Abstract
In order to identify biomarkers for early diagnosis and/or for therapeutic targets in the delayed health effects of ionizing radiation, we analyzed the subgroups of lymphocytes, serum protein levels and gene expression profiles in the peripheral blood of three 60Co γ-ray accidentally exposed persons during the three years after irradiation. Flow cytometry analyses and agarose gel electrophoresis were applied to investigate the subgroups of lymphocytes and the composition of serum proteins, respectively. Gene expression profiling was obtained using a whole genome gene expression chip assay. Both the percentage of CD4+ T lymphocytes and the ratio of Th to Ts were reduced compared with the normal control values. The percentage of albumin decreased whereas beta globulin increased. There were 285 up-regulated and 446 down-regulated genes in irradiated samples relative to the control samples. The expression of KDR, CEACAM8 and OSM was validated by RT-PCR. The majority of the differentially expressed genes encode proteins associated with the immune response, inflammation, oncogenesis, cell structure, oxidative stress, neuro-hormone regulation, reproduction, susceptibility to psychiatric disorders, or transcriptional regulation. We have identified a number of promising novel candidates that have potential for serving as biomarkers for delayed damage. Furthermore, the changes in the immunological indicator CD4+ T cells, and the ratio of CD4+ T to CD8+ T cells may be biomarkers for the prediction of delayed damage by ionizing radiation. The findings of our study are useful for forming a comprehensive understanding of the mechanisms underlying the delayed effects of ionizing radiation.
Keywords: accidental exposure, gene expression profile, ratio of Th to Ts, radiation biomarkers
INTRODUCTION
It is clear that exposure to ionizing radiation can cause delayed effects, such as tumorigenesis, cataracts, infertility, chronic inflammation, and diffuse fibrosis of the affected organ and tissue. Although the human risks of cardiovascular diseases and neuro-psychiatric diseases caused by ionizing radiation have not been validated by definitive epidemiological studies, several reports have described the effects of ionizing radiation on the incidence and prevalence of such diseases [1–7]. Understanding cellular and molecular pathological changes, and establishing biomarkers for cancer and non-cancer effects of chronic low-dose or acute high-dose radiation have become the focus in the field of radiation protection in recent years [8–11]. Such knowledge will guide the prevention, early diagnosis and treatment of ionizing radiation-induced damage, as well as the related drug design. It will also provide the theoretical basis for adopting efficient measures in the practice of radiation protection (e.g. health surveillance of radiation workers), medical treatment and therapeutic strategies in radiation emergency medicine, and for providing health services for long-term cancer survivors after radiotherapy.
According to the National Cancer Prevention office, there were 2 million new cancer occurrences in the year 2000 in China. Among them, 400 000 cases received radiotherapy. After secondary malignancies, cardiovascular disease is the leading cause of late morbidity and death among cancer survivors in the United States [12]. Similarly, we can speculate that the number of cancer survivors is increasing in China, even though there is no up-to-date statistical data available, and that non-cancer diseases will occupy an important role in late morbidity and death in cancer patients after radiotherapy and chemotherapy. From 1994–1998 in China there were 139 cases of radiation accidents and 238 persons were overexposed to radiation to some degree. There were 230 000 radiation workers in China up to the year 2009. Among them, 1718 were identified as sustaining ‘radiation damage’, and 464 were diagnosed as having ‘suspected radiation disease’. Therefore, there is a great need to investigate delayed radiation effects in the above-mentioned three categories of persons, particularly since the use of the emerging tools of bioinformatics technology has shown that the effects of large doses are distinguishable from those of low doses. The early identification of risk groups and the diagnosis of oncological and degenerative diseases will be highly beneficial. Therefore, we performed analyses using blood samples from three persons who were accidentally exposed to high-dose radiation, simulating the biological condition of cancer survivors who have been treated with high-dose radiation, and those who have been accidentally exposed to radiation in some other way.
The three accidentally exposed persons followed up in the present study were exposed in a radiation-sterilization facility accident that occurred on 12 May 2008. Five male workers from 38–45 years of age were exposed to Co-60 source (activity, 1.6415 × 104 Ci) γ-rays for nearly 20 minutes. The Co-60 source was being used for sterilization of Chinese herbs. The initial cause of this accident was a loss of function of the facility safety interlock. The five workers entered the radiation room to move out the irradiated herbs and move in new herbs for irradiation, while failing to check the radiation meter count. Additionally, the audio alarm sound of the portable radiation meter was masked by the noise of the ventilation system in the radiation room. Only when one person noticed that the steel line controlling the up-and-down movement of the radiation source was very loose did they become aware that the source was still in exposure status. They evacuated the site after they dropped the source down to the safe position. The five persons were sent to the Affiliated Hospital of China Institute for Radiation Protection 40 min later. Physical doses were estimated using the thermoluminescence dosimetry system in the Dosimetry Laboratory of the institute, according to the simulated exposure scenario. The physical doses were 12.0, 11.2, 1.82, 1.82 and 1.0 Gy, respectively. The five persons were sent to the Affiliated Hospital of Beijing Institute of Radiation Medicine for further treatment. They were diagnosed with acute radiation syndrome, one with gastrointestinal syndrome, and the other four with hematopoietic syndrome with either a high or medium level of severity. Four of the five persons received bone marrow transplantation from their brothers or son. Two persons with physical doses of 12.0 and 1.82 Gy died on Day 60 and 17 months after the accident, respectively.
The three remaining persons (here named A, B and C) were followed annually with clinical examinations, laboratory tests and biodosimetric estimations. Among them, A and B received bone marrow transplants on Day 14 and 21 after the accident, respectively. Patient A presented with the most serious injuries, manifesting acute double pneumonia, upper respiratory infection and middle ear infection during the treatment. His lymphocyte percentage went back to the normal level more than two years after the accident. Patient B experienced acute pneumonia and upper respiratory infection. His lymphocyte percentage went back to the normal level within six months. Patient C was treated with G-CSF 48 h after the accident and manifested upper respiratory infection within two months. The three patients were monitored for delayed health effects. We noticed some diseases appeared gradually in the exposed persons, including opacity of the lens, skin disorders, sperm abnormalities, and abnormalities in lymphocyte subgroups, as well as osteopenia. The mechanisms underlying these delayed effects of exposure to a high dose of irradiation are not well known. Previous studies demonstrated that oxidative stress is associated with the acute skin reactions caused by high doses of irradiation [13–15], and changes in immunological indicators were found in the ‘liquidators’ at the Chernobyl accident and in the atomic bomb survivors [1–3]. Therefore, the aim of the present study was to undertake follow-up studies of the immunological status and gene expression profiles of the three accidentally exposed persons in order to establish candidate biomarkers for the early identification of persons at risk or for the diagnosis of oncological diseases and other delayed harmful effects caused by ionizing radiation.
