SUMMARY
Lysyl-tRNA synthetase (LysRS), a component of the translation apparatus, is released from the cytoplasmic multi-tRNA synthetase complex (MSC) to activate the transcription factor MITF in stimulated mast cells through undefined mechanisms. Here we show that Ser207-phosphorylation provokes a new conformer of LysRS that inactivates its translational, but activates its transcriptional function. The crystal structure of an MSC sub-complex established that LysRS is held in the MSC by binding to the N-terminus of the scaffold protein p38/AIMP2. Phosphorylation-created steric clashes at the LysRS domain interface disrupt its binding grooves for p38/AIMP2, releasing LysRS and provoking its nuclear translocation. This alteration also exposes the C-terminal domain of LysRS to bind to MITF and triggers LysRS-directed production of the second messenger Ap4A that activates MITF. Thus our results establish that a single conformational change triggered by phosphorylation leads to multiple effects driving an exclusive switch of LysRS function from translation to transcription.
Keywords: tRNA synthetase, Structure, Phosphorylation, H/D Exchange, Hydrogen/Deuterium Exchange, Mass Spectrometry, Fourier Transform, Ion Cyclotron Resonance, FTMS, Conformational Change
INTRODUCTION
Production of a specific set of proteins at any given time is critical for achieving the appropriate phenotype in response to a changing environment. This process, carried out by both the transcription and the translation machineries, is subject to robust and rapid regulation involving crosstalk between these two processes (Preker and Jensen, 2010). Aminoacyl-tRNA synthetases (aaRSs) are essential for translation, where they catalyze the attachment of amino acids to their cognate tRNAs, generating aminoacyl-tRNAs that are transferred to the ribosome for translation (Carter, 1993; Ibba and Soll, 2000). Because the cognate tRNAs harbor the anticodon triplets of the genetic code, the specific aminoacylations catalyzed by the aaRSs establish the rules of the genetic code (Woese et al., 2000). Higher eukaryotes have uniquely evolved a high molecular weight multi-tRNA synthetase complex (MSC) (Deutscher, 1984; Kellermann et al., 1982), which is regarded as a reservoir for almost half of the cellular tRNA synthetases and which controls the flow of synthetases between their canonical functions and those beyond translation (Park et al., 2005; Ray et al., 2007). In previous studies of immune activation in mast cells, we have shown that lysyl-tRNA synthetase (LysRS) exhibits an additional transcriptional function that is similarly associated with its dissociation from the MSC (Lee and Razin, 2005; Yannay-Cohen et al., 2009).
Antigen-IgE treatment of mast cells robustly induces production of allergic mediators that are controlled by microphthalmia associated transcription factor (MITF). