Abstract
Momordica charantia is a well known medicinal plant used in the traditional medicinal system for the treatment of various diseases including diabetes mellitus. Recently, a novel protein termed as ADMc1 from the seed extract of M. charantia has been identified and isolated showing significant antihyperglycemic activity in type 1 diabetic rats in which diabetes was induced. However, the structure of this protein has not yet been analyzed. Homology modeling approach was used to generate a high quality protein 3D structure for the amino acid sequence of the ADMc1 protein in this study. The comparative assessment of secondary structures revealed ADMc1 as an all-alpha helix protein with random coils. Tertiary structure predicted on the template structure of Napin of B. Napus (PDB ID: 1SM7) with which the ADMc1 showed significant sequence similarity, was validated using protein structure validation tools like PROCHECK, WHAT_CHECK, VERIFY3D and ProSA. Arrangement of disulfide bridges formed by cysteine residues were predicted by the Dianna 1.1 server. The presence of multiple disulfide bond confers the stable nature of the ADMc1 protein. Further, the biological activity of the ADMc1 was assessed in non-obese diabetic (NOD) mice which are spontaneous model of type 1 diabetes. Significant reduction in the blood glucose levels of NOD mice was observed up to 8 h post administration of the rADMc1 protein. Overall, the structural characterizations with antihyperglycemic activity of this seed protein of Momordica charantia demonstrate its potential as an antidiabetic agent.
Keywords: Momordica charantia, homology modeling, structure validation
Background
Diabetes represents one of the most challenging health problems in the 21st century due to the tremendous increase in the number of diabetic patients in past few years as announced by International Diabetes Federation (IDF) [1]. According to World Health Organization (WHO) the increase in the prevalence of diabetes is occurring more in the developing countries than in the developed countries [2]. India, China and USA are the top three countries with highest number of diabetic patients [2]. Diabetes is defined as a group of heterogeneous metabolic disorders with the common symptoms of hyperglycaemia and glucose intolerance, due to insulin deficiency, impaired insulin action, or both [1]. Based on the insulin dependence, diabetes has been classified into two main types i.e. Type I diabetes (T1DM) or insulin dependent diabetes mellitus (IDDM) and Type 2 diabetes or non-insulin dependent diabetes mellitus (NIDDM).
T1DM is an autoimmune disorder resulting from a complex interaction of both genetic and environmental factors. In The T1DM condition, inflammation of the pancreatic β cells occur which leads to impaired insulin secretion to an absolute deficit of insulin in the body [1]. The impaired insulin secretion results in disrupted glucose homeostasis, and if not treated properly it further cause micro- and macrovascular complications [3]. Although only 5-10% of total diabetic patients suffer from T1DM, it affects more than 5.0 million people all over the world. Moreover, there is marked rise in the incidence of T1DM in both developed and developing nations, and there is a clear indication of a shift towards T1DM occurring in children at earlier ages [4].
Approximately 60–80% of the global population relies on plantbased medicines as their primary health care system [5].
Particularly, in the area of diabetes more than 1200 plants are being used worldwide in traditional system of medicine as antihyperglycemic therapeutics with very less or no adverse side effects. The bioactive phytochemical components that exert specific biomedical effects on the human body are the reasons behind the high medicinal value of these plants [6]. Momordica charantia (Mc) or bitter gourd is one such plant that has been extensively used and reported for its antihyperglycemic effects [7–10]. It has been commonly used as a traditional remedy in Asia, Africa, England, India and Sri Lanka [8]. Many studies have been carried out reporting its anti-diabtic potential using extracts from different parts of the plant such as fruit pulp, leaves and whole plants in experimental diabetic animal models such as alloxan/ streptozotocin (STZ)-induced diabetic [8–10].
