Abstract
Follicle stimulating hormone (FSH) is a glycoprotein secreted by gonadotrophs of the anterior pituitary gland that regulates reproduction in mammals. FSH targets its receptor (FSHR) expressed only on grannulosa cells and induce the maturation of ovarian follicles in females. The levels of both FSH and FSHR rise until the middle of estrus cycle and then falls on level at the time of ovulation. It is associated with stimulated sertoli cell proliferation in testes and supports spermatogenesis in males. The interaction between the polypeptide FSH hormone and its corresponding receptor is highly selective. Therefore, it is of interest to inhibit FSH in the context of infertility. The structure of FSH (PDB ID: 1XWD) is screened using molecular docking techniques against the ZINC database (a database of 2.7 million compounds) with reference to known standard compounds. This exercise identifies compounds with better binding and ADMET (Absorption, Digestion, Metabolism, Excretion and Toxicity) properties compared to known standard compounds. These observations find application for the consideration of such compounds for further validation towards inhibiting the FSH.
Keywords: FSH, Virtual screening, ADMET Prediction, Antifertility, 1XWD
Background
Follicle stimulating hormone (FSH) found in humans and other animals as crucial molecule in regularion of reproductive organs and physical characteristics of the gender [1]. It is synthesized and secreted by gonadotrops of the anterior pituitary gland which regulates the growth and reproductive process of the body. Structurally, it is a glycoprotein with two polypeptides such as alpha and beta subunits having 92 and 111 amino acids respectively, which contributes to the biological function and is reliable for interaction with FSH receptor [2]. In females, FSH helps to control the menstrual cycle and the production of eggs by the ovaries. The amount of FSH varies throughout a women's menstrual cycle and is highest just before she releases an egg [3]. In men, FSH helps to control the production of sperm and its level is normally remains constant. The ability of fertilization leading to conception of a woman can be determined by measuring levels of sex hormones in both couple along with FSH [4]. Several attempts are made to build models by evaluating the binding ability of different FSH related proteins with their inhibitors. Moreover, these types of encouraging studies disclose the molecular mechnism behind the male or female infertility and stratagies would come into view for design of authentic medicine to regulate the process. In this paper, we report virtual screening studies to screen inhibitors for the selected protein to investigate the influence of molecular structure and biological activity with its receptor. Several investigations are reported on drug-receptor interactions, which states that their compatibility is always depends on robustness and domain applicability of the screened compounds (drug/ligand). However, Molecular docking approaches are commonly used in modern drug design process to understand the drug–receptor interactions. The three dimensional structure of the protein-ligand composite could be served as a considerable source of understanding the way of protein interact with one another and perform biological functions [5]. Therefore, it is worthwhile to know the comprehensive structure of protein-ligand and its complexes at atomic level and is one of the significant subjects in biological sciences. Infact, it is observed that the conformational changes occur in proteins up on binding of ligand during the docking studies using data bank proteins (PDB). Moreover, the binding sites for regulatory ligands in the counter part protien are often located at the boundary between individual polypeptide chains. This arrangement may occupy small side chain rotations to enhance interaction between ligand and receptor. Prediction, Molecular Docking and Virtual Screening based studies at molecular level have become an integral part of many modern structure-based drug discovery efforts. Therefore, our molecular docking studies on the protein–ligand interactions with the specific drugs may provide a significant insight into the binding interactions and relativeness of the drugs.
