiles in CNG channel-deficient retinas. Total retinal RNAs were prepared from Cnga3−/−/Nrl−/−, Cngb3−/−/Nrl−/− and Nrl−/− mice at postnatal 30 days (P30) and used in the assays. We chose P30 days as a suitable time-point because the retinal defective phenotype in CNG channel-deficient mice, including impaired cone function, apoptotic death, ER stress and opsin mis-localization, has been well characterized at this age (16–20). The microarray data were compared between two classes of unpaired data to identify statistically significant differentially expressed genes at a 5% false discovery rate with a minimum 1.5 fold change. Comparison of Cnga3−/−/Nrl−/− retinal RNA with Nrl−/− controls identified a total of 105 genes significantly altered, with 44 downregulated and 61 upregulated in Cnga3−/−/Nrl−/− retinas. Compared with Nrl−/− controls, Cngb3−/−/Nrl−/− retinas had 92 genes significantly altered, with 46 downregulated and 46 upregulated. Tables 1 and 2 show the 30 most down and upregulated genes identified in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas, respectively. As it has been reported that clustered gene expression changes flank-targeted gene loci in knockout mice (30), we examined the chromosome regions to determine whether some gene expression changes could be due to close proximity to the Cnga3 or Cngb3 genes. Cnga3 is localized in murine chromosome 1B and Cngb3 is localized in chromosome 4A3. There are four differentially expressed genes (Arhgef4, Coq1, Hspd1 and Nop58) in Chromosome 1 in Cnga3−/−/Nrl−/− mice and three genes (Bach2, Dani1 and Tdrd7) in Chromosome 4 in Cngb3−/−/Nrl−/− mice. None of these genes are located within a 2 mb up and downstream region of the chromosome. The chromosomal proximity analysis also showed that the 22 up and 132 downstream genes for Cnga3 and the 38 up and 17 downstream genes for Cngb3 are not differentially expressed. Thus, it is less likely that the gene expression alterations identified are due to close chromosomal proximity to the Cnga3 or Cngb3 genes. Among the differentially expressed genes identified, there were 27 genes having directional matches in both genotypes relative to Nrl−/−. The sharing rate for the top 20 (fold-change) downregulated genes was 60 and 45% in Cnga3−/−/Nrl−/− retinas and Cngb3−/−/Nrl−/− retinas, respectively, and was ∼35% for the top 20 (fold-change) upregulated genes in both genotypes.
Table 1.
The 30 most down and upregulated genes in Cnga3−/−/Nrl−/− retinas
| Symbol | Unique ID | Name | Fold change |
|---|---|---|---|
| Arr3 | ILMN_2726271 | Arrestin 3, retinal | −4.58 |
| Pde6b | ILMN_2595543 | Guanylate cyclase Activator 1B | −4.54 |
| Guca1b | ILMN_2754287 | Guanylate cyclase activator 1B | −3.80 |
| Erdr1 | ILMN_1246153 | Erythroid differentiation regulator 1 | −2.99 |
| Casp7 | ILMN_2648548 | Caspase 7 | −2.87 |
| Rilad1 | ILMN_1222196 | Regulatory subunit of type II pka R-subunit domain containing 1 | −2.58 |
| Actb | ILMN_2588055 | Actin, beta | −2.48 |
| Abi3 | ILMN_1223041 | ABI family, member 3 | −2.40 |
| 2610034M16Rik | ILMN_2980212 | RIKEN cDNA 2610034M16 gene | −2.30 |
| Nr6a1 | ILMN_2739599 | Nuclear receptor subfamily 6, Group A, Member 1 | −1.97 |
| Tuft1 | ILMN_2419185 | tuftelin 1 | −1.94 |
| Acadm | ILMN_2810473 | Acyl-Coenzyme A dehydrogenase, medium chain | −1.85 |
| Fam114a2 | ILMN_1232976 | Family with sequence similarity 114, member A2 | −1.81 |
| C2orf71 | ILMN_1236750 | Chromosome 2 open reading frame 71 | −1.80 |
| Gckr | ILMN_2710811 | Glucokinase regulatory protein | −1.76 |
| C20orf7 | ILMN_2747677 | Chromosome 20 open reading frame 7 | −1.76 |
| Fscn2 | ILMN_2655965 | Fascin homolog 2, actin-bundling protein, retinal (S. purpuratus) | -1.75 |
| Mat2a | ILMN_2978838 | Methionine adenosyltransferase II, alpha | −1.75 |
| Ehd4 | ILMN_1224768 | EH-domain containing 4 | −1.74 |
| Arhgef4 | ILMN_2760548 | Rho guanine nucleotide exchange factor 4 | −1.72 |
| Abhd14b | ILMN_3007862 | Abhydrolase domain containing 14b | −1.69 |
| Spry3 | ILMN_2869082 | Sprouty homolog 3 (Drosophila) | −1.66 |
| Tnc | ILMN_2463181 | Tenascin C | −1.65 |
| Mettl17 | ILMN_2534921 | Methyltransferase like 17 | −1.65 |
| Krt18 | ILMN_2711267 | Keratin 18 | −1.64 |
| Plekhf2 | ILMN_2798694 | Pleckstrin homology domain containing, Family F (with FYVE domain) member 2 | −1.60 |
| Xpo7 | ILMN_1240092 | Exportin 7 | −1.59 |
| Mett11d1 | ILMN_2856668 | Methyltransferase 11 domain containing 1 | −1.58 |
| Lgals4 | ILMN_2968211 | Lectin, galactose binding, soluble 4 | −1.57 |
| Alox5ap | ILMN_2863837 | Arachidonate 5-lipoxygenase activating protein | −1.57 |
| Drd4 | ILMN_1217098 | Dopamine receptor 4 | 4.05 |
| Gdi1 | ILMN_2632299 | Guanosine diphosphate (GDP) dissociation inhibitor 1 | 2.39 |
| Gkap1 | ILMN_2682279 | G kinase anchoring protein 1 | 2.20 |
| Tspan3 | ILMN_1251499 | Tetraspanin 3 | 2.19 |
| Twistnb | ILMN_2466121 | TWIST neighbor | 1.91 |
| Dlg4 | ILMN_2710764 | Discs, large homolog 4 (Drosophila) | 1.90 |
| Eif4g2 | ILMN_1213167 | Eukaryotic translation initiation factor 4 gamma, 2 | 1.83 |
| Hif1a | ILMN_2852034 | Hypoxia inducible factor 1, alpha subunit | 1.83 |
| Ttyh1 | ILMN_1216021 | Tweety homolog 1 (Drosophila), transcript variant 2 | 1.82 |
| Rab6 | ILMN_1242802 | RAB6, member RAS oncogene family | 1.82 |
| LOC674706 | ILMN_2610798 | Similar to Zinc finger protein 341 | 1.82 |
| Eif5 | ILMN_3072536 | Eukaryotic translation initiation factor 5, transcript variant 1 | 1.80 |
| Pabpc1 | ILMN_1259482 | Poly A binding protein, cytoplasmic 1 | 1.79 |
| Btg1 | ILMN_2705119 | B-cell translocation gene 1, antiproliferative | 1.79 |
| Rcvrn | ILMN_2661924 | Recoverin | 1.77 |
| Ppp1cb | ILMN_2647628 | Protein phosphatase 1, catalytic subunit, beta isoform | 1.76 |
| Coq10b | ILMN_3031099 | Coenzyme Q10 homolog B (Saccharomyces cerevisiae), transcript variant 1 | 1.76 |
| Rnf11 | ILMN_2654403 | Ring finger protein 11 | 1.73 |
| Gnaz | ILMN_3161289 | Guanine nucleotide binding protein, alpha z subunit | 1.73 |
| C12orf43 | ILMN_1247760 | Chromosome 12 open reading frame 43 | 1.70 |
| Hnrnpk | ILMN_1245987 | Heterogeneous nuclear ribonucleoprotein K | 1.70 |
| Rab5a | ILMN_2786764 | RAB5A, member RAS oncogene family | 1.69 |
| Tob1 | ILMN_1250011 | Transducer of ErbB-2.1 | 1.69 |
| Ucbm2 | ILMN_2512043 | Similar to ubiquitin-conjugating enzyme UbcM2 | 1.69 |
| Gnao1 | ILMN_2773191 | Guanine nucleotide-binding protein, alpha O, transcript variant A | 1.68 |
| G3bp2 | ILMN_2691815 | GTPase activating protein (SH3 domain) binding protein 2, transcript variant 1 | 1.68 |
| Atp2b1 | ILMN_2878021 | ATPase, Ca++ transporting, plasma membrane 1 | 1.67 |
| Atp6v0d1 | ILMN_2608429 | ATPase, H+ transporting, lysosomal V0 subunit D1 | 1.66 |
| Gnas | ILMN_2632206 | Guanine nucleotide-binding protein, alpha stimulating complex locus, transcript variant 3 | 1.66 |
| Tmed2 | ILMN_1231873 | Transmembrane emp24 domain trafficking protein 2 | 1.65 |
Genes shared by Cngb3−/−/Nrl−/− are shown in bold.
Table 2.