MATERIALS AND METHODS
Reagents
Cy3 NHS ester was from GE, Healthcare (Connecticut, USA) and aaUTP was from Ambion (California, USA). The fluorochrome-conjugated antibodies CD4-FITC/CD8-PE/CD3-PC5, CD3-FITC/CD(16 + 56)-PE and CD19-PC5, and the isotypic antibodies for control IgG1-FITC/IgG1-PE/IgG1-PC5 were from Immunotech, Beckman, France. Ethanol and β-diathiothretol were from Sigma. RNase-free water, and Nuclease-free water were products of HyClone, (Logan, UT, USA). Water was double distilled and deionized in an ultra-pure water apparatus (Millipore).
Blood sampling and preparation
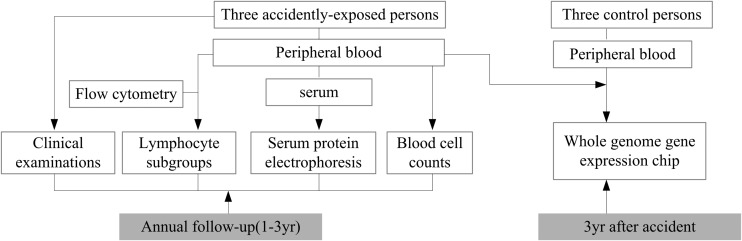
Fresh blood samples were collected from the three persons accidentally exposed to 60Co-γ-rays (three years post-irradiation) and three age-matched, non-irradiated healthy donors. Leukocytes were isolated and total RNA was extracted. Four RNA samples, one from each of the three accidentally exposed persons, along with a RNA mixture extracted from a combination of three healthy donors' leukocytes (ratio 1:1:1, each 3.3 × 106 leukocytes), were subjected to Agilent 4 × 44 k whole genome gene expression microarray assay. Fresh blood samples collected from the three exposed persons were used for the leukocyte count, measurement of the subgroups of lymphocytes by flow cytometry, and separation and quantification of serum proteins by agarose gel electrophoresis. The measurements were conducted in The Second Affiliated Hospital of Shan Xi Medical College. The experimental systems with the control reference levels that we used in the present study are routinely used for clinical diagnosis in the hospital. All the assays were performed following a single blood draw during yearly follow up study. The overview of the experimental design is shown in Fig. 1.
Fig. 1.
Overview of experimental design.
Whole genome gene expression microarray assay
Total RNA was extracted using the QIAamp RNA Blood Minikit (QIAGEN GmbH, Hilden, Germany) following the manufacturer's instructions. An RNase-Free DNase Set (QIAGEN) was used to digest DNA efficiently during RNA purification. RNA quality was assessed by spectrophotometry (NanoDrop, ND1000, Massachusetts, USA) and gel electrophoresis. To be considered high quality, the RNA had to have a 260/280 ratio > 1.7 and an 18S/28S rRNA ratio ∼2.
Labeling was done using a Low RNA Input Linear Amplification Kit (Agilent, California, USA) according to the manufacturer's instructions, followed by RNAeasy Column Purification (QIAGEN, Hilden, Germany). Briefly, cDNA was generated using a T7 promoter element coupled oligo dT primer. From the cDNA, labeled cRNA was generated via an in vitro transcription reaction using T7 RNA polymerase and aa-UTP. aa-UTP incorporated into cRNA was dyed with Cy3 NHS ester. Dye incorporation and cRNA yield were checked with the NanoDrop ND-1000 Spectrophotometer.
Labeled cRNA (825 ng) from the respective sample was fragmented and hybridized onto Agilent whole human genome microarrays (4 × 44 K G4112F, slide number, 66634, California, USA ) using a Gene Expression Hybridization Kit (5188-5242, Agilent Technologies). Hybridization was performed for 17 h, rotating at a speed of 10 rpm at 65°C in an Agilent hybridization oven (G2545A). After hybridization, microarrays were washed for 1 min at room temperature with GE Wash Buffer 1, and 1 min with 37°C GE Wash buffer 2 (5188-5327, Agilent Technologies).
The slides were scanned with Agilent microarray scanner (G2505C) using one color setting for 4 × 44 K array slides (scan resolution 5 µm, dye channel set to green, and green PMT set to 100%).
Images were processed with Agilent Feature Extraction Version 10.7.1.1 to obtain raw data files. The raw data files that contained ‘Processed Signal’ were processed using the Limma package developed within the Bioconductor project in the R statistical programming environment. After log transformation, the data were normalized using the ‘quantile’ method. The gene expression data were filtered according to the values of AFE quantification flags of gIsFound, IsWellAboveBG, gIsSaturated and the computing results of WellAboveNeg. After combinations of the replicated probes, the processed data was used to analyze the differentially expressed genes using a threshold of absolute log2 fold change value cut off at 1.
RT-PCR analysis
The quantitative RT-PCR was carried out with a 7500 Real-Time PCR System (Applied Biosystems, California, USA), with primers for KDR, OSM, CEACAM8 and ACTB, as shown in Table 1. The primers were designed using Primer Express 2.0. The synthesis of cDNA and fluorescence PCR amplification were performed using an one-step QuantiFast SYBR Green RT-PCR Kit (QIAGEN). Relative quantification of mRNA expression was performed by the delta delta Ct (Ct0) method using the level of β-actin mRNA for normalization of mRNA abundances across the samples. 2−Ct(Ct0) was used for calculation of the relative expression amount:
Table 1.