Earlier studies from our laboratory resulted in identification of an active anti-hyperglycemic proteinaceous fraction from the seed extract of Momordica charantia, significantly reducing the fasting blood glucose levels in alloxan induced type 1 diabetic rats on 24 h basis (once daily intraperitonial administration) [11]. Further fractionation of this proteinaceous fraction has led to the identification of a novel protein designated as ADMc1 [12]. The detailed structural analysis of this protein has not been done so far. Also, the antihyperglycemic activity of this protein was established by inducing diabetes in animals, and not in non-obese diabetic (NOD) mice, the spontaneous model of type 1 diabetes. The NOD mice represent one of the primary experimental models of T1DM for screening of therapeutics as these mice share many immunological and pathophysiological similarities to human insulin-dependent diabetes mellitus [13]. The present study reports comparative sequence analysis, prediction of secondary structural components, 3-dimensional structural model using in silico homology modeling and antihyperglycemic activity of the rADMc1 in non-obese diabetic (NOD) mice.
Methodology
Sequence analysis and structure prediction of ADMc1:
The amino acid sequence of the ADMc1 protein of M. charantia was used for homology modeling [12], the gene sequence submitted to GenBank with WEBIN ID Hx2000033195]. The ADMc1 genes encodes for a protein of 138 amino acid residues which after processing gives rise to a protein of 96 amino acid residues [12]. The secondary structures of full length (96 amino acid sequence) ADMc1 protein were predicted by PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) and JPRED [14]. Prediction of putative disulfide bonds was performed using the Dianna 1.1 web server (http://clavius.bc.edu/~clotelab/DiANNA/). The NCBI BlastP server was used to find structural templates for ADMc1 protein. PSI Blast was carried out against Protein Data Bank (PDB) (http://www.rcsb.org /pdb /home /home. do). Suitable templates were found for homology modeling on the basis of high score, lower e-value and maximum sequence identity. To ensure the high accuracy of the structure, sequence alignment between query and template was done using ClustalW [15]. Homology based modeling of the ADMc1 protein of Momordica charantia was performed using automated homology modeling server EsyPred 3D [16] using the template structure PDB ID: 1SM7 of the corresponding protein napin of B. Napus [17]. The generated 3D structural models were visualized by Swiss PDB Viewer (SPDBV) [18].
Structure validation:
The modeled structure predicted using EsyPred 3D was submitted to different servers for structure validation. The stereo-chemical qualities of the modeled structure and Ramachandran plots were generated using PROCHECK [19]. PROCHECK assesses the various parameters of the generated model such as bond length, bond angle, residue by residue properties, main chain properties, side chain properties, RMS distance from planarity and distorted geometry plots. The Root Mean Square Deviation (RMSD) value was computed by the superposition of the predicted model and the template protein structures to analyze the structural similarities between the two structures, and overall quality of predicted model. SPDBV server [18] was used to generate, analyze and represent the superposed structures. ProSA [20] was employed for the analysis of structural errors and making plot of residue scores and calculating Z scores. The residue profiles of the three dimensional model were further checked by VERIFY3D [21] and stereochemical parameters of the residues were evaluated by WHAT_CHECK [22].
Evaluation of blood glucose lowering ability in diabetic animals:
NOD mice, (25–30 g), were obtained from Centre for Cellular and Molecular Biology (CCMB), Hyderabad, India. The animals were kept under standard environment conditions at 12-h light:dark cycle at a temperature of 22–24°C, in the Small Animal Facility of the Jawaharlal Nehru University, New Delhi, India and given laboratory chow diet and water ad libitum. Spontaneously diabetic NOD mice of either sex showing blood glucose levels above 250 mg/dl (diabetic condition) were selected for study and grouped into two groups of 5-8 animals per group. Animals were treated with the recombinant ADMc1 (rADMc1; intraperitoneal administration, i.p.) in PBS (produced in Pichia pastoris as described earlier) [12] at a dose of 15 mg/kg body weight. The control animals were administered with equivalent volume of PBS. Before administration of the recombinant protein, food was removed from the cage and mice were fasted for ~ 6h and during the course of the experiment. Drinking water was provided to mice throughout the experiment. Fasting glucose concentrations in blood drawn from tail vein were measured at indicated time intervals using an Accu-Chek glucometer. Statistical analysis of the data to determine the significance of the effect of the treatments was carried out by Student's t-test.