The 30 most down and upregulated genes in Cngb3−/−/Nrl−/− retinas
| Symbol | Unique ID | Entrez Gene Name | Fold change |
|---|---|---|---|
| Arr3 | ILMN_2717844 | Arrestin 3, retinal | −3.46 |
| Guca1b | ILMN_2754287 | Guanylate cyclase activator 1B | −3.45 |
| Cngb3 | ILMN_2722268 | CNG channel beta 3 | −2.75 |
| Casp7 | ILMN_2648548 | Caspase 7 | −2.54 |
| Pde6b | ILMN_2595543 | Phosphodiesterase 6B, cGMP-specific, rod, beta | −2.49 |
| Rilad1 | ILMN_1222196 | Regulatory subunit of type II pka R-subunit domain containing 1 | −2.37 |
| Bach2 | ILMN_2819558 | BTB and CNC homology 2 | −2.24 |
| Nxnl2 | ILMN_2702547 | Nucleoredoxin-like 2 | −1.91 |
| Egr1 | ILMN_2662926 | Early growth response 1 | −1.91 |
| 2610034M16Rik | ILMN_2980212 | RIKEN cDNA 2610034M16 gene | −1.87 |
| Pygm | ILMN_1220498 | Muscle glycogen phosphorylase | −1.83 |
| Gulo | ILMN_2631948 | Gulonolactone (l-) oxidase | −1.77 |
| Abi3 | ILMN_1223041 | ABI family, member 3 | −1.76 |
| Ccl9 | ILMN_2776603 | Chemokine (C–C motif) ligand 9 | −1.71 |
| Scnm1 | ILMN_2711714 | Sodium channel modifier 1 | −1.70 |
| Pde6d | ILMN_3158668 | Phosphodiesterase 6d, cGMP-specific, rod, delta | −1.69 |
| Dusp11 | ILMN_2711705 | Dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) | −1.69 |
| Gckr | ILMN_2710811 | Glucokinase regulatory protein | −1.68 |
| Atg16l1 | ILMN_2620574 | Autophagy-related 16-like 1 (yeast), transcript variant b | −1.67 |
| Vtn | ILMN_1234111 | Vitronectin | −1.66 |
| Fam114a2 | ILMN_1232976 | Family with sequence similarity 114, Member A2 | −1.66 |
| Rd3 | ILMN_1242556 | Retinal degeneration 3 | −1.58 |
| Arid3b | ILMN_1238558 | AT rich interactive domain 3B (BRIGHT-like) | −1.58 |
| Trim3 | ILMN_3134607 | Tripartite motif-containing 3 | −1.58 |
| Rpl29 | ILMN_2857570 | Ribosomal protein L29 | −1.58 |
| Znrd1 | ILMN_2971486 | Zinc ribbon domain containing, 1 | −1.58 |
| C20orf7 | ILMN_2747677 | Chromosome 20 open reading frame 7 | −1.57 |
| Kif22 | ILMN_2762326 | Kinesin family member 22 | −1.56 |
| Rgs9bp | ILMN_2795473 | Regulator of G-protein signaling 9 binding protein | −1.56 |
| Tulp1 | ILMN_1236774 | Tubby-like protein 1 | −1.55 |
| Ednrb | ILMN_2589640 | Endothelin receptor type B | 4.78 |
| Drd4 | ILMN_1217098 | Dopamine receptor 4 | 3.80 |
| Dnaic1 | ILMN_2744879 | Dynein, axonemal, intermediate chain 1 | 2.96 |
| Chac1 | ILMN_2617468 | ChaC, cation transport regulator-like 1 (Escherichia coli) | 2.05 |
| Esrrb | ILMN_1228497 | Estrogen-related receptor, beta | 2.04 |
| Slc24a1 | ILMN_2618257 | Solute carrier family 24, member 1 | 2.04 |
| Gkap1 | ILMN_2682279 | G kinase anchoring protein 1 | 2.00 |
| Lgr5 | ILMN_2650008 | Leucine-rich repeat containing G protein-coupled receptor 5 | 1.92 |
| Lrfn2 | ILMN_2651081 | Leucine-rich repeat and fibronectin type III domain containing 2 | 1.92 |
| Pon1 | ILMN_2676379 | Paraoxonase 1 | 1.89 |
| Ttyh1 | ILMN_1216021 | Tweety homolog 1 (Drosophila), transcript variant 2 | 1.87 |
| Cox6a2 | ILMN_2629581 | Cytochrome c oxidase, subunit VI a, polypeptide 2, nuclear gene encoding mitochondrial protein | 1.81 |
| Aqp4 | ILMN_2757232 | Aquaporin 4 | 1.79 |
| Eif5 | ILMN_3072536 | Eukaryotic translation initiation factor 5, transcript variant 1 | 1.77 |
| Ttr | ILMN_2443330 | Transthyretin | 1.76 |
| Ralyl | ILMN_1236932 | RALY RNA-binding protein-like | 1.72 |
| Tdrd7 | ILMN_2818678 | Tudor domain containing 7 | 1.72 |
| Atp6v1a | ILMN_1255570 | ATPase, H+ transporting, lysosomal V1 subunit A | 1.70 |
| Twistnb | ILMN_2466121 | TWIST neighbor | 1.70 |
| Trib3 | ILMN_1225528 | tribbles homolog 3 (Drosophila) | 1.69 |
| Atp2b1 | ILMN_2878021 | ATPase, Ca2+ transporting, plasma membrane 1 | 1.68 |
| Cadps2 | ILMN_2998313 | Ca2+-dependent activator protein for secretion 2 | 1.66 |
| Mat2a | ILMN_2593368 | Methionine adenosyltransferase II, alpha | 1.66 |
| Mdga2 | ILMN_1241332 | MAM domain containing glycosylphosphatidylinositol anchor 2 | 1.65 |
| Rbms2 | ILMN_3111877 | RNA-binding motif, single stranded interacting protein 2, transcript variant 2 | 1.63 |
| Mpp6 | ILMN_3007680 | Membrane protein, palmitoylated 6 | 1.62 |
| Npm3 | ILMN_3020829 | Nucleoplasmin 3 | 1.62 |
| Slc7a3 | ILMN_1257987 | Solute carrier family 7, Member 3 | 1.62 |
| Eif4g2 | ILMN_1213167 | Eukaryotic translation initiation factor 4 gamma, 2 | 1.60 |
| Mmpde8 | ILMN_1255207 | Similar to cAMP-specific cyclic nucleotide phosphodiesterase PDE8 | 1.57 |
Genes shared by Cnga3−/−/Nrl−/− are shown in bold.
Validations of differentially expressed genes by quantitative real-time PCR
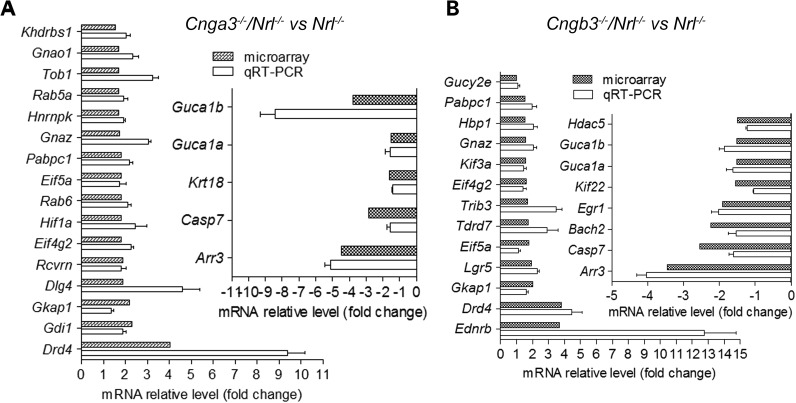
Quantitative real-time PCR (qRT–PCR) was performed to validate the microarray data. A total of 24 differentially expressed genes for Cnga3−/−/Nrl−/− and 21 for Cngb3−/−/Nrl−/− mice, based on the fold-change values and their biological function, were examined. The qRT–PCR data indicated a good agreement between the two methods of analysis. The direction of expression was validated in 21 of the 24 genes tested in Cnga3−/−/Nrl−/− retinas (88% validation rate) and 20 of the 21 genes in Cngb3−/−/Nrl−/− retinas (95% validation rate). Figure 1 shows the fold-change values calculated from the microarray and qRT–PCR results in Cnga3−/−/Nrl−/− (Fig. 1A) and Cngb3−/−/Nrl−/− (Fig. 1B) retinas.
Figure 1.
qRT–PCR validations of differentially expressed genes identified in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas. Shown are predicted fold changes from microarray analysis and relative fold changes from qRT-PCR in Cnga3−/−/Nrl−/− (A) and Cngb3−/−/Nrl−/− (B) retinas. Data are represented as means ± SEM of three independent analyses using cDNAs prepared from 3–5 mice.
Functional categories of differentially expressed genes in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retina
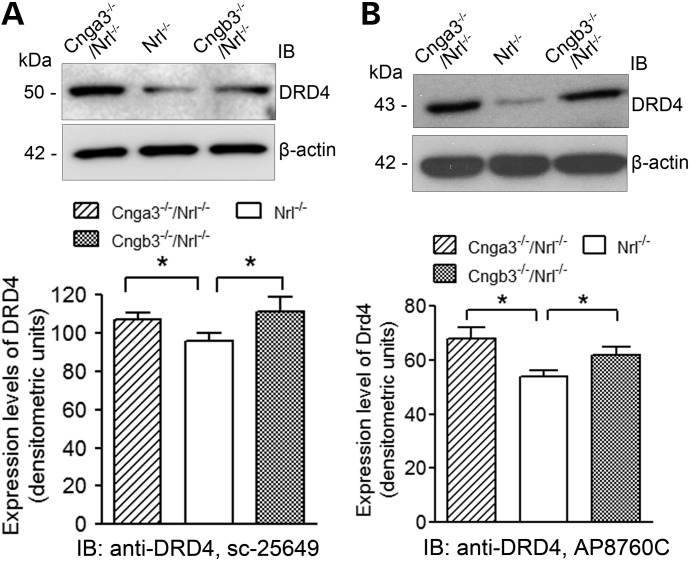
The differentially expressed genes were shown to primarily encode proteins associated with cell signaling, cellular function maintenance, transportation and gene expression, including enzymes, ion channels/receptors/transporters, transcription/translation regulators and other regulatory molecules (Table 3). Among the 18 altered genes encoding enzymes identified in Cnga3−/−/Nrl−/− and 14 in Cngb3−/−/Nrl−/− retina, 5 (Acadm, Mat2a, Pde6b, Nop58 and Gnaz) were shared by both genotypes. Genes for the G protein-coupled receptors, Drd4 (encoding the dopamine receptor D4, DRD4) and Ednrb (encoding endothelin receptor type B, EDNRB), were the most upregulated relative to Nrl−/− mice. We further examined retinal expression levels of DRD4 by western blotting using two different antibodies against DRD4 (see Materials and Methods). Both antibodies gave a similar result and detected increased expression levels of DRD4 in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas, compared with the Nrl−/− controls (Fig. 2). Of note, the two antibodies recognize proteins at slightly different sizes; the Santa Cruz antibody labels the protein band migrated at a position close to 50 kDa, whereas the Abgent antibody labels the band migrated at a position around 43 kDa. The reason for detection of two bands with two different anti-DRD4 antibodies is not known, but it might be associated with DRD4 polymorphic variants. The DRD4 polymorphic variants have been well characterized in human (31,32). The human DRD4 gene contains extensive polymorphisms consisting of variable numbers of tandem repeats in sequences corresponding to the third cytoplasmic loop of the protein, and about 27 different haplotypes encoding 20 different protein variants were documented (31,32). The DRD4 polymorphic variants have also been reported in nonhuman primates (33,34) and several other species including canine (35–38), horse (39) and chicken (40). To our knowledge, no research regarding DRD4 variants in mouse has been documented. A recent study investigating the influence of DRD4 gene on longevity in human and mice (41) implies a possibility of DRD4 variants in the mouse receptor. Here, we detected DRD4 at different sizes using two different anti-DRD4 antibodies, an observation favoring the presence of DRD4 variants in the mouse receptor. Nevertheless, Drd4 variants in mice merit further investigation. There were four genes encoding ion channels shown to be altered; one (Ttyh1, encoding a probable chloride channel) was shared by both genotypes. Among the three genes encoding kinases, Mpp6 was altered in both genotypes. There were 6 and 10 genes involved in transcriptional regulation altered in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas, respectively; none were shared. In contrast, among the four genes involved in translational regulation identified in Cnga3 deficiency, three (Eif4g2, Eif5 and Pabpc1) were shared. There were 11 genes involved in transportation of nutrients and related small molecules identified in Cnga3−/−/Nrl−/− retina (∼10%, 11 of 105) and 10 in Cngb3−/−/Nrl−/− retinas (∼10%, 10 of 92); only 2, Aps18 and Atp2b1, were shared. Among the remaining genes identified, ∼50% were grouped into the enzyme regulators and other function category with ∼20% shared by both genotypes.
Table 3.
Categories of differentially expressed genes identified in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas
| Gene type | Cnga3−/−/Nrl−/− | Cngb3−/−/Nrl−/− | Common genes |
|---|---|---|---|
| Cytokine | 0 | 1 | 0 |
| Enzyme | 18 | 14 | 5 |
| G-protein-coupled receptor | 2 | 3 | 1 |
| Ion channel | 2 | 3 | 1 |
| Kinase | 2 | 2 | 1 |
| Ligand-dependent nuclear receptor | 1 | 1 | 0 |
| Peptidase | 2 | 1 | 1 |
| Phosphatase | 4 | 2 | 1 |
| Transcription regulator | 6 | 10 | 0 |
| Translation regulator | 4 | 3 | 3 |
| Transporter | 11 | 10 | 2 |
| Enzyme regulators and other functions | 53 | 42 | 12 |
| Total | 105 | 92 | 27 |
Figure 2.
Enhanced expression of DRD4 in cone CNG channel-deficient retinas. Retinal expression levels of DRD4 in Cnga3−/−/Nrl−/−, Cngb3−/−/Nrl−/− and Nrl−/−mice at P30 were analyzed by immunoblotting using antibodies against DRD4 from Santa Cruz Biotechnology, Inc. (sc-25649, A) and from Abgent Inc. (AP8760C, B). Shown are representative images of the western blot detections (upper panels) and densitometric analysis of the relative expression levels of DRD4 in Cnga3−/−/Nrl−/−, Cngb3−/−/Nrl−/− and Nrl−/− retinas (lower panels). Data are represented as means ± SEM of measurements from four independent experiments using retinas from 4–5 mice. Unpaired Student's t-test was used for determination of the significance (*, P < 0.05).
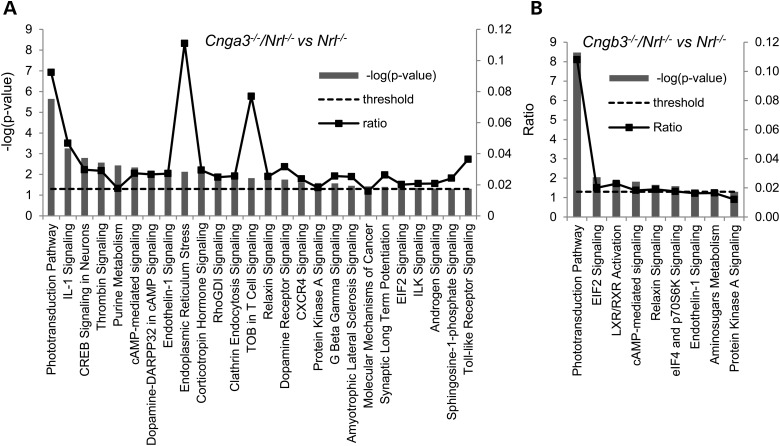
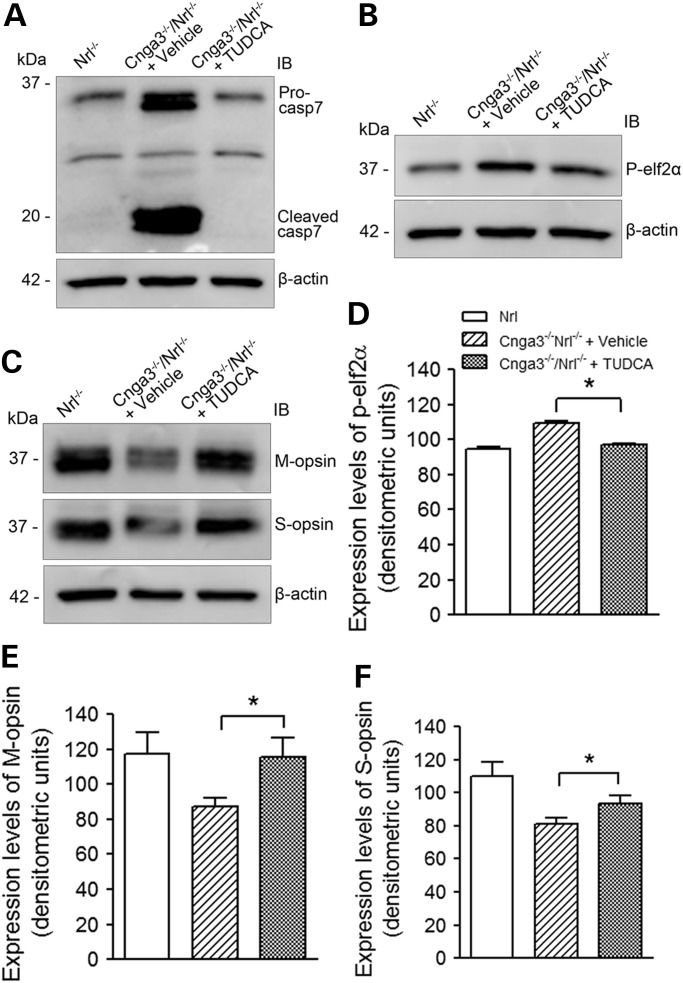
Differentially expressed genes were input to Ingenuity Pathway Analysis (IPA) software to better understand the nature of the effects of these CNG channel deficiencies. We identified 26 canonical pathways in Cnga3−/−/Nrl−/− (Fig. 3A) and 9 in Cngb3−/−/Nrl−/− (Fig. 3B) retinas at the threshold of P-value <0.05. Table 4 shows a list of genes identified in the canonical pathways. Among these, six pathways, including phototransduction, cAMP-mediated signaling, PKA-mediated signaling, endothelin signaling, EIF2/EIF4 ER stress signaling and relaxin signaling, were shared by both genotypes, while pathways, including IL-1 signaling, CREB signaling in neurons, thrombin signaling and purine metabolism, were shown to be altered specifically in Cnga3−/−/Nrl−/− retinas. The phototransduction pathway had the lowest P-values (Fig. 3). The ER stress pathway was identified as a regulated canonical pathway with the highest ratio (11%) (Fig. 3A). Indeed, we recently documented the unfolding protein response and ER stress in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas by showing the enhanced levels of phospho-eIF2α, accumulation of CHOP, and processing of caspase-12 and caspase-7 (19). The present work provided evidence showing the ER stress at the gene expression levels. We further evaluated whether ER stress contributes to cone degeneration in CNG channel deficiency by treating Cnga3−/−/Nrl−/− mice with the chemical chaperone tauroursodeoxycholate (TUDCA). This drug has been shown to be effective in alleviating ER stress and preventing cell death in a variety of animal disease models including retinal degeneration (42–44). Analysis of the mice following TUDCA administrations showed that the treatment significantly reduced levels of phospho-eIF2α and processing of caspase-7, and increased levels of cone opsin in Cnga3−/−/Nrl−/− mice, compared with the vehicle-treated controls (Fig. 4). The levels of phospho-eIF2α and cleaved caspase-7 in TUDCA-treated Cnga3−/−/Nrl−/− mice were reduced to the levels seen in Nrl−/− mice (Fig. 4A, B and D), and the levels of M-opsin and S-opsin in TUDCA-treated Cnga3−/−/Nrl−/− mice were increased by ∼30 and ∼15.6%, respectively, compared with the vehicle-treated controls (Fig. 4C, E and F). These results demonstrated a role of ER stress in cone degeneration.
Figure 3.
Canonical pathways identified in Cnga3 and Cngb3 deficiency. IPA identified 26 canonical pathways in Cnga3−/−/Nrl−/− (A) and 9 in Cngb3−/−/Nrl−/− retina (B). The Fisher's exact test was set at a P-value threshold of 0.05 and the ratio of a given pathway was obtained by dividing the number of genes by the total number of genes that make up the particular canonical pathway in Ingenuity Knowledge Database.
Table 4.