Primers for genes of KDR, OSM, CEACAM8 and ACTB
| Gene | Sequence | Length of product (bp) |
|---|---|---|
| KDR | forward: 5'CTCAAACCAGACAAGCGGCTA3′ | 121 |
| backward: 5'GCTACCGGTTTGCACTCCAAT3′ | ||
| OSM | forward: 5'CCAGACTTCCTCCTTTCCGTG3′ | 137 |
| backward: 5'ACACCCTGCCGCTGTTACAG3′ | ||
| CEACAM8 | forward: 5'TCAGTGACCCAGTCACCCTGA3′ | 122 |
| backward: 5'TGGATTAGAGGCCGCATGG3′ | ||
| ACTB | forward: 5'AAGTACTCCGTGTGGATCGGC3′ | 125 |
| backward: 5'GCCTAGAAGCATTTGCGGTG3′ |
Measurement of serum proteins by agarose gel electrophoresis
The serum proteins were separated and quantified by agarose gel electrophoresis using the REP SP Applicator Kit (Helena, BioSciences Europe). After programming the parameters into the automatic instrument Spife Combo (Helena Laboratories, USA), 40 µl of the control/sample serum was pipetted into the appropriate sample well and placed in the correct position on the sample tray. Following electrophoresis, the gel was stained with acid blue stain for 10 min, destained with 5% glacial acetic acid and dried at 65°C in a drying oven. The REP Applicator kit separates proteins into five main classes (albumin, alpha1-globulin, alpha2-globulin, beta-globulin and gamma globulin) according to charge in an agarose gel. The stained gel was subjected to visual and quantitative interpretation.
Measurement of subgroups of lymphocytes by flow cytometry
Three reaction tubes were designated for every blood sample, and contained OptiClone monoclonal antibodies CD4-FITC/CD8-PE/CD3-PC5, CD3-FITC/CD(16 + 56)-PE and CD19-PC5, and corresponding isotypic controls, respectively. The 12 × 75 mm tubes contained 20 µl of OptiClone monoclonal antibodies, and 100 µl of anticoagulated whole blood was added and briefly vortexed to mix. The reactions were incubated for 15 min at room temperature (18–25°C), in the dark. The red blood cells were lysed using Optilyse lysing solution, and incubated for 10 min at room temperature, protected from light. Following centrifugation for 5 min at 150 × g at room temperature, the cell pellets were washed twice with 3 ml of PBS. The resulting cell pellets were dissolved in 0.5 ml PBS. Acquisition and analysis were performed with a Cytometric FC 500 (Beckman Coulter, California, USA) flow cytometer equipped with CXP Software.
RESULTS
Changes in the subgroups of lymphocytes
The subgroups of lymphocytes in the peripheral blood of the three exposed persons changed during the three years following the accident. The leukocyte count was elevated in all three individuals within hours after the exposure but returned to normal after one year. The percentage of lymphocytes was depressed in all three individuals within hours after the exposure and remained depressed in one individual for two years (Table 2). Changes in lymphocyte subgroups are shown in Table 3. The percentages of helper T (Th, CD3 + CD4+) cells were reduced in all three persons. The ratios of Th to Ts (CD3 + CD8 + , Suppressor T) were also reduced compared with the reference level. The total T cells decreased or were near the lower limit of the reference level.
Table 2.
Leukocytes and lymphocyte percentage in the peripheral blood of accidentally exposed persons
| Cell type | Time after radiation | A | B | C | Reference value |
|---|---|---|---|---|---|
| (11.2 Gy) | (1.82 Gy) | (1.0 Gy) | |||
| Leukocyte (×109/l) | 2 h | 9.89 | 7.97 | 10.4 ↑ | 4–10 109/l |
| 4 h | 12.75 ↑ | 12.09 ↑ | 15.86 ↑ | ||
| 1 year | 5.73 | 4.61 | 5.01 | ||
| 2 year | 7.19 | 4.4 | 5.52 | ||
| 3 year | 5.46 | 4.67 | 5.57 | ||
| Lymphocyte (%) | 2 h | 8.92 ↓ | 14.62 ↓ | 12.82 ↓ | 20–40% |
| 4 h | 3.52 ↓ | 4.82 ↓ | 4.62 ↓ | ||
| 1 year | 18.02 ↓ | 25.8 | 25.7 | ||
| 2 year | 16.02 ↓ | 27 | 24.5 |
Table 3.
Lymphocyte sub-groups distribution (%) in the accidentally exposed persons
| Cell classification | Time after irradiation (year) | A (11.2 Gy) | B (1.82 Gy) | C (1.0 Gy) | Reference value (%) |
|---|---|---|---|---|---|
| Th (CD3 + CD4 + ) | 1 | 18.7 ↓ | 25.9 ↓ | 26.7 ↓ | 30.09–40.41 |
| 2 | 19.0 ↓ | 27.1 ↓ | 28.0 ↓ | ||
| 3 | 19.5 ↓ | 28.2 ↓ | 27.2 ↓ | ||
| Ts (CD3 + CD8 + ) | 1 | 41.7 ↑ | 27.8 | 29.6 | 20.74–29.42 |
| 2 | 34.6 ↑ | 25.5 | 30.9 ↑ | ||
| 3 | 35.9 ↑ | 24.3 | 29.3 | ||
| Th/Ts | 1 | 0.45 ↓ | 0.93 ↓ | 0.90 ↓ | 0.98–1.94 |
| 2 | 0.55 ↓ | 1.06 | 0.91 ↓ | ||
| 3 | 0.54 ↓ | 1.16 | 0.93 ↓ | ||
| Total T cell (CD3+) | 1 | 62.1 | 57.6 ↓ | 59.9 ↓ | 60.0–80.0 |
| 2 | 56.1 ↓ | 56.4 ↓ | 61.8 | ||
| 3 | 56.6 ↓ | 56.0 ↓ | 60.1 | ||
| NK cell {CD3–CD(16 + 56) + } | 1 | 5.50 ↓ | 24.5 | 27.8 ↑ | 8.1–25.6 |
| 2 | 5.0 ↓ | 28.3 ↑ | 22.2 | ||
| 3 | 5.3 ↓ | 23.6 | 17.2 | ||
| CD3 + CD(16 + 56) + | 1 | 0.30 | 0.10 | 0.30 | <5 |
| 2 | 8.6 ↑ | 5.4 ↑ | 3.4 | ||
| 3 | 4.8 | 3.0 | 1.4 | ||
| CD19 + | 1 | 24.6 ↑ | 9.40 | 13.9 | 9–15 |
| 2 | 38.1 ↑ | 14.8 | 15.1 | ||
| 3 | 34.5 ↑ | 16.6 ↑ | 11.5 |
Changes in serum protein levels
The percentage of albumin was lower, whereas Beta globulin was higher compared with the reference values (Table 4).