Methodology
Protein-ligand docking:
It is a process for promising and consistent scoring scheme to evaluate the protein-ligand complex in order to select the best binding conformations in which two molecules fit together in 3D space and it is a key tool in structural biology as well as in computer-aided drug design [6, 7]. The goal of ligand and protein docking is mainly to predict the major binding mode(s) of a ligand with a protein of known three-dimensional structure [8]. Schrodinger 9.3 is used for molecular docking analysis and QikProp tool is for screening of pharmacological and pharmacokinetic parameters. Receptor docking is done by Glide [Grid-Based Ligand Docking with Energetics] in Schrodinger suite [9]. Glide is an integrated platform and a systematic approach for searching conformations, orientations and positions of ligand in the receptor site using a series of hierarchical filters which improves the binding affinities by lowering the penalties
Virtual screening:
It is an in-silico tool for drug designing and widely used for lead identification in drug discovery programs [10]. Experimental efforts to carry out the biological screening of many compounds are still considerably high and therefore, computer-aided drug design approaches have become attractive alternatives. Structure hits of FSH are found from PDB [11], out of which 1XWD having resolution 2.92 Å with respective ligands is retrieved for docking studies and for searching. Filtering of chemical structures screening is done with database namely ZINC [12]. Protein preparation of 1XWD is done by using protein preparation wizard in maestro 9.3. Desolvation is done by removing crystallized free water molecules beyond 5Å which hinders the mobility of ligand and results in entropy gain by applying OPLS-2001 Force Field. Finally, optimization and free energy minimization is done after adding bond orders to Hydrogens.
ADMET prediction analysis:
QikProp module of the Schrödinger 9.3 software is used to predict pharmacokinetic properties. This is standard as being dissimilar to other 95% of the known drugs. Predicted significant ADMET properties in accordance with Lipinski's rule of five, QikProp is used to evaluate the bioavailability of the lead molecules by assessing their physicochemical properties to observe the range of the Lipinski rule for induced molecules [13]. These compounds are also evaluated for their chemical behavior through analysis of pharmacokinetic parameters required for absorption, distribution, metabolism, excretion and toxicity (ADMET). Compounds obtained after ADMET analyses are to be prepared for docking using glide xp mode. LigPrep tool is used for preparing Ligands by optimizing geometries through OPLS-2001 Force Field. Prepared Ligands are rigidly docked to receptors of 1XWD using Glide extra precision function. Initially, a set of ligand poses which are generated by torsional minima are clustered and docked as a single object. Ligands with more than 300 atoms or 50 rotatable groups are not docked. Further post dock minimization is done for the molecules having 5-10 Glide score lowest energy poses and threshold for rejecting is 0.5 Kcal/mol. Finally minimized poses are rescored by Glide scoring function and visualized and data recorded through XP-Visualizer.
Results & Discussion
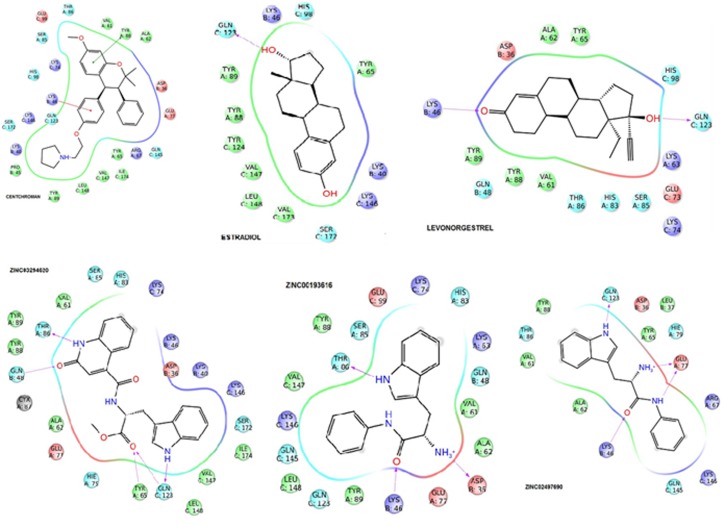
Virtual screening has become a vital part of contemporary drug research. Flexible ligand based high-throughput virtual screening (HTVS) mode of Glide is carried out and identified inhibitors against the pdb. Then, all these ligands are docked with the original protein 1XWD. The process of docking using the Receptor Grid Generation protocol with centroid at the active site of the enzyme generated grid file represented the shape and properties of receptor on a grid for more accurate scoring of ligand pose. Docked ligands with the (HTVS) mode and obtained molecules which are subjected to the Glide extra precision (XP) mode of docking performed extensive sampling and provides reasonable binding poses those interacted with the residues that bind substrate analogs in the active site. Docked poses with original three ligands resulted with the hydrogen bond and ligand interactions with amino acid residues such as serine, aspartic acid, glutamine, histidine, tyrosine respectively. Where as in zinc compounds (ZINC database of 2.7 million compounds), screened against original protein resulted that the interactions of H-Bonds formed by the Ligand with the active residues of 1XWD i.e. serine, aspartic acid, glutamine, histidine, and tyrosine with modest discrepancy with its amino acid positions (Figure 1). These amino acid positions play a vital role to determine the activity of the screened ligands when compared with the original ligand. Binding affinity glide scores [G-scores] are better than the reference i.e. standard score as in Table 1 (see supplementary).