Canonical pathways detected in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas
| Ingenuity canonical pathways | Genes |
|---|---|
| Cnga3−/−/Nrl−/− retina | |
| Phototransduction pathway | Arr3,Guca1a,Guca1b,Pde6b,Opn1sw,Rcvrn |
| IL-1 signaling | Tollip,Myd88,Gnao1,Adcy1,Gnaz |
| CREB signaling in neurons | Polr2e,Gnao1,Adcy1,Atf4,Gnaz,Opn1sw |
| Thrombin signaling | Arhgef4,Gnao1,Adcy1,Ppp1cb,Gnaz,Opn1sw |
| Purine metabolism | Mpp6,Polr2e,Impdh1,Atp5a1,Adcy1,Hspd1,Pde6b |
| cAMP-mediated signaling | Dusp6,Gnao1,Adcy1,Atf4,Drd4,Pde6b |
| Dopamine-DARPP32 in cAMP signaling | Adcy1,Atf4,Ppp1cb,Drd4,Opn1sw |
| Endothelin-1 signaling | Gnao1,Adcy1,Gnaz,Casp7,Opn1sw |
| ER stress | Atf4,Casp7 |
| Corticotropin hormone signaling | Gnao1,Adcy1,Atf4,Opn1sw |
| RhoGDI signaling | Gdi1,Arhgef4,Actb,Gnao1,Gnaz |
| Clathrin endocytosis signaling | Rab5a,Actb,Rab7a,Snap91,Fgf5 |
| Antiproliferative role of TOB in T cells | Pabpc1,Tob1 |
| Relaxin signaling | Gnao1,Adcy1,Gnaz,Pde6b |
| Dopamine receptor signaling | Adcy1,Ppp1cb,Drd4 |
| CXCR4 signaling | Gnao1,Adcy1,Gnaz,Opn1sw |
| Protein kinase A signaling | Adcy1,Anapc5,Atf4,Ppp1cb,Pde6b,Opn1sw |
| G beta gamma signaling | Gnao1,Adcy1,Gnaz |
| Amyotrophic sclerosis signaling | Rab5a,Slc1a2,Casp7 |
| Molecular mechanisms of cancer | Arhgef4,Gnao1,Adcy1,Hif1a,Gnaz,Casp7 |
| Synaptic long-term potentiation | Adcy1,Atf4,Ppp1cb |
| EIF2 signaling | Pabpc1,Eif4g2,Eif5,Ppp1cb |
| ILK signaling | Krt18,Actb,Atf4,Hif1a |
| Androgen signaling | Polr2e,Gnao1,Gnaz |
| Sphingosine- 1-phosphate signaling | Adcy1,Casp7,Opn1sw |
| Toll-like receptor signaling | Tollip, Myd88 |
| Cngb3−/−/Nrl−/− retina | |
| Phototransduction pathway | Arr3,Guca1a,Guca1b,Rgs9 bp,Cngb3,Pde6b,Pde6d |
| EIF2 signaling | Pabpc1,Eif4g2,Rps5,Eif5 |
| LXR/RXR activation | Pon1,Ttr,Vtn |
| cAMP-mediated signaling | Cngb3,Drd4,Pde6b,Pde6d |
| Relaxin signaling | Gnaz,Pde6b,Pde6d |
| eIF4 and p70S6 K signaling | Pabpc1,Eif4g2,Rps5 |
| Endothelin-1 signaling | Ednrb,Gnaz,Casp7 |
| Aminosugars metabolism | Pde6b,Pde6d |
| Protein kinase A signaling | Pygm, Cngb3, Pde6b, Pde6d |
Figure 4.
Reduced ER stress and increased levels of cone opsin in retinas of Cnga3−/−/Nrl−/− mice following treatment with TUDCA. Cnga3−/−/Nrl−/− mice were treated with TUDCA (500 mg/kg body weight) or vehicle (0.15 M NaHCO3, pH 7.0) by subcutaneous injection every three days for three weeks, starting at P8. At the end of the experiments, retinas were isolated and analyzed by western blotting. (A–C). Shown are representative images of the western blot detection of caspase-7 cleavage (A), phospho-eIF2α (B), and cone opsin (C). (D–F). Shown are results of densitometric analysis of the relative expression levels of phospho-eIF2α (D), M-opsin (E), and S-opsin (F). Data are represented as means ± SEM of measurements from four independent experiments using retinas from 4–5 mice. Unpaired Student's t test was used for determination of the significance (*, P < 0.05).
IPA also showed many major biological function and disease categories shared in Cnga3 and Cngb3 deficiency. The top five categories identified by this software from the differentially expressed genes in Cnga3−/−/Nrl−/− retinas were genetic disorder, neurological disease, skeletal and muscular disorders, nucleic acid metabolism and small-molecule biochemistry, and the top five in Cngb3−/−/Nrl−/− retinas contain genetic disorder, neurological disease, skeletal and muscular disorders, cellular compromise and ophthalmic disease (see Supplementary Material, Fig. S1).
Network analysis
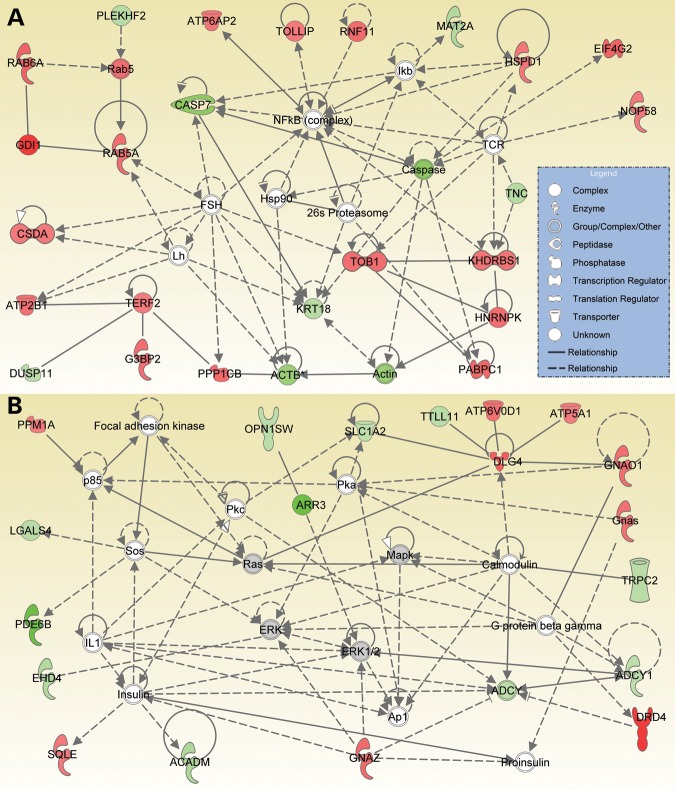
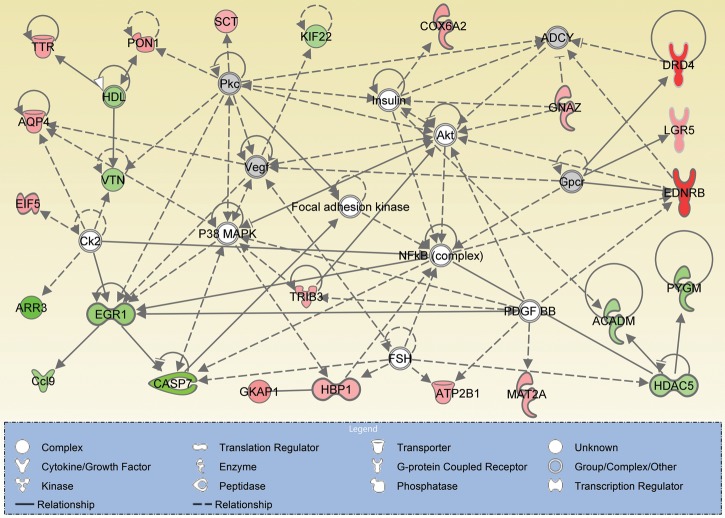
We also performed network analysis to further understand the cellular responses to CNG channel deficiency. A number of networks that illustrate the interactions of products of differentially expressed genes relevant to Cnga3 and Cngb3 deficiency were generated using IPA. These networks revealed that the overall cellular processes that were altered in response to CNG channel deficiency mainly involve cell survival/death, functional maintenance and gene expression. Figure 5A shows the networks with the highest score in Cnga3−/−/Nrl−/− retinas. Known interactions of gene products are illustrated in the networks and microarray changes of 10 of these genes (Tob1, Khdrbs1, Casp7, Eif4g2, Gdi1, Krt18, Pabpc1, Hnrnpk, Rab6a and Rab5a) were validated by qRT–PCR (also see Fig. 1A). The following gene products were detected as hubs: NFκB complex (the nuclear factor kappa B complex, involved in cellular responses to stimuli such as stress, including those caused by cytokines, free radicals, oxidized lipids and infectious organisms) and FSH (the follicle stimulating hormone, involved in proliferation, development, growth and maturation). Figure 5B shows the networks with the second-highest score in Cnga3−/−/Nrl−/− retinas. The microarray changes of five genes (Drd4, Arr3, Gnaz, Gnao1 and Dlg4) were validated by qRT–PCR (see Fig. 1A). The following gene products served as hubs: MAPK/ERKS, insulin, PKA, PKC, RAS and AP1. Similar to the identifications in CNGA3 deficiency, the top networks identified in CNGB3 deficiency illustrated the main functionalities including cell survival/death, gene expression and cellular function/maintenance. Figure 6 shows the networks with the highest score in Cngb3−/−/Nrl−/− retinas. Known interactions of gene products are illustrated in the networks and microarray changes of 10 genes (Drd4, Ednrb, Gkap1, Lgr5, Eif5a, Trib3, Gnaz, Hbp1, Casp7 and Arr3) were validated by qRT–PCR (see Fig. 1B). The following gene products served as hubs: p38MAPK, AKT, NFκB complex, PKC, PDGFB (the platelet-derived growth factor subunit B), EGR1 (the early growth response 1), VEGF (the vascular endothelial growth factor) and Insulin.
Figure 5.
Known interactions among differentially expressed genes in Cnga3 deficiency. IPA identified a number of networks altered in Cnga3−/−/Nrl−/− retinas. Shown are the two most significant networks. The gene symbols that occur in our data set are shown in red (upregulated) or green (downregulated). The gene entries from Ingenuity Knowledge Database inserted to connect all relevant molecules are shown in gray. The solid lines between molecules indicate direct physical relationships between molecules (such as regulating and interacting protein domains) while the dotted lines indicate indirect functional relationships (such as co-regulation of expression of both genes in cell lines). (A) The networks with the highest score show main functionalities associated with cell death, cell development and function. (B) The networks with second-highest score show main functionalities associated with cell growth and death, nucleic acid metabolism and small molecule biochemistry.
Figure 6.