Table 4.
Serum proteins distribution (%) in the three accidentally exposed persons
| Component | Time after irradiation (year) | A (11.2 Gy) | B (1.82 Gy) | C (1.0 Gy) | Reference value (%) |
|---|---|---|---|---|---|
| Albumin | 1 | 58.65 | 46.89↓ | 62.74 | 52.30–66.00 |
| 2 | 42.75↓ | 50.89↓ | 42.26↓ | ||
| 3 | 49.49↓ | 45.59↓ | 49.25↓ | ||
| Alpha 1 | 1 | 3.45 | 6.43 | 2.97↓ | 3.30–7.00 |
| 2 | 5.06 | 2.55↓ | 4.36 | ||
| 3 | 5.99 | 4.41 | 2.55↓ | ||
| Alpha2 | 1 | 7.59 | 7.23 | 4.96↓ | 6.30–11.70 |
| 2 | 15.71↑ | 8.78 | 10.79 | ||
| 3 | 10.42 | 6.83 | 9.19 | ||
| Beta | 1 | 16.04↑ | 12.58 | 12.81 | 7.80–14.00 |
| 2 | 18.26↑ | 14.83↑ | 16.44↑ | ||
| 3 | 17.37↑ | 16.27↑ | 17.06↑ | ||
| Gamma | 1 | 14.27 | 26.87↑ | 16.72 | 11.10–20.40 |
| 2 | 18.21 | 22.96 | 26.16↑ | ||
| 3 | 16.73 | 26.90↑ | 21.49↑ | ||
| Ratio | 1 | 1.42 | 0.88 | 1.68 | |
| 2 | 0.75 | 1.04 | 0.73 | ||
| 3 | 0.98 | 0.84 | 0.97 |
Differentially expressed genes
There were 731 differentially expressed genes in the exposed persons. Among these genes, 285 were up-regulated and 446 were down-regulated. The raw data had been accepted by the GEO database of NCBI (http://www.ncbi.nlm.nih.gov/geo/ Access number GSE34131). Tables 5–7 show some of the up-regulated, down-regulated and immunological function-associated genes, along with protein-associated information including subcellular localization, biological processes, molecular function and/or association with diseases (see Supplementary Materials). The absolute log2 ratio of the gene lists was cut off at 2.0.
Table 5.
Partial list of upregulated genes
| Accession number | Gene | Protein | Log2Ratio |
|---|---|---|---|
| NM_018702 | ADARB2 | adenosine deaminase B2 | 6 |
| NM_001925 | DEFA4 | defensin, alpha 4 | 3 |
| NM_006671 | SLC1A7 | solute carrier family 1 (glutamate transporter), member 7 | 3.5 |
| NM_004171 | SLC1A2 | solute carrier family 1, member 2 | 2.7 |
| NM_000853 | GSTT1 | glutathione S-transferase theta 1 | 4.7 |
| NM_002424 | MMP8 | matrix metallopeptidase 8 | 3.9 |
| NM_005143 | HP | haptoglobin | 4.3 |
| NM_020995 | HPR | haptoglobin-related protein | 4.0 |
| NM_005980 | S100P | S100 calcium binding protein P | 3.7 |
| NM_001972 | ELANE | elastase | 2.5 |
| NM_000494 | COL17A1 | collagen, type XVII, alpha 1 | 3.0 |
| NM_004496 | FOXA1 | forkhead box A1 | 3.9 |
| NM_015714 | GOS2 | G0/G1switch 2 | 3.7 |
| NM_004430 | EGR3 | early growth response 3 | 3.0 |
| NM_000963 | PTGS2 | inducible cyclooxygenase | 2.5 |
| NM_001549 | IFIT3 | interferon-induced protein with tetra-tricopeptide repeats 3 | 2.0 |
RT-PCR analysis
The differentially expressed genes CEACAM8, OSM and KDR were validated by real time RT-PCR analysis. The expression analysis showed an up-regulation of CEACAM8 and OSM, and a down-regulation of KDR, relative to control levels (Supplementary Table 8). Thus the expressions of the three genes were consistent with that of whole genome gene expression CHIP assay.
DISCUSSION
There are few validated biomarkers for the prediction or early diagnosis of delayed health effects caused by exposure to ionizing radiation. The knowledge of characteristic alterations occurring at the level of the tissue, cell and molecule after radiation will help identify the pathological processes involved in delayed harmful effects. Radiation biomarkers are indicative components that play important roles in the signaling pathways underlying the pathological processes. There are few reports in the literature on radiation biomarkers in accidentally exposed persons. The goals for identifying radiation biomarkers are: (i) to characterize the mechanisms underlying delayed radiation damage; (ii) early diagnosis or prediction of delayed effects; (iii) assessment of the risk of secondary health effects in cancer patients treated with radiotherapy. Generally, radiation biomarkers are categorized by application into indicators for diagnosis, dose estimation, sensitivity and risk assessment.
In the present study we followed biomarkers for three years in three accidentally exposed persons. Our results showed that the percentage of CD4+ T lymphocytes remained lower and the ratio of CD4+ T lymphocytes to CD8+ T lymphocytes decreased compared with the reference levels. The level of albumin (known to be involved in the non-specific immune response) decreased, whereas the level of beta globulin increased. These changes were persistent for the three years after the accident. Further, gene expression profiles in the leukocytes indicated that the differentially expressed genes were involved in the immune response, inflammation, oncogenesis, cell structure, oxidative stress, neuro-hormone regulation, reproduction, susceptibility to psychiatric diseases, and regulation of gene expression, which were summarized by reviewing the Geneontology, PubmedOMIM and Pubmedgene databases and related original research papers. Down-regulated genes predominated among the differentially expressed genes.
Epidemiological analysis of the immune status in the ‘liquidators’ of the Chernobyl accident showed that the percentage of CD4+ T lymphocytes decreased, and the ratio of CD4+ to CD8+ T lymphocytes decreased; the absolute number of CD8+ lymphocytes increased over one to three years before a cancer-confirming diagnosis [8]. Investigation of the immune status and inflammatory response in the atomic bomb survivors showed that the immunity of the T cells was compromised, including an impaired mitogen-stimulated proliferation, a decrease in the quantity of naïve T cells, an increase in the function-weak memory CD4+ T cells, and an increase in CD25 + /CD127– Reg T (Regulatory T) cells in a dose-dependent way [9]. The roles of CD4+ and CD8+ T cells, B-lymphocytes and natural killer cells in the prediction of radiation-induced delayed toxicity in cervical cancer have been reported [10]. That research data, combined with our results showing alterations in subgroups of lymphocytes, serum proteins and gene expression in the accidentally exposed persons, suggests that persistent immunological change is one of the main processes after a high dose of radiation, and it may also be an important focus for revealing the mechanisms underlying the delayed effects of ionizing radiation. Moreover, the decrease in CD4+ T cells and the decrease in the ratio of CD4+ to CD8+ T cells may be biomarkers for prediction of delayed damage from ionizing radiation.
Our results from the agarose gel electrophoresis analysis showed that beta globulin increased. Lactotransferrin (LTF) (Table 7) and haptoglobin (HP) (Table 5), two components of the beta globulin band area, are 2.5 and 4.0 times higher in mRNA levels relative to the control levels. This suggests that the increase in LTF and HP production may contribute to the increase in protein production in the beta band area, indicative of activation of the non-specific immune response and inflammatory response.
Table 7.
Partial list of immunological function-associated genes
| Accession number | Gene | Protein | Log2Ratio |
|---|---|---|---|
| NM_000588 | IL3 | interleukin 3 | –3.7 ↓ |
| BC040272 | IL16 | interleukin 16 | –2.9 ↓ |
| NM_002253 | KDR | kinase insert domain receptor | –3.5 ↓ |
| NM_002175 | IFNA21 | interferon, alpha 21 | –2.5 ↓ |
| NM_030788 | TM7SF4 | transmembrane 7 superfamily member 4 | +2.5 ↑ |
| NM_020530 | OSM | oncostatin M | +2.6 ↑ |
| NM_001816 | CEACAM8 | carcinoembryonic antigen-related cell adhesion molecule 8 | +2.5 ↑ |
| NM_002343 | LTF | lactotransferrin | +2.5 ↑ |
| NM_000547 | TPO | thyroid peroxidase | +2.8 ↑ |
| NM_080657 | RSAD2 | radical S-adenosyl methionine domain containing 2 | +2.6 ↑ |
Except for genes EGR3, IL3, IL16, KDR, GSTT1, ADARB2, and PTGS2 that have been reported as radiation-associated [16–21], most of the genes shown in Tables 5–7 are shown for the first time to be involved in radiation-induced responses.
IL3, IL16 and KDR are involved in the regulation of the proliferation, growth and differentiation of hematopoietic cells, T cells and endothelial cells. The down-regulation of those genes, as shown in Table 7, suggests a cell-growth inhibitory effect has been induced by radiation. Similarly, the down-regulation of NRG2 and FOSL1 (shown in Table 6) reflects the inhibitory effect on cell growth. NRG2 induces growth and differentiation of epithelial, neuronal and glial cells [22]. FOSL is heterodimerized with proteins of the JUN family forming transcription factor complex AP-1, and is implicated as a regulator of cell proliferation, differentiation and transformation [23]. The up-regulation of EGR3 (shown in Table 5) may present negative or positive regulation of cell growth and differentiation [17].
Table 6.
Partial list of down-regulated genes
| Accession number | Gene | Protein | Log2Ratio |
|---|---|---|---|
| NM_198129 | LAMA3 | laminin, alpha 3 | –4.3 |
| NM_001464 | ADAM2 | ADAM metallopeptidase domain 2 | –4.7 |
| NM_001079858 | GPR64 | G protein-coupled receptor 64 | –4.6 |
| NM_194250 | ZNF804A | zinc finger protein 804A | –4.1 |
| NM_145740 | GSTA1 | glutathione S-transferase alpha 1 | –4.0 |
| NM_016931 | NOX4 | NADPH oxidase 4 | –4.3 |
| NM_001137669 | RGSL1 | regulator of G-protein signaling like 1 | –4.1 |
| NM_005438 | FOSL1 | FOS-like antigen 1 | –3.4 |
| NM_020868 | DPP10 | dipeptidyl peptidase 10 | –3.9 |
| NM_000817 | GAD1 | glutamate decarboxylase 1 | –3.4 |
| NM_153699 | GSTA5 | glutathione S-transferase alpha 5 | –3.9 |
| NM_004883 | NRG2 | neuregulin2 | –3.4 |
| NM_003551 | NME5 | non-metastatic cells 5 | –3.2 |
| NM_001836 | CMA1 | chymase 1 | –3.4 |
| NM_001040105 | MUC17 | mucin17 | –3.4 |
| NM_080801 | COL13A1 | collagen, type XIII, alpha 1 | –3.0 |
| NM_002538 | OCLN | occludin | –3.4 |
CEACAM8 is known as the activation marker of human granulocytes and regulates adhesion and activation of human eosinophils [24]. TM7SF4, preferentially expressed by dendritic cells, presents as an antigen to naïve T cells [25]. OSM and OSMR function to regulate cytokine production (including IL-6, G-CSF and GM-CSF) by endothelial cells, and are also known as inflammatory markers [26]. Thus, the up-regulation of those genes in our results suggests activation of granulocytes, T cells and immune responses in the accidentally exposed persons.
The up-regulation of HP, HPR, PTGS2, IFIT3, OSM, OSMR and MMP8, which is associated with inflammation, reflects the occurrence of the inflammatory response in the accidentally exposed persons.
The alterations to the glutathione metabolic pathway are intriguing. The function of the glutathione-s-transferase family is to catalyze the conjugation of reduced glutathiones and a variety of electrophiles, such as mutagens and carcinogens, thereby playing roles in detoxification. The changes in expression levels of these family members in the accidentally exposed persons might be attributed to effects resulting in perturbations in oxidative metabolism. Such effects may lead to excess generation of reactive oxygen species, including free radicals. The latter effect may be associated with altered expression or activity of antioxidants. Among the family members, GSTT1 was up-regulated whereas GSTA1 and GSTA5 were down-regulated. One study showed that deletion of GSTT1 increased the risk of senile cataracts in Asian people [27]. The up-regulation of GSTT1 in the accidentally exposed persons might be accounted for as a functional compensatory increase to protect the body from forming cataracts, and this is supported by the occurrences of opacity of the lens in the three accidentally exposed persons at different times after the accident. Glutamate transporters of SLC1A7 and SLC1A2 were up-regulated, and they might serve to clear the excitatory neurotransmitter glutamate from the extracellular space of synapses in the central nervous system [28]. GAD1 was down-regulated in our results, and according to the literature [29, 30], down-regulation of this gene has been seen in Parkinson's disease and schizophrenia.
The up-regulation of ADARB2, a RNA editing enzyme, and FOXA1, a transcriptional activator for liver-specific transcripts such as albumin and transthyretin, suggests enhancement of the expression of genes that regulate a multitude of responses, which when perturbed leads to tumorigenesis.
CEACAM8 was up-regulated; its high expression has been found in acute lymphoblastic leukemias [31]. MUC17 was down-regulated; its low expression has been found in inflammation and neoplastic disease in the digestive tract [32]. RGSL1 was down-regulated, and its function is to increase the GTPase activity of the Gαsubunit to attenuate signaling from the G-protein-coupled receptor, located in 1q25.3, the prostate cancer region (HPC1) associated with breast cancer and prostate cancer [33]. S100P was up-regulated; its high expression has been reported to be associated with prostate cancer progression [34]. A high level of expression has also been found in pancreatic ductal adenocarcinoma and S100P may be a marker for early diagnosis [35]. OCLN was down-regulated; it encodes an integral membrane protein required for the cytokine-induced regulation of the tight junction paracellular permeability barrier, and one report has shown that loss of OCLN leads to the progression of human breast cancer [36].
GPR64, NME5 and ADAM2 were down-regulated. GPR64 is an epididymis-specific transmembrane protein, NME5 exhibits a nucleoside diphosphate kinase activity [37] and is involved in spermiogenesis and flagellar movement [38], and ADAM2 is a subunit of an integral sperm membrane glycoprotein called fertilin that plays an important role in sperm–egg interactions. The down-regulation of those three genes was shown to coincide with the occurrences of poor sperm motility, low sperm count, or semen with no sperm, as shown by the clinical examinations of the accidentally exposed persons.
DEFA4 was up-regulated in our results. It is found in the granules of neutrophils and has corticostatic activity, inhibiting corticotrophin-stimulated corticosterone production. It functions via the renin-angiotensin system pathway. CMA1 was down-regulated in our results. It is expressed in mast cells, and is largely responsible for converting angiotensin to the vasoactive peptide angiotensin. Angiotensin II has been implicated in blood pressure control, and a perturbation of its yard is associated with the pathogenesis of hypertension, cardiac hypertrophy, and heart failure. The changes in expression level of these two genes suggest a response of the renin-antiotensin system to ionizing radiation in the accidentally exposed persons. SLCA2 was up-regulated and its encoded protein functions to clear the excitatory neurotransmitter glutamate from the extracellular space at synapses in the central nervous system. Glutamate clearance is necessary for proper synaptic activation and to prevent neuronal damage from excessive activation of glutamate receptors. Decreased expression of this protein or loss of its function is associated with amyotrophic lateral sclerosis, Parkinson's disease, Alzheimer's disease, autism spectrum disorders and psychiatric disorders [28]. Taken together, those data suggest the disturbance of the neuro-hormone regulation system and a susceptibility to neuro-psychiatric disorders in the accidentally exposed persons.
LAMA3 was down-regulated in our results. Its encoded protein is a component of basement membrane, involved in cell adhesion, signal transduction and differentiation of keratinocytes. COL17A1 was up-regulated. It encodes a transmembrane protein, a structural component of hemidesmosomes, multiprotein complexes at the dermal–epidermal basement membrane zone that mediate adhesion of keratinocytes to the underlying membrane. Mutations in LAMA3 and COL17A1 have been associated with junctional epidermolysis bullosa [39, 40]. Skin disorders manifesting pigmentation in the face, desquamation and itching in the skin of the arms and lower legs, thickening of the fingernails, or psoriasis that appeared consecutively after the exposure accident might be related the alterations of those two genes.
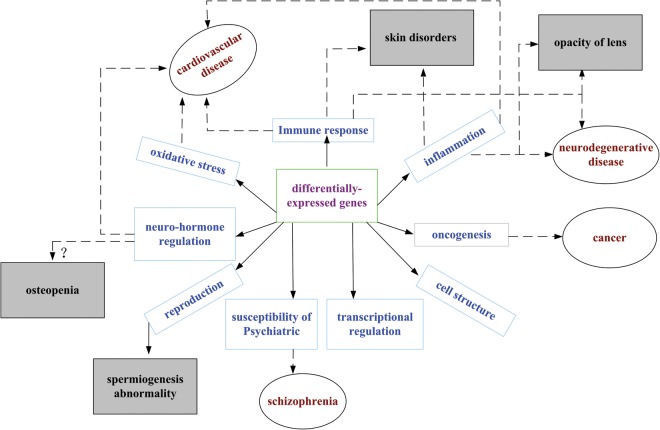
We have discussed the immune response, the inflammatory response, oxidative stress, tumorigenesis, spermiogenesis abnormality, skin disorders and disturbance of neuro-hormone regulation axes in the accidentally exposed persons in the above paragraphs. Additionally, our results suggest that CD4+ T cells, and the ratio of CD4+ to CD8+ T cells may be biomarkers for radiation-caused delayed damage. Collectively, we propose the following candidate genes as biomarkers for delayed damage: (i) inflammation and immunity associated genes: IL3, IL6, HP, HPR, PTGS2, IFIT3, ELANE, OSM, OSMR and MMP8; (ii) oncogenesis associated genes: CEACAM8, S100P, MUC17, RGSL1 and OCLN; (iii) the glutathione-s-transferase family and the metabolism of glutamate associated genes: GSST1, GSTA1, GSTA5, SLC1A2, SLC1A7 and GAD1; (iv) spermatogenesis associated genes: GPR64, NME5 and ADAM2; (v) skin disease associated genes: LAMA3 and COL17A1; (vi) neuro-hormone regulation and susceptibility of psychiatric disorders associated genes: DEFA4, CMA1, GAD1 and SLCA2. The alterations of expression of the above genes eventually lead to the onset of delayed injuries. Therefore, those genes are candidates for the prediction of delayed injuries. The relationship between differentially expressed genes, related pathological processes, and delayed effects (diseases) is outlined in Fig. 2.
Fig. 2.
Differentially expressed genes, related pathological processes and delayed effects of ionizing radiation. The diseases (late effects) in the shaded rectangles indicate the diseases already occurring in the exposed individuals, while the diseases noted in black ellipses are as yet non-occurring. Solid arrows show the results of gene expressions, and dotted arrows connect the pathological processes indicated by the results of gene expressions to the delayed effects (diseases).
One issue we should note is that two of the three accidentally exposed persons were given bone marrow transplantation, the other one was administered with G-CSF. The influence of those treatments on the delayed effects is unknown. It is clear, however, that those appropriate treatments improve survival in LD50/30 exposed persons. Moreover, it should be recognized that the gene expression chip assay was not repeated for confirmatory purposes and thus generation of an indicator of statistical significance has not been provided.
The candidate genes noted above are members of signaling pathways in processes responding to a high dose of gamma rays. Their biological functions and association with diseases will become clearer with increasing time. This will ultimately allow characterization of biomarkers that will permit prediction or early diagnosis of delayed harmful effects of radiation to be established more accurately.
SUPPLEMENTARY DATA
Supplementary data is available at the Journal of Radiation Research online.
FUNDING
This work was financially supported by Shanxi Center for Disease Control and Prevention and China Institute for Radiation Protection(No.201106).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Mitchel, Researcher Emeritus, from Atomic Energy of Canada Limited for his helpful comments and language revision.
REFERENCES
- 1.Yamada M, Naito K, Kasagi F, et al. Prevalence of atherosclerosis in relation to atomic bomb radiation exposure: an RERF Adult Health Study. Int J Radiat Biol. 2005;81:821–6. doi: 10.1080/09553000600555504. [DOI] [PubMed] [Google Scholar]
- 2.Stewart FA, Hoving S, Russell NS. Vascular damage as an underlying mechanism of cardiac and cerebral toxicity in irradiated cancer patients. Radiat Res. 2010;174:865–9. doi: 10.1667/RR1862.1. [DOI] [PubMed] [Google Scholar]
- 3.Picano E, Vano E, Domenici L, et al. Cancer and non-cancer brain and eye effects of chronic low-dose ionizing radiation exposure. BMC Cancer. 2012;12:157. doi: 10.1186/1471-2407-12-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darby SC, Cutter DJ, Boerma M, et al. Radiation-related heart disease: current knowledge and future prospects. Int J Radiat Oncol Biol Phys. 2010;76:656–65. doi: 10.1016/j.ijrobp.2009.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromet EJ, Havenaar JM, Guey LT. A 25 year retrospective review of the psychological consequences of the Chernobyl accident. Clin Oncol. 2011;23:297–305. doi: 10.1016/j.clon.2011.01.501. [DOI] [PubMed] [Google Scholar]
- 6.Loganovsky KN, Loganovskaja TK. Schizophrenia spectrum disorders in persons exposed to ionzing radiation as a result of the Chernobyl accident. Schizophr Bull. 2000;26:751–73. doi: 10.1093/oxfordjournals.schbul.a033492. [DOI] [PubMed] [Google Scholar]
- 7.Yamada M, Kasagi F, Mimori Y, et al. Incidence of dementia among atomic-bomb survivors – Radiation Effects Research Foundation Adult Health Study. J Neurol Sci. 2009;281:11–4. doi: 10.1016/j.jns.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Oradovskaia IV, Pashchenkova IuG, Feoktistov VV, et al. The epidemiological analysis of monitoring of the immune status in liquidators of consequences of the Chernobyl accident for early identification of risk groups and diagnostics of oncological diseases. Report 2. Dependence of frequency and changes in the immune status on risk factors of radiation accident. Radiats Biol Radioecol. 2011;51:117–33. [PubMed] [Google Scholar]
- 9.Kusunoki Y, Yamaoka M, Kubo Y, et al. T-cell immunosenescence and inflammatory response in atomic bomb survivors. Radiat Res. 2010;174:870–6. doi: 10.1667/RR1847.1. [DOI] [PubMed] [Google Scholar]
- 10.Bordón E, Henríquez-Hernández LA, Lara PC, et al. Role of CD4 and CD8 T-lymphocytes, B-lymphocytes and Natural Killer cells in the prediction of radiation-induced late toxicity in cervical cancer patients. Int J Radiat Biol. 2011;87:424–31. doi: 10.3109/09553002.2010.537433. [DOI] [PubMed] [Google Scholar]
- 11.Fachin AL, Mello SS, Sandrin-Garcia P, et al. Gene expression profiles in radiation workers occupationally exposed to ionizing radiation. J Radiat Res. 2009;50:61–71. doi: 10.1269/jrr.08034. [DOI] [PubMed] [Google Scholar]
- 12.Daher IN, Daigle TR, Bhatia N, et al. The prevention of cardiovascular disease in cancer survivors. Tex Heart Inst J. 2012;39:190–8. [PMC free article] [PubMed] [Google Scholar]
- 13.Chi C, Hayashi D, Nemoto M, et al. Vitamin E-deficiency did not exacerbate partial skin reactions in mice locally irradiated with X-rays. J Radiat Res. 2011;52:32–8. doi: 10.1269/jrr.10042. [DOI] [PubMed] [Google Scholar]
- 14.Chi C, Ozawa T, Anzai K. In vivo nitric oxide production and iNOS expression in X-ray irradiated mouse skin. Biol Pharm Bull. 2006;29:348–53. doi: 10.1248/bpb.29.348. [DOI] [PubMed] [Google Scholar]
- 15.Chi C, Tanaka R, Okuda Y, et al. Quantitative measurement of oxidative stress in mouse skin induced by X-ray irradiation. Chem Pharm Bull (Tokyo) 2005;53:1411–5. doi: 10.1248/cpb.53.1411. [DOI] [PubMed] [Google Scholar]
- 16.Katoh O, Tauchi H, Kawaishi K, et al. Expression of the vascular endothelial growth factor (VEGF) receptor gene, KDR, in hematopoietic cells and inhibitory effect of VEGF on apoptotic cell death caused by ionizing radiation. Cancer Res. 1995;55:5687–92. [PubMed] [Google Scholar]
- 17.Vares G, Uehara Y, Ono T, et al. Transcription factor-recognition sequences potentially involved in modulation of gene expression after exposure to low-dose-rate γ-rays in the mouse liver. J Radiat Res. 2011;52:249–56. doi: 10.1269/jrr.10110. [DOI] [PubMed] [Google Scholar]
- 18.Pawlik A, Delmar P, Bosse S, et al. Changes in transcriptome after in vivo exposure to ionising radiation reveal a highly specialised liver response. Int J Radiat Biol. 2009;85:656–71. doi: 10.1080/09553000903020024. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Guo C, Zhang H, et al. Synergistic protecting effect of cord blood CD34+ cells over-expressing both interleukin-3 and Flt3 ligand on lethally irradiated mice. Int J Hematol. 2009;90:64–73. doi: 10.1007/s12185-009-0348-8. [DOI] [PubMed] [Google Scholar]
- 20.Niu N, Qin Y, Fridley BL, et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 2010;20:1482–92. doi: 10.1101/gr.107672.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang HJ, Youn H, Seong KM, et al. Psoralidin, a dual inhibitor of COX-2 and 5-LOX, regulates ionizing radiation (IR)-induced pulmonary inflammation. Biochem Pharmacol. 2011;82:524–34. doi: 10.1016/j.bcp.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 22.Carraway KL, Weber JL, Unger MJ, et al. Neuregulin-2, a new ligand of ErbB3/ErbB4-receptor tyrosine kinases. Nature. 1997;387:512–6. doi: 10.1038/387512a0. [DOI] [PubMed] [Google Scholar]
- 23.Teutschbein J, Haydn JM, Samans B, et al. Gene expression analysis after receptor tyrosine kinase activation reveals new potential melanoma proteins. BMC Cancer. 2010;10:386. doi: 10.1186/1471-2407-10-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon J, Terada A, Kita H. CD66b regulates adhesion and activation of human eosinophils. J Immunol. 2007;179:8454–62. doi: 10.4049/jimmunol.179.12.8454. [DOI] [PubMed] [Google Scholar]
- 25.Eleveld-Trancikova D, Sanecka A, van Hout-Kuijer MA, et al. DC-STAMP interacts with ER-resident transcription factor LUMAN which becomes activated during DC maturation. Mol Immunol. 2010;47:1963–73. doi: 10.1016/j.molimm.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Fossey SL, Bear MD, Kisseberth WC, et al. Oncostatin M promotes STAT3 activation, VEGF production, and invasion in osteosarcoma cell lines. BMC Cancer. 2011;11:125. doi: 10.1186/1471-2407-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L, Xi B, Yu L, et al. Association of glutathione S-transferases polymorphisms (GSTM1 and GSTT1) with senile cataract: a meta-analysis. Invest Ophthalmol Vis Sci. 2010;51:6381–6. doi: 10.1167/iovs.10-5815. [DOI] [PubMed] [Google Scholar]
- 28.Kim K, Lee SG, Kegelman TP, et al. Role of excitatory amino acid transporter-2 (EAAT2) and glutamate in neurodegeneration: opportunities for developing novel therapeutics. J Cell Physiol. 2011;226:2484–93. doi: 10.1002/jcp.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanoue AC, Dumitriu A, Myers RH, et al. Decreased glutamic acid decarboxylase mRNA expression in prefrontal cortex in Parkinson's disease. Exp Neurol. 2010;226:207–17. doi: 10.1016/j.expneurol.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson Ray M, Weickert CS, Wyatt E, et al. Decreased BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders. J Psychiatry Neurosci. 2011;36:195–203. doi: 10.1503/jpn.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lasa A, Serrano E, Carricondo M, et al. High expression of CEACAM6 and CEACAM8 mRNA in acute lymphoblastic leukemias. Ann Hematol. 2008;87:205–11. doi: 10.1007/s00277-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 32.Senapati S, Ho SB, Sharma P, et al. Expression of intestinal MUC17 membrane-bound mucin in inflammatory and neoplastic diseases of the colon. J Clin Pathol. 2010;63:702–7. doi: 10.1136/jcp.2010.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiechec E, Overgaard J, Hansen LL. A fragile site within the HPC1 region at 1q25.3 affecting RGS16, RGSL1, and RGSL2 in human breast carcinomas. Genes Chromosomes Cancer. 2008;47:766–80. doi: 10.1002/gcc.20578. [DOI] [PubMed] [Google Scholar]
- 34.Basu GD, Azorsa DO, Kiefer JA, et al. Functional evidence implicating S100P in prostate cancer progression. Int J Cancer. 2008;123:330–9. doi: 10.1002/ijc.23447. [DOI] [PubMed] [Google Scholar]
- 35.Levy M, Lin F, Xu H, et al. S100P, von Hippel-Lindau gene product, and IMP3 serve as a useful immunohistochemical panel in the diagnosis of adenocarcinoma on endoscopic bile duct biopsy. Hum Pathol. 2010;41:1210–9. doi: 10.1016/j.humpath.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Martin TA, Mansel RE, Jiang WG. Loss of occludin leads to the progression of human breast cancer. Int J Mol Med. 2010;26:723–34. doi: 10.3892/ijmm_00000519. [DOI] [PubMed] [Google Scholar]
- 37.Munier A, Serres C, Kann ML, et al. Nm23/NDP kinases in human male germ cells: role in spermiogenesis and sperm motility? Exp Cell Res. 2003;289:295–306. doi: 10.1016/s0014-4827(03)00268-4. [DOI] [PubMed] [Google Scholar]
- 38.Depa-Martynów M, Kempisty B, Lianeri M, et al. Association between fertilin beta, protamines 1 and 2 and spermatid-specific linker histone H1-like protein mRNA levels, fertilization ability of human spermatozoa, and quality of preimplantation embryos. Folia Histochem Cytobiol. 2007 45 Suppl 1:S79–85. [PubMed] [Google Scholar]
- 39.Wu Y, Li G, Zhu X. A novel homozygous point mutation in the COL17A1 gene in a Chinese family with generalized atrophic benign epidermolysis bullosa. J Dermatol Sci. 2002;28:181–6. doi: 10.1016/s0923-1811(01)00163-3. [DOI] [PubMed] [Google Scholar]
- 40.Hamill KJ, Paller AS, Jones JC. Adhesion and migration, the diverse functions of the laminin alpha3 subunit. Dermatol Clin. 2010;28:79–87. doi: 10.1016/j.det.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.