The animal experiment presented in the study has the due approval of the the Institutional Animal Ethics Committee of the University, and the guidelines prescribed by the Institutional Animal Ethics Committee, JNU, New Delhi, were followed while handling animals.
Results & Discussion
Comparative sequence analysis and secondary structure of ADMc1:
BlastP analysis of the amino acid sequence of the ADMc1 with the entries in the Protein Data Bank (PDB) revealed it to share maximum similarity with the napin of Brassica napus (PDB entry 1SM7). Amino acid sequence alignment of the ADMc1 and napin of B. napus is shown in (Figure 1A). Detail analysis of amino acid sequence of the ADMc1 indeed shows it to contain the characteristic features of 2S albumin family of proteins such as presence of eight conserved cysteine residues in a common pattern of CC/CCCXCCC (Figure 1A). In the 2S albumin family of proteins these cysteine residues are involved in the formation of two disulfide linkage between the two chains of the protein derived from a single polypeptide precursor and two additional ones between cysteine residues in the large chain [23]. The common pattern of eight cysteine residues has also been reported in other small seed proteins unrelated sequentially with napins. These include nonspecific lipid transfer proteins (ns-LTP) [24] and the hydrophobic protein from soybean (HPS) [25], which represent the diversity of the 2S albumin family of seed storage proteins. Amino acid composition analysis revealed that the protein does not contain any lysine, threonine, and tyrosine. The secondary structure prediction of the ADMc1 protein showed it as an all-helix protein (~ 61% alpha helices) with loops or turn like conformations (~ 39%) (Figure 1B). Since the protein has been reported to have anti-hyperglycemic activity, its sequence was also compared with the amino acid sequence of insulin. However, no significant similarity was found between the amino acid sequence of the two (data not shown).
Figure 1.
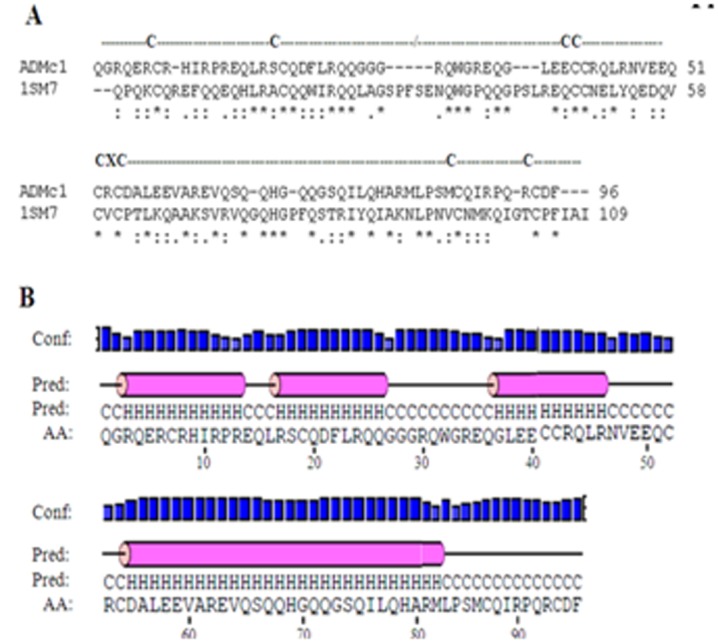
(A) CLUSTALW multiple sequence alignment between amino acid sequences of ADMc1 of M. charantia and napin of B. Napus showing common cysteine residues pattern. (*) represent single fully conserved residues whereas conservation of strong and weak groups are denoted by (:) and (.), respectively; (B) Predicted secondary structural components using ADMc1 amino acid sequence, Blue bars represent confidence of prediction (conf); pink cylinders and “H” denote alpha helix; and the solid line and “C” denote coils.