Known interaction among differentially expressed genes in Cngb3 deficiency. IPA identified a number of networks altered in Cngb3−/−/Nrl−/− retinas and shown is the most significant networks. The gene symbols that occur in our data set are shown in red (upregulated) or green (downregulated). The gene entries from Ingenuity Knowledge Database inserted to connect all relevant molecules are shown in gray. The solid lines between molecules indicate direct physical relationships between molecules (such as regulating and interacting protein domains) while the dotted lines indicate indirect functional relationships (such as co-regulation of expression of both genes in cell lines). The main functionalities given by Ingenuity for the networks are cell survival/death, gene expression, and cellular function maintenance.
DISCUSSION
This work investigated the gene expression profiles in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas in order to understand the cellular responses at the gene expression levels in CNG channel deficiency. The microarray analysis showed that Cnga3 and Cngb3 deficiency shared many differentially regulated genes and all shared genes had the same direction. Using a minimum 1.5-fold change between groups, a total of 105 altered genes were identified in Cnga3−/−/Nrl−/− and 92 in Cngb3−/−/Nrl−/− retinas, relative to Nrl−/− retinas, with 27 genes changed in both genotypes. These reflect only a portion of the overall differences detected. When a minimum 1.2-fold-change filter was used, 265 differentially regulated genes were identified in Cnga3−/−/Nrl−/− and 328 in Cngb3−/−/Nrl−/− retinas, with 114 genes being shared (see Supplementary Material, Tables S1 and S2). An example of such high sharing rates was seen in genes involved in translational regulation. Among the four genes identified (using a minimum 1.5-fold change), three (Eif4g2, Eif5 and Pabpc1) were shared by both genotypes. In contrast, comparing the genes differentially expressed in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas with the data from a mouse line with deficiency of an unrelated photoreceptor-specific gene, encoding the rhodopsin kinase (GRK1) (Grk1−/−/Nrl−/− mice) (45), we identified only 4 and 11 common genes, respectively. The similar regulation pattern of gene expression in Cnga3 and Cngb3 deficiency was also demonstrated by a number of shared canonical and functional pathways identified in the two genotypes. Hence, the neuroretinal cells responded quite similarly to Cnga3 and Cngb3 deficiency at the gene expression levels, which is in line with the function of the two proteins in cone photoreceptors.
Among the genes differentially downregulated in both genotypes were several photoreceptor-specific genes, including Arr3, Guca1a, Guca1b and Pde6b. This is consistent with the overall suppression/loss of photoreceptor function and photoreceptor degeneration. The downregulation of photoreceptor-specific genes was also shown previously in other mouse models of retinal degeneration, including cpfl1 mice (with a mutation in Pde6c), Rpe65−/− and Bbs4−/− mice (models of Leber congenital amaurosis) (46–48). Arr3 was the most downregulated gene in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas (see Tables 1 and 2, Fig. 1), confirming our previous finding of a reduced cone arrestin (encoded by Arr3) level in the channel-deficient retinas (19,20). The microarray and qRT–PCR data demonstrated that the reduced cone arrestin is caused (at least in part) by downregulation at the transcriptional level. It remains to be determined how Arr3 is downregulated in CNG channel deficiency. Is it related to deficiency of the two channel subunits specifically or a general response in an abnormal cone? As the downregulation of Arr3 was also identified in other mouse models of cone degeneration [including Rpe65−/− (47), cpfl1 (46), and Bbs4−/− mice (49)], it likely represents a general response. The observation that the downregulation of Arr3 in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− mice was already detected at young age (postnatal 15 days) (see Supplementary Material, Fig. S2) suggests a response to impaired phototransduction rather than a consequence of photoreceptor degeneration. Nevertheless, we know little about Arr3 expression regulation, though an upregulation of Arr3 by retinoic acid was described previously in a cell culture system (50). It is worth noting that only one photoreceptor-specific gene, Rcvrn (encoding recoverin), was found to be upregulated in Cnga3 deficiency (see Table 1). Recoverin binds to GRK1 and plays a key role in the inhibition of this enzyme and regaining function of rhodopsin. It is unclear whether the upregulation of Rcvrn detected in Cnga3-deficient retinas is associated with the downregulation of Arr3 in response to impaired cone phototransduction.
Using IPA software, we identified cAMP/PKA signaling as one of the significantly regulated canonical pathways shared in both Cnga3 and Cngb3 deficiency. The genes involved include Drd4, Gnao1, Adcy1, Atf4, and Ppp1cb (see Fig. 1 and Table 4). Drd4 was the most upregulated gene in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas. Upregulation of Drd4 was also shown at the protein level as detected by western blot analyses. To assure the detection of DRD4 (an integral membrane protein), we used two different antibodies and both antibodies gave a similar result (see Fig. 2). We also performed a heterologous expression experiment to determine the antibody specificity. In this experiment, plasmids harboring hDRD4 complementary DNA (cDNA) (kindly provided by Dr Marc Caron at Duke University) and control vectors were transfected into HEK293 cells and expression of DRD4 following transfection was examined by western blotting. The assays showed that the expression of hDRD4 was detected only in cells transfected with hDRD4 cDNA but not control vectors (see Supplementary Material, Fig. S3). In the retina, dopamine is a retinal neuromodulator secreted from amacrine and interplexiform cells (51,52), and DRD4 receptors are expressed in the photoreceptor layer where they affect cAMP metabolism and gene expression (53,54). Activation of DRD4/cAMP/PKA pathway is known to modulate numerous ocular functions, including light and dark adaptation, circadian rhythms, retinal development and ocular growth (55–57). It is worth noting that DRD4-mediated circadian oscillations in photoreceptors modulate CNG channel function, and circadian control of CNG channel function has been well studied in chick cones (58–61). The apparent affinity of the CNG channel for cGMP was significantly greater during the subjective night than during the subjective day, such that considerable changes in channel gating would be expected to occur at physiological concentrations of cGMP (60,61). Moreover, tyrosine phosphorylation of cone CNG channel was shown to be under the circadian control (62). Based on these findings, one may presume that the loss of functional CNG channel/cone phototransduction renders a feedback effect on the DRD4-mediated signaling. In addition, the cAMP/PKA pathway has also been shown to be involved in photoreceptor degeneration caused by rhodopsin mutants or mis-localized phototransduction (63), and to mediate the opsin mis-localization-induced rod reactive neuritic sprouting (64). Whether the cAMP/PKA pathway plays a role in opsin mis-localization-induced cone death would be interesting to explore.
The ER stress was identified as a regulated canonical pathway with the highest ratio (11%) in CNG channel deficiency (see Fig. 3). We recently showed the enhanced levels of ER stress marker proteins in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas (19). Hence, the ER stress in CNG channel-deficient mice occurs at both protein and gene expression levels. In this work, we further showed that alleviating ER stress by using chemical chaperone effectively decreased levels of ER stress markers and preserved cones (as shown by increased levels of cone opsin) in Cnga3−/−/Nrl−/− mice, demonstrating a role of ER stress in cone degeneration. The cellular mechanism(s) underlying the unfolded protein response and ER stress in CNG channel deficiency remain(s) to be elucidated. It likely involves an impaired calcium homeostasis and opsin mis-trafficking/mis-localization (17,21).
Endothelin signaling was shown as another canonical pathway altered in CNG channel deficiency. The genes involved were Ednrb, Gnaz, Gnao1, Adcy1 and Casp7 (see Table 4), and Ednrb (encoding endothelin receptor type B, EDNRB) was the most upregulated (with highest fold-change) gene in Cngb3−/−/Nrl−/− retinas (see Table 2 and Fig. 1B). Endothelin signaling from photoreceptors to activate EDNRB on Müller cells (the most abundant glial cells in the retina) is the major component of glial activation in response to photoreceptor stress or injury (65–67). Regardless of proximal causes, photoreceptor injury or disease almost invariably leads to the activation of Müller cells. Similar to other types of retinal degeneration animal models, Cnga3−/− mice display activation of Müller cells (manifested as enhanced glial fibrillary acidic protein labeling in the retinas (17)). Identification of a regulated endothelin pathway is consistent with the glial activation in the channel-deficient retina. Though glial activation is thought to limit or repair neuronal damage, excessive activation may inhibit regeneration and functional recovery (65,68). The functional significance of the endothelin signaling and glial activation in CNG channel deficiency merits further explorations. Of note, the androgen signaling pathway is among the other pathways identified in this work. To our knowledge, the role of androgen signaling pathway in photoreceptor degeneration has not been documented. However, a cross-talk between the androgen signaling and Wnt signaling pathway has been well recognized (69,70), and the latter is known to play a role in photoreceptor degeneration (71). The examination of the androgen signaling pathway in degenerating retinas should be undertaken in the future.
Although lack of Cnga3 and Cngb3 shares many common differentially altered genes and pathways, deficiency of CNGA3 caused numerous alterations beyond those detected in CNGB3 deficiency. This is evidenced by the larger number of differentially expressed genes and many more regulatory canonical pathways identified in Cnga3−/−/Nrl−/− retina (26 pathways) compared with that in Cngb3−/−/Nrl−/− retina (9 pathways). The more striking response observed in Cnga3−/−/Nrl−/− retina corresponds to the more severe phenotype seen in these mice. Cnga3 deficiency leads to a complete loss of cone function, whereas mice lacking Cngb3 still retain residual cone light response (16,18,19). Consequently, the cellular responses to deficiency of the two subunits are similar but distinct. Cnga3-deficient mice showed a faster cone degeneration, higher levels of ER stress and much more striking accumulation of cGMP, compared with that in Cngb3-deficient mice (17,19,20). Indeed, studies have shown a trend of a more severe clinical phenotype in achromatopsia patients with CNGA3 mutations, compared with CNGB3 defect (9,15,72), though a clear genotype–phenotype correlation remains to be determined (this might also depend on the type of mutation, i.e. loss of function vs. gain of function). Nevertheless, there were ∼70% of unshared differentially expressed genes identified in the two genotypes, indicating that deficiency of the two channel subunits affects retinal gene expression quite differently. There are two examples. (i) There were 6 and 10 genes involved in transcriptional regulation altered in Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− retinas, respectively; none were shared, suggesting a high-differentially regulated transcriptional process in response to Cnga3 deficiency versus Cngb3 deficiency. (ii) There were 11 genes involved in transportation of nutrients and related small molecules identified in Cnga3−/−/Nrl−/− retina and 10 in Cngb3−/−/Nrl−/− retinas; only two were shared, which may imply a highly regulated cellular transportation process in CNG channel deficiency with a substantial difference between deficiency of Cnga3 and Cngb3.