Figure 1.
Images showing the interactions of ligands (standard and zinc database compounds) with amino acid residues.
Properties based on ADMET analysis assessed for their chemical properties of these ligands with their molecular weights are < 500 Daltons with < 5 hydrogen bond donors, < 10 hydrogen bond acceptors and QPlogPo/w < 5; these properties are well within the acceptable range of the Lipinski rule for drug-like molecules. Bioavailability of these compounds resulted in the partition coefficient (QPlogPo/w) ranges from - 2.0 to 6.5 and water solubility (QPlogS), critical for estimation of absorption and distribution of drugs within the body, ranged between -6.5 and 0.5, cell permeability (QPPCaco), a key factor governing drug metabolism and its access to biological membranes ranged from < 25 poor to >500 great. Overall, the percentage human oral absorption for the compounds tested ranged from 82% to 89%. All these pharmacokinetic parameters are within the acceptable range Table 2 (see supplementary material).
Conclusion
There is an increasing interest to inhibit FSH in the context of infertility. Hence, the structure of FSH (PDB ID: 1XWD) is screened using molecular docking techniques against the ZINC database in reference to known compounds given in (Table 1). Data depicted in Table 1 with corresponding ADMET data in (Table 2) shows compounds with better binding and ADMET properties compared to known standard compounds. These observations find application for the consideration of such compounds for further validation towards inhibiting FSH.
Supplementary material
Footnotes
Citation:Bhogireddy et al, Bioinformation 9(15): 788-791 (2013)
References
- 1.Daya S. Treat endocrinol. 2004;3:161. doi: 10.2165/00024677-200403030-00004. [DOI] [PubMed] [Google Scholar]
- 2.Biome I, Ben-Menahem D. Recent Prog Horm Res. 1999;54:271. [PubMed] [Google Scholar]
- 3.Bäckström CT, et al. Clin Endocrinol (Oxf) 1982;17:29. doi: 10.1111/j.1365-2265.1982.tb02631.x. [DOI] [PubMed] [Google Scholar]
- 4.Fan QR, Hendrickson WA. Nature. 2005;433:269. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lengauer T, Rarey M. Curr Opin Struct Biol. 1996;6:402. doi: 10.1016/s0959-440x(96)80061-3. [DOI] [PubMed] [Google Scholar]
- 6.Walters PW, et al. Drug Discovery Today. 1998;3:160. [Google Scholar]
- 7. http://www.pharmainfo.net/reviews/computer-aideddrug- design-and-bioinformatics-current tool-designing.
- 8.Orgensen WL, et al. Science. 1991;254:954. doi: 10.1126/science.1719636. [DOI] [PubMed] [Google Scholar]
- 9.Friesner RA, et al. J Med Chem. 2004;47:1739. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 10.Schapira M, et al. J Med Chem. 2003;46:3045. doi: 10.1021/jm0300173. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein FC, et al. J Mol Biol. 1977;112:535. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 12.Irwin JJ, Shoichet BK. J Chem Inf Model. 2005;45:177. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombardo F, et al. Mini Rev Med Chem. 2003;3:861. doi: 10.2174/1389557033487629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.