3D structure model of rADMc1 protein:
Determination of the three-dimensional structure of ADMc1 constitutes a fundamental first step in the direction of structural characterization of this protein with a diverse function from the other proteins that share sequence similarity with napins. Figure 2A shows the predicted structure in the form of ribbons as a Swiss PDB viewer representation. The predicted 3D model of ADMc1 also confirmed it as all-alpha helix linked with beta turn. The template 1SM7 is made up of two chains with disulfide bridges between the cysteines present in the two chains. Therefore, the presence of 8 cysteine residues in the sequence prompts us to identify the disulfide bond arrangements in the protein. The prediction of disulphide bonds by Diaana server showed the ADMc1 to contain four predicted disulfide bonds, between cysteines 7-52, 19-41, 42-87, 54-94. The front and side views of the predicted model showing the disulfide bonds between the two chains are shown in (Figure 2B & Figure 2C), respectively. The physicochemical analysis on the protein shown that, presence of disulfide bridges in the protein provide much more stability to the helical structure and without these bonds the helical structure unfolds into a random, disoriented strand in the solution. The presence of four disulfide bonds in ADMc1 protein makes it more stable in solution form (unpublished data).
Figure 2.
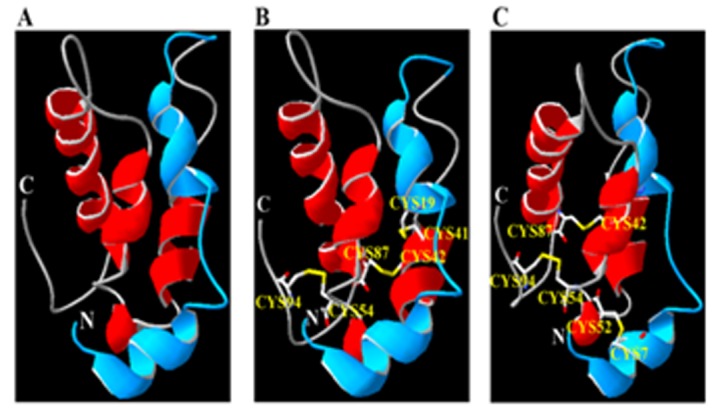
(A) The homology-modeled structure (Ribbon structure) of ADMc1 linked with two chains (Short chain; blue, long chain; red) showing N and C terminal using NMR structure of Napin of B. Napus (PDB entry 1SM7_A), as a template as visualized by Swiss PDB Viewer. (B & C) The predicted disulfide bonds between the two chains of the homology modeled structure of ADMc1. Panels B and C shows the front and side views, respectively.
Assessment of model quality:
Validation of the quality of the predicted model of the ADMc1 using PROCHECK showed the model to be good. The Ramachandran plot for the model of ADMc1 shows that the modeled ADMc1 has 81.9% residues in most favorable regions, 16.8% residues occurring in allowed regions and 1.2% residue (only one residue i.e. Glu 49) were found in the disallowed regions . The quality of the predicted model is also assessed by determining the G-factor which takes in account the observed distribution of stereochemcial parameters and is a log odds score based on the distribution. A predicted model with a score for G- factor above -0.5 is considered reliable. The observed overall G-factor score for the present model was -0.24. Also, all the planar groups were within the limits. A small root mean square deviation (RMSD) of 0.6Å, as resulted by superposition of the modeled structure of ADMc1 protein and the NMR structure of the template napin of B. Napus (PDB ID: 1SM7) showed the high structural similarities between target and template as well as good quality of generated structure model (Figure 3A). The quality of the generated model of ADMc1 was also evaluated by ProSA provided a z-score of -5.57, which falls within the range of values, observed for the experimentally determined structures of similar lengths (Figure 3B). The quality of the predicted model of ADMc1 was also verified by other structure verification servers like WHAT_CHECK and Verify-3D. Verify-3D showed 100 % of the amino acid residues had an averaged 3D-1D score > 0.2 in the modeled structure of ADMc1 which also passed the quality parameters assessed by WHAT_CHECK and overall results suggested that the generated model of ADMc1 can be used for further studies in the absence of experimentally driven structure.
Figure 3.
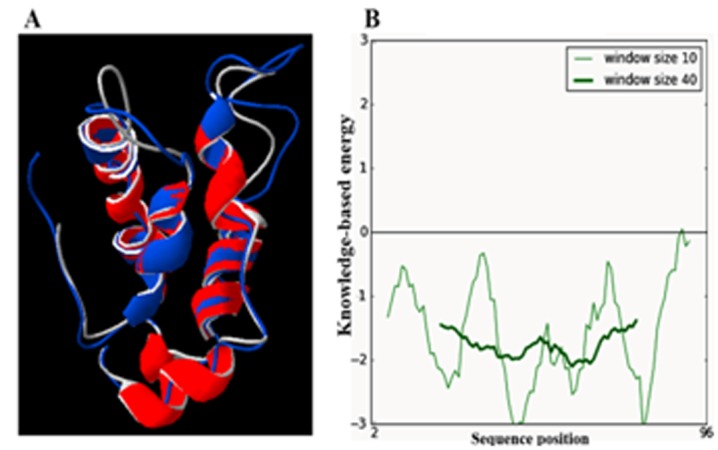
(A) Superimposition of homology modeled structure of ADMc1 of M. charantia onto a napin of B. Napus (PDB entry 1SM7). ADMc1 is shown in red and Napin of B. Napus in blue. (B) Protein Structure Analysis (ProSA) energy plot of homology model of ADMc1of M. charantia.
Blood glucose in NOD mice treated with rADMc1:
Blood glucose levels were determined prior to and after administration of a single dose of the recombinant ADMc1 at different time intervals. As evident from the (Figure 4), a significant decrease in the blood glucose levels in NOD mice treated with the recombinant ADMc1 was observed after 2 h of treatment. The blood glucose levels reduced by ~ 50% as compared with the initial levels prior to treatment. The control animals administered with PBS showed only a very slight decrease from initial blood glucose levels. It is interesting to note that the blood glucose lowering effect of the rADMc1 protein in NOD mice was persistent and up to ~ 75% reduction in blood glucose levels (from the pre-treatment levels) was observed 8 h post administration, in comparison to control mice treated with vehicle which showed only a reduction of ~5-8% at 8 h when compared to the blood glucose levels in this group prior to treatment. Unlike insulin that needs to be administered before every meal, the rADMc1 maintains the lower blood glucose level for longer periods and needs to be administered only once a day. It is important to note that ADMc1 brings down the glucose levels slowly and maintains them for longer period. Thus, unlike some other known hypoglycemic/antihyperglycemic agents, there is no risk of sudden hypoglycemia with ADMc1.
Figure 4.
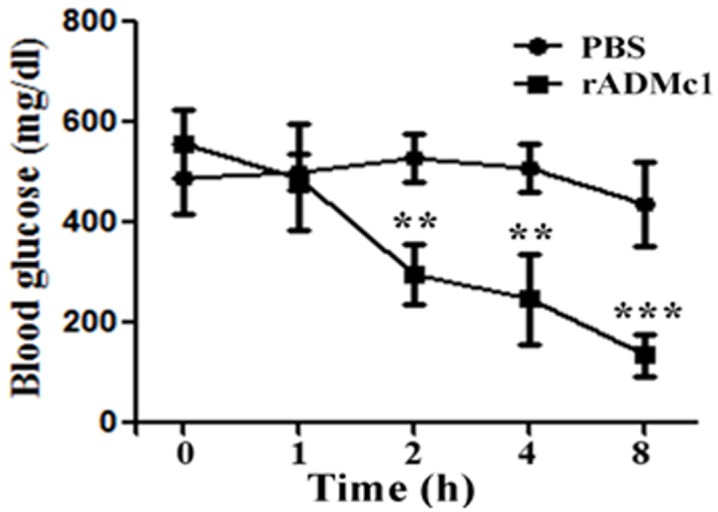
Effect of rADMc1 protein of M. charantia seed on blood glucose levels (mg/dL) in non-obese diabetic (NOD) mice in comparison to control NOD mice. Data represent mean±S.D for 5-8 mice per group. **, p<0.01; ***, p<0.001.