In summary, deficiency of Cnga3 and Cngb3 differentially regulates expression of a wide range of retinal genes with 25–30% of altered genes shared in both genotypes. Those that directly or indirectly affect cell processes such as phototransduction, cellular survival, cell function maintenance and gene expression play crucial roles in the retinal adaptation to impaired cone phototransduction. Though lack of Cnga3 and Cngb3 share many altered genes and signaling pathways, deficiency of Cnga3 indeed causes more significant gene alterations. This work sheds light on how cones respond to impaired phototransduction at the gene expression levels.
MATERIALS AND METHODS
Mice
The Cnga3−/− and Cngb3−/− mouse lines on a C57BL/6 background were generated as described previously (16,18). The Nrl−/− mouse line on a C57BL/6 background was provided by Dr Anand Swaroop (National Eye Institute, Bethesda, MD, USA). The Cnga3−/−/Nrl−/− and Cngb3−/−/Nrl−/− mouse lines were generated by cross-mating (19). All mice were maintained under cyclic light (12 h light–dark) conditions; cage illumination was ∼seven foot candles during the light cycle. All animal maintenance and experiments were approved by the local Institutional Animal Care and Use Committee (Oklahoma City, OK, USA) and conformed to the guidelines on the care and use of animals adopted by the Society for Neuroscience and the Association for Research in Vision and Ophthalmology (Rockville, MD, USA).
Antibodies and other reagents
Affinity purified rabbit polyclonal antibodies against mouse M-opsin and cone arrestin were provided by Dr Cheryl Craft (University of Southern California, Keck School of Medicine). Rabbit polyclonal antibody against mouse S-opsin was provided by Dr Muna Naash (University of Oklahoma Health Sciences Center). Antibodies against DRD4 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, catalog number: sc-25649, lot number: C1560) and Abgent Inc. (San Diego, CA, catalog number: AP8760C, lot number: SA100222AA); anti-phospho-eIF2α (catalog number: 33985, lot number: 2) and anti-caspase-7 (catalog number: 94925, lot number: 6) were obtained from Cell Signaling Technology (Beverly, MA, USA); and anti-β-actin was obtained from Abcam, Inc. (Cambridge, MA, USA). Horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse secondary antibodies were purchased from Kirkegaard & Perry Laboratories Inc. (Gaithersburg, MD, USA). All other reagents were purchased from Sigma Aldrich (St Louis, MO, USA), Bio-Rad (Hercules, CA, USA) and Invitrogen (Carlsbad, CA, USA).
Retinal RNA isolation, microarray assay and data analyses
Five mice at P30 were used in each group. Retinas were collected at the same time of day (2:00–3:00 pm) to avoid any circadian rhythm effects (73). Total RNA was isolated from mouse retinas as described previously (18,21). Briefly, two retinas from a single mouse were stored in Trizol (Invitrogen, Carlsbad) as one sample immediately after their isolation and frozen at −80°C until use. Retinas were homogenized and total RNA was isolated using a PureLinkTM RNA Mini kit (Invitrogen) following the manufacturer's instructions. RNA quality was verified by capillary gel electrophoresis (Agilent Technologies, Santa Clara, CA, USA).
RNA samples were labeled using an Illumina Total Prep RNA Amplification kit according to the manufacturer's directions (Invitrogen), hybridized to mouse Ref-8 version 2.0 Expression BeadChips (Illumina, San Diego, CA) and scanned using an Illumina iScan array scanner. Data from the scanner were obtained with GenomeStudio software (Illumina), quantile normalized (Matlab) and analyzed for differentially expressed genes between groups using a 5% false discovery rate (BRB-ArrayTools v 4.2.0 beta-2 release developed by Dr Richard Simon and BRB-ArrayTools Development Team, National Cancer Institute, Bethesda, MD, USA). The microarray results have been submitted to the Gene Expression Omnibus (reference series number: GSE40982).
Ingenuity pathway analyses
All differentially regulated genes were imported into IPA (Ingenuity Systems, Redwood City, CA; www.ingenuity.com) for canonical pathway analyses and generation of networks. The data set containing all significant gene identifiers, along with the corresponding expression and significance values, was uploaded into the application. The significance of the association between the data set and the canonical pathway was measured in two ways. One is the ratio of the number of molecules from the data set that map to the pathway, which was obtained by dividing the number of genes identified with the total number of genes that make up the particular canonical pathway in Ingenuity Knowledge Database. The other one is Fisher's exact test (with a P-value threshold of 0.05) to determine the probability that the association between the genes in the data set and in the canonical pathway is explained by chance alone.
For network generation, each identifier was mapped to its corresponding object in Ingenuity's Knowledge Base and overlaid onto a global molecular network developed from information contained in Ingenuity's Knowledge Base. Networks were then algorithmically generated based on their connectivity. The functional analysis of a network identified the biological functions, using a right-tailed Fisher's exact test (with a P-value threshold of 0.05) that was most significant to the molecules in the network.
Reverse transcription and quantitative real-time PCR (qRT–PCR)
A qRT–PCR was performed to validate microarray data as described previously (18,21). Briefly, cDNAs were prepared from RNA samples. Three micrograms of total RNA was reverse-transcribed into first-strand cDNA using an oligo-dT primer and SuperScript III reverse transcriptase (Invitrogen) as per the manufacturer's instructions, and RNase H (Invitrogen) treatment was applied in all cDNA synthesis processes. The cDNA products were dissolved in DEPC-treated water at a final concentration of 10 ng/µl of cDNA and stored at −20°C before use.
The qRT–PCR assays were performed using fluorescence reagents (IQTM SYBR® Green Supermix, Bio-Rad) and a thermal cycler platform (iCycler; Bio-Rad Laboratories, Hercules, CA, USA). Primers (see Supplementary Material, Table S3) used for qRT–PCR were designed to generate amplicons of 120–150 bp with similar melting temperature (Tm). The primer efficiency was determined and primers with efficiency at 90–105% were used in qRT–PCR assays. The cycling condition was 95°C for 3 min, followed by 40 cycles of 95°C for 15 and 58°C for 40 s for quantification. The melting curve was calculated using one cycle of 95°C for 1 s and 65°C for 10 s × 70, followed by continuous acquisition at 95°C and cooling at 4°C. Melting curve analysis confirmed the absence of primer dimers. A relative gene expression value was calculated against the mouse neuronal housekeeping gene hypoxanthine phosphoribosyltransferase (Hprt) using a ΔΔCT method as described previously (18,74).
Retinal protein sample preparation, SDS–PAGE and western blot analysis
Retinal protein sample preparation, SDS–PAGE and western blotting were performed as described previously (19,75). Briefly, retinas were homogenized in homogenization buffer containing protease cocktail (Sigma Aldrich Co.). The homogenate was centrifuged at 1000g for 10 min at 4°C to pellet down nuclei and cell debris. The supernatant of the homogenate was further centrifuged at 16 000g for 30 min at 4°C to separate cytosolic (supernatant) and membrane fractions (pellet). Membrane protein concentration was measured using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories). The membrane and cytosolic protein samples were solubilized in SDS–PAGE sample buffer and separated on 10% SDS–PAGE gel and transferred onto polyvinylidene difluoride membrane. After 1 h blocking in 5% milk containing Tris-buffered saline with 0.1% Tween (v/v) (TBST) at room temperature, the membranes were incubated with primary antibodies (anti-DRD4-sc-25649, 1:400; anti-DRD4-AP8760C, 1:500; anti-M-opsin, 1:2000; anti-S-opsin, 1:1000; anti-phospho-eIF2α, 1:500; anti-caspase-7, 1:250; anti-cone arrestin, 1:2000; anti-actin, 1:2000) overnight at 4°C and washed with TBST three times and incubated with HRP-conjugated secondary anti-rabbit or anti-mouse immunoglobulin for 1 h at room temperature followed by three washes. Images were captured using a KODAK Image Station 4000R Digital Imaging System (Carestream Molecular Imaging, New Haven, CT, USA). Densitometry analysis was performed by quantifying the intensities of the bands of interest using KODAK Molecular Imaging software with β-actin as a loading control. Four independent western blot experiments were performed using retinas prepared from four to five mice.
TUDCA treatment
Treatment of mice with TUDCA was conducted as described by Zhang et al. (44). Briefly, TUDCA (TCI America, 500 mg/kg, body weight) or vehicle (0.15 M NaHCO3, pH 7.0) was given to Cnga3−/−/Nrl−/− mice by subcutaneous injection every 3 days for 3 weeks, starting at P8. Retinas of mice were collected at the end of the experiments and analyzed for expression levels of cone opsin and phospho-eIF2α and for caspase-7 cleavage.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by NIH grants P20RR017703, P30EY12190 and R01EY019490; a research grant from the Oklahoma Center for the Advancement of Science & Technology (OCAST) and by grants from the Deutsche Forschungsgemeinschaft (DFG).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Anand Swaroop, Cheryl Craft, Muna Naash and Marc Caron for providing Nrl−/− mouse line, antibodies for M-opsin, S-opsin and cone arrestin, and hDRD4 cDNA. We thank Dr Jianhua Xu for providing excellent technical assistance.
Conflict of Interest statement. None declared
REFERENCES
- 1.Kaupp U.B., Seifert R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 2002;82:769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 2.Gerstner A., Zong X., Hofmann F., Biel M. Molecular cloning and functional characterization of a new modulatory cyclic nucleotide-gated channel subunit from mouse retina. J. Neurosci. 2000;20:1324–1332. doi: 10.1523/JNEUROSCI.20-04-01324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuart N.G., Haitin Y., Camp S.S., Black K.D., Zagotta W.N. Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels. Nat. Commun. 2011;2:457. doi: 10.1038/ncomms1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding X.Q., Matveev A., Singh A., Komori N., Matsumoto H. Biochemical characterization of cone cyclic nucleotide-gated (CNG) channel using the infrared fluorescence detection system. Adv. Exp. Med. Biol. 2012;723:769–775. doi: 10.1007/978-1-4614-0631-0_98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J., Trudeau M.C., Zagotta W.N. Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron. 2002;36:891–896. doi: 10.1016/s0896-6273(02)01099-1. [DOI] [PubMed] [Google Scholar]
- 6.Weitz D., Ficek N., Kremmer E., Bauer P.J., Kaupp U.B. Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron. 2002;36:881–889. doi: 10.1016/s0896-6273(02)01098-x. [DOI] [PubMed] [Google Scholar]
- 7.Zhong H., Molday L.L., Molday R.S., Yau K.W. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature. 2002;420:193–198. doi: 10.1038/nature01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohl S., Baumann B., Broghammer M., Jagle H., Sieving P., Kellner U., Spegal R., Anastasi M., Zrenner E., Sharpe L.T., et al. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum. Mol. Genet. 2000;9:2107–2116. doi: 10.1093/hmg/9.14.2107. [DOI] [PubMed] [Google Scholar]
- 9.Nishiguchi K.M., Sandberg M.A., Gorji N., Berson E.L., Dryja T.P. Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum. Mutat. 2005;25:248–258. doi: 10.1002/humu.20142. [DOI] [PubMed] [Google Scholar]
- 10.Wissinger B., Gamer D., Jagle H., Giorda R., Marx T., Mayer S., Tippmann S., Broghammer M., Jurklies B., Rosenberg T., et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am. J. Hum. Genet. 2001;69:722–737. doi: 10.1086/323613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidjanin D.J., Lowe J.K., McElwee J.L., Milne B.S., Phippen T.M., Sargan D.R., Aguirre G.D., Acland G.M., Ostrander E.A. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum. Mol. Genet. 2002;11:1823–1833. doi: 10.1093/hmg/11.16.1823. [DOI] [PubMed] [Google Scholar]
- 12.Kohl S., Varsanyi B., Antunes G.A., Baumann B., Hoyng C.B., Jagle H., Rosenberg T., Kellner U., Lorenz B., Salati R., et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur. J. Hum. Genet. 2005;13:302–308. doi: 10.1038/sj.ejhg.5201269. [DOI] [PubMed] [Google Scholar]
- 13.Varsanyi B., Somfai G.M., Lesch B., Vamos R., Farkas A. Optical coherence tomography of the macula in congenital achromatopsia. Invest. Ophthalmol. Vis. Sci. 2007;48:2249–2253. doi: 10.1167/iovs.06-1173. [DOI] [PubMed] [Google Scholar]
- 14.Andersen M.K., Christoffersen N.L., Sander B., Edmund C., Larsen M., Grau T., Wissinger B., Kohl S., Rosenberg T. Oligocone trichromacy: clinical and molecular genetic investigations. Invest. Ophthalmol. Vis. Sci. 2010;51:89–95. doi: 10.1167/iovs.09-3988. [DOI] [PubMed] [Google Scholar]
- 15.Thiadens A.A., Somervuo V., van den Born L.I., Roosing S., van Schooneveld M.J., Kuijpers R.W., van Moll-Ramirez N., Cremers F.P., Hoyng C.B., Klaver C. Progressive loss of cones in achromatopsia. An imaging study using spectral-domain optical coherence tomography. Invest. Ophthalmol. Vis. Sci. 2010;51:5952–5957. doi: 10.1167/iovs.10-5680. [DOI] [PubMed] [Google Scholar]
- 16.Biel M., Seeliger M., Pfeifer A., Kohler K., Gerstner A., Ludwig A., Jaissle G., Fauser S., Zrenner E., Hofmann F. Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc. Natl Acad. Sci. USA. 1999;96:7553–7557. doi: 10.1073/pnas.96.13.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalakis S., Geiger H., Haverkamp S., Hofmann F., Gerstner A., Biel M. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Invest. Ophthalmol. Vis. Sci. 2005;46:1516–1524. doi: 10.1167/iovs.04-1503. [DOI] [PubMed] [Google Scholar]
- 18.Ding X.Q., Harry C.S., Umino Y., Matveev A.V., Fliesler S.J., Barlow R.B. Impaired cone function and cone degeneration resulting from CNGB3 deficiency: down-regulation of CNGA3 biosynthesis as a potential mechanism. Hum. Mol. Genet. 2009;18:4770–4780. doi: 10.1093/hmg/ddp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thapa A., Morris L., Xu J., Ma H., Michalakis S., Biel M., Ding X.Q. Endoplasmic reticulum stress-associated cone photoreceptor degeneration in cyclic nucleotide-gated channel deficiency. J. Biol. Chem. 2012;287:18018–18029. doi: 10.1074/jbc.M112.342220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J., Morris L., Fliesler S.J., Sherry D.M., Ding X.Q. Early-onset, slow progression of cone photoreceptor dysfunction and degeneration in CNG channel subunit CNGB3 deficiency. Invest. Ophthalmol. Vis. Sci. 2011;52:3557–3566. doi: 10.1167/iovs.10-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho L.S., Xu J., Pearson R.A., Smith A.J., Bainbridge J.W., Morris L.M., Fliesler S.J., Ding X.Q., Ali R.R. Long-term and age-dependent restoration of visual function in a mouse model of CNGB3-associated achromatopsia following gene therapy. Hum. Mol. Genet. 2011;20:3161–3175. doi: 10.1093/hmg/ddr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haverkamp S., Michalakis S., Claes E., Seeliger M.W., Humphries P., Biel M., Feigenspan A. Synaptic plasticity in CNGA3−/− mice: cone bipolar cells react on the missing cone input and form ectopic synapses with rods. J. Neurosci. 2006;26:5248–5255. doi: 10.1523/JNEUROSCI.4483-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 24.Daniele L.L., Lillo C., Lyubarsky A.L., Nikonov S.S., Philp N., Mears A.J., Swaroop A., Williams D.S., Pugh E.N., Jr. Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest. Ophthalmol. Vis. Sci. 2005;46:2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikonov S.S., Daniele L.L., Zhu X., Craft C.M., Swaroop A., Pugh E.N., Jr. Photoreceptors of Nrl−/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J. Gen. Physiol. 2005;125:287–304. doi: 10.1085/jgp.200409208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida S. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum. Mol. Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 27.Mustafi D., Kevany B.M., Genoud C., Okano K., Cideciyan A.V., Sumaroka A., Roman A.J., Jacobson S.G., Engel A., Adams M.D., et al. Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. FASEB J. 2011;25:3157–3176. doi: 10.1096/fj.11-186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks M.J., Rajasimha H.K., Roger J.E., Swaroop A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl−/− retinal transcriptomes. Mol. Vis. 2011;17:3034–3054. [PMC free article] [PubMed] [Google Scholar]
- 29.Corbo J.C., Myers C.A., Lawrence K.A., Jadhav A.P., Cepko C.L. A typology of photoreceptor gene expression patterns in the mouse. Proc. Natl Acad. Sci. USA. 2007;104:12069–12074. doi: 10.1073/pnas.0705465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valor L.M., Grant S.G. Clustered gene expression changes flank targeted gene loci in knockout mice. PLoS One. 2007;2:e1303. doi: 10.1371/journal.pone.0001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Tol H.H., Wu C.M., Guan H.C., Ohara K., Bunzow J.R., Civelli O., Kennedy J., Seeman P., Niznik H.B., Jovanovic V. Multiple dopamine D4 receptor variants in the human population. Nature. 1992;358:149–152. doi: 10.1038/358149a0. [DOI] [PubMed] [Google Scholar]
- 32.Lichter J.B., Barr C.L., Kennedy J.L., Van Tol H.H., Kidd K.K., Livak K.J. A hypervariable segment in the human dopamine receptor D4 (DRD4) gene. Hum. Mol. Genet. 1993;2:767–773. doi: 10.1093/hmg/2.6.767. [DOI] [PubMed] [Google Scholar]
- 33.Livak K.J., Rogers J., Lichter J.B. Variability of dopamine D4 receptor (DRD4) gene sequence within and among nonhuman primate species. Proc. Natl Acad. Sci. USA. 1995;92:427–431. doi: 10.1073/pnas.92.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seaman M.I., Chang F.M., Quinones A.T., Kidd K.K. Evolution of exon 1 of the dopamine D4 receptor (DRD4) gene in primates. J. Exp. Zoolog. 2000;288:32–38. [PubMed] [Google Scholar]
- 35.Niimi Y., Inoue-Murayama M., Kato K., Matsuura N., Murayama Y., Ito S., Momoi Y., Konno K., Iwasaki T. Breed differences in allele frequency of the dopamine receptor D4 gene in dogs. J. Hered. 2001;92:433–436. doi: 10.1093/jhered/92.5.433. [DOI] [PubMed] [Google Scholar]
- 36.Niimi Y., Inoue-Murayama M., Murayama Y., Ito S., Iwasaki T. Allelic variation of the D4 dopamine receptor polymorphic region in two dog breeds, Golden retriever and Shiba. J. Vet. Med. Sci. 1999;61:1281–1286. doi: 10.1292/jvms.61.1281. [DOI] [PubMed] [Google Scholar]
- 37.Hejjas K., Vas J., Kubinyi E., Sasvari-Szekely M., Miklosi A., Ronai Z. Novel repeat polymorphisms of the dopaminergic neurotransmitter genes among dogs and wolves. Mamm. Genome. 2007;18:871–879. doi: 10.1007/s00335-007-9070-0. [DOI] [PubMed] [Google Scholar]
- 38.Hejjas K., Vas J., Topal J., Szantai E., Ronai Z., Szekely A., Kubinyi E., Horvath Z., Sasvari-Szekely M., Miklosi A. Association of polymorphisms in the dopamine D4 receptor gene and the activity-impulsivity endophenotype in dogs. Anim. Genet. 2007;38:629–633. doi: 10.1111/j.1365-2052.2007.01657.x. [DOI] [PubMed] [Google Scholar]