Conclusion
M. charantia is a well known medicinal plant used for antidiabetic treatment and isolation of various active compounds with antidiabetic activity. The amino acid sequence of the protein ADMc1 isolated from the seed extract of M. charantia was used for comparative sequence analysis and homology modeling. The results showed common disulfide linkage pattern and structural conservation with 2S albumin family proteins. The protein ADMc1 was predicted as a novel small protein of two chains consisting of four disulfide bonds. The proteins of this family primarily act as storage proteins but diverse functions have been reported for many proteins of this family. The present investigations demonstrate the blood glucose lowering ability of the ADMc1 in NOD mice.
Acknowledgments
The Department of Biotechnology, New Delhi is acknowledged for the research grant to AD. The Indian Council of Medical Research, New Delhi is acknowledged for senior research fellowship to GC.
Footnotes
Citation:Chhabra & Dixit, Bioinformation 9(15): 766-770 (2013)
References
- 1. http://www.idf.org/sites/default/files/The_Global_Burd en.pdf.
- 2. http://www.who.int/diabetes/facts/en/diabcare0504.pdf.
- 3.Bresson D, von Herrath M. Autoimmun Rev. 2007;6:315. doi: 10.1016/j.autrev.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patterson CC, et al. Lancet. 2009;373:2027. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 5. www.who.int/mediacentre/factsheets/fs134/cn/
- 6.Brusotti G, et al. J Pharm Biomed Anal. 2013;13:00117. [Google Scholar]
- 7.Grover JK, Yadav SP. J Ethnopharmacol. 2004;93:123. doi: 10.1016/j.jep.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes NP, et al. BMC Complement Altern Med. 2007;7:29. doi: 10.1186/1472-6882-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali L, et al. Planta Med. 1993;59:408. doi: 10.1055/s-2006-959720. [DOI] [PubMed] [Google Scholar]
- 10.Day C, et al. Planta Med. 1996;56:426. doi: 10.1055/s-2006-961003. [DOI] [PubMed] [Google Scholar]
- 11.Choudhary SK, et al. Evid Based Complement Alternat Med. 2012;2012:293650. doi: 10.1155/2012/293650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixit A, et al. European patent no. 2132222. [Google Scholar]
- 13.Kachapati K, et al. Methods Mol Biol. 2012;933:3. doi: 10.1007/978-1-62703-068-7_1. [DOI] [PubMed] [Google Scholar]
- 14.Cole C, et al. Nucleic Acids Res. 2008;36:W197. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson JD, et al. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert C, et al. Bioinformatics. 2002;18:1250. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 17.Pantoja-Uceda D, et al. Biochemistry. 2004;43:16036. doi: 10.1021/bi048069x. [DOI] [PubMed] [Google Scholar]
- 18. http://www.expasy.org/spdbv.
- 19.Laskowski RA, et al. J Appl Cryst. 1993;26:283. [Google Scholar]
- 20.Sippl MJ. Proteins. 1993;17:355. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 21.Eisenberg D, et al. Methods Enzymol. 1997;277:396. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 22.Vriend G. J Mol Graph. 1990;8:52. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 23.Moreno FJ, Clemente A. Open Biochem J. 2008;2:16. doi: 10.2174/1874091X00802010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kader JC, et al. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:627. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- 25.Odani S. Eur J Biochem. 1987;162:485. doi: 10.1111/j.1432-1033.1987.tb10666.x. [DOI] [PubMed] [Google Scholar]