- 39.Fan C.Y., Cheng J.B., Li J.L., Wang Q.H., Manglai D. Polymorphism analysis of the horse dopamine receptor D4 gene (DRD4) sequence. J. Anim. Vet. Adv. 2011;10:1855–1858. [Google Scholar]
- 40.Sugiyama A., Inoue-Murayama M., Miwa M., Ohashi R., Kayang B.B., Mizutani M., Nirasawa K., Odai M., Minezawa M., Watanabe S., et al. Polymorphism of dopamine receptor D4 exon I corresponding region in chicken. Zoolog. Sci. 2004;21:941–946. doi: 10.2108/zsj.21.941. [DOI] [PubMed] [Google Scholar]
- 41.Grady D.L., Thanos P.K., Corrada M.M., Barnett J.C., Jr., Ciobanu V., Shustarovich D., Napoli A., Moyzis A.G., Grandy D., Rubinstein M., et al. DRD4 genotype predicts longevity in mouse and human. J. Neurosci. 2013;33:286–291. doi: 10.1523/JNEUROSCI.3515-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantopoulos D., Murakami Y., Comander J., Thanos A., Roh M., Miller J.W., Vavvas D.G. Tauroursodeoxycholic acid (TUDCA) protects photoreceptors from cell death after experimental retinal detachment. PLoS One. 2011;6:e24245. doi: 10.1371/journal.pone.0024245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez-Sanchez L., Lax P., Pinilla I., Martin-Nieto J., Cuenca N. Tauroursodeoxycholic acid prevents retinal degeneration in transgenic P23H rats. Invest. Ophthalmol. Vis. Sci. 2011;52:4998–5008. doi: 10.1167/iovs.11-7496. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T., Baehr W., Fu Y. Chemical chaperone TUDCA preserves cone photoreceptors in a mouse model of Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 2012;53:3349–3356. doi: 10.1167/iovs.12-9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yetemian R.M., Brown B.M., Craft C.M. Neovascularization, enhanced inflammatory response, and age-related cone dystrophy in the Nrl−/−Grk1−/− mouse retina. Invest. Ophthalmol. Vis. Sci. 2010;51:6196–6206. doi: 10.1167/iovs.10-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaeferhoff K., Michalakis S., Tanimoto N., Fischer M.D., Becirovic E., Beck S.C., Huber G., Rieger N., Riess O., Wissinger B., et al. Induction of STAT3-related genes in fast degenerating cone photoreceptors of cpfl1 mice. Cell Mol. Life Sci. 2010;67:3173–3186. doi: 10.1007/s00018-010-0376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cottet S., Michaut L., Boisset G., Schlecht U., Gehring W., Schorderet D.F. Biological characterization of gene response in Rpe65−/− mouse model of Leber's congenital amaurosis during progression of the disease. FASEB J. 2006;20:2036–2049. doi: 10.1096/fj.06-6211com. [DOI] [PubMed] [Google Scholar]
- 48.Znoiko S.L., Rohrer B., Lu K., Lohr H.R., Crouch R.K., Ma J.X. Downregulation of cone-specific gene expression and degeneration of cone photoreceptors in the Rpe65−/− mouse at early ages. Invest. Ophthalmol. Vis. Sci. 2005;46:1473–1479. doi: 10.1167/iovs.04-0653. [DOI] [PubMed] [Google Scholar]
- 49.Swiderski R.E., Nishimura D.Y., Mullins R.F., Olvera M.A., Ross J.L., Huang J., Stone E.M., Sheffield V.C. Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Invest. Ophthalmol. Vis. Sci. 2007;48:3329–3340. doi: 10.1167/iovs.06-1477. [DOI] [PubMed] [Google Scholar]
- 50.Li A., Zhu X., Craft C.M. Retinoic acid upregulates cone arrestin expression in retinoblastoma cells through a Cis element in the distal promoter region. Invest. Ophthalmol. Vis. Sci. 2002;43:1375–1383. [PubMed] [Google Scholar]
- 51.Nguyen-Legros J., Berger B., Vigny A., Alvarez C. Tyrosine hydroxylase-like immunoreactive interplexiform cells in the rat retina. Neurosci. Lett. 1981;27:255–259. doi: 10.1016/0304-3940(81)90439-0. [DOI] [PubMed] [Google Scholar]
- 52.Versaux-Botteri C., Martin-Martinelli E., Nguyen-Legros J., Geffard M., Vigny A., Denoroy L. Regional specialization of the rat retina: catecholamine-containing amacrine cell characterization and distribution. J. Comp. Neurol. 1986;243:422–433. doi: 10.1002/cne.902430311. [DOI] [PubMed] [Google Scholar]
- 53.Klitten L.L., Rath M.F., Coon S.L., Kim J.S., Klein D.C., Moller M. Localization and regulation of dopamine receptor D4 expression in the adult and developing rat retina. Exp. Eye Res. 2008;87:471–477. doi: 10.1016/j.exer.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen A.I., Todd R.D., Harmon S., O'Malley K.L. Photoreceptors of mouse retinas possess D4 receptors coupled to adenylate cyclase. Proc. Natl Acad. Sci. USA. 1992;89:12093–12097. doi: 10.1073/pnas.89.24.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witkovsky P. Dopamine and retinal function. Doc. Ophthalmol. 2004;108:17–40. doi: 10.1023/b:doop.0000019487.88486.0a. [DOI] [PubMed] [Google Scholar]
- 56.Jackson C.R., Chaurasia S.S., Zhou H., Haque R., Storm D.R., Iuvone P.M. Essential roles of dopamine D4 receptors and the type 1 adenylyl cyclase in photic control of cyclic AMP in photoreceptor cells. J. Neurochem. 2009;109:148–157. doi: 10.1111/j.1471-4159.2009.05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson C.R., Chaurasia S.S., Hwang C.K., Iuvone P.M. Dopamine D(4) receptor activation controls circadian timing of the adenylyl cyclase 1/cyclic AMP signaling system in mouse retina. Eur. J. Neurosci. 2011;34:57–64. doi: 10.1111/j.1460-9568.2011.07734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ko G.Y., Ko M.L., Dryer S.E. Circadian regulation of cGMP-gated channels of vertebrate cone photoreceptors: role of cAMP and Ras. J. Neurosci. 2004;24:1296–1304. doi: 10.1523/JNEUROSCI.3560-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ko G.Y., Ko M., Dryer S.E. Circadian and cAMP-dependent modulation of retinal cone cGMP-gated channels does not require protein synthesis or calcium influx through L-type channels. Brain Res. 2004;1021:277–280. doi: 10.1016/j.brainres.2004.05.072. [DOI] [PubMed] [Google Scholar]
- 60.Ko G.Y., Ko M.L., Dryer S.E. Circadian regulation of cGMP-gated cationic channels of chick retinal cones. Erk MAP Kinase and Ca2+/calmodulin-dependent protein kinase II. Neuron. 2001;29:255–266. doi: 10.1016/s0896-6273(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 61.Ko G.Y., Ko M.L., Dryer S.E. Circadian phase-dependent modulation of cGMP-gated channels of cone photoreceptors by dopamine and D2 agonist. J. Neurosci. 2003;23:3145–3153. doi: 10.1523/JNEUROSCI.23-08-03145.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chae K.S., Ko G.Y., Dryer S.E. Tyrosine phosphorylation of cGMP-gated ion channels is under circadian control in chick retina photoreceptors. Invest. Ophthalmol. Vis. Sci. 2007;48:901–906. doi: 10.1167/iovs.06-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakao T., Tsujikawa M., Notomi S., Ikeda Y., Nishida K. The role of mislocalized phototransduction in photoreceptor cell death of retinitis pigmentosa. PLoS One. 2012;7:e32472. doi: 10.1371/journal.pone.0032472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J., Zhang N., Beuve A., Townes-Anderson E. Mislocalized opsin and cAMP signaling: a mechanism for sprouting by rod cells in retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2012;53:6355–6369. doi: 10.1167/iovs.12-10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rattner A., Nathans J. The genomic response to retinal disease and injury: evidence for endothelin signaling from photoreceptors to glia. J. Neurosci. 2005;25:4540–4549. doi: 10.1523/JNEUROSCI.0492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torbidoni V., Iribarne M., Suburo A.M. Endothelin receptors in light-induced retinal degeneration. Exp. Biol. Med. (Maywood) 2006;231:1095–1100. [PubMed] [Google Scholar]
- 67.Joly S., Lange C., Thiersch M., Samardzija M., Grimm C. Leukemia inhibitory factor extends the lifespan of injured photoreceptors in vivo. J. Neurosci. 2008;28:13765–13774. doi: 10.1523/JNEUROSCI.5114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suburo A.M., Torbidoni V., Iribarne M., Prasanna G. Expression of endothelin and its receptors in light-injured retina. Invest. Ophthalmol. Vis. Sci. 2004;45:5295. [Google Scholar]
- 69.Yang F., Li X., Sharma M., Sasaki C.Y., Longo D.L., Lim B., Sun Z. Linking beta-catenin to androgen-signaling pathway. J. Biol. Chem. 2002;277:11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi S., Watanabe T., Okada M., Inoue K., Ueda T., Takada I., Watabe T., Yamamoto Y., Fukuda T., Nakamura T., et al. Noncanonical Wnt signaling mediates androgen-dependent tumor growth in a mouse model of prostate cancer. Proc. Natl Acad. Sci. USA. 2011;108:4938–4943. doi: 10.1073/pnas.1014850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hackam A.S. The Wnt signaling pathway in retinal degenerations. IUBMB Life. 2005;57:381–388. doi: 10.1080/15216540500137586. [DOI] [PubMed] [Google Scholar]
- 72.Thiadens A.A., Roosing S., Collin R.W., van Moll-Ramirez N., van Lith-Verhoeven J.J., van Schooneveld M.J., den Hollander A.I., van den Born L.I., Hoyng C.B., Cremers F.P., et al. Comprehensive analysis of the achromatopsia genes CNGA3 and CNGB3 in progressive cone dystrophy. Ophthalmology. 2010;117:825–830 e821. doi: 10.1016/j.ophtha.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 73.Storch K.F., Paz C., Signorovitch J., Raviola E., Pawlyk B., Li T., Weitz C.J. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu J., Morris L.M., Michalakis S., Biel M., Fliesler S.J., Sherry D.M., Ding X.Q. CNGA3 deficiency affects cone synaptic terminal structure and function and leads to secondary rod dysfunction and degeneration. Invest. Ophthalmol. Vis. Sci. 2012;53:1117–1129. doi: 10.1167/iovs.11-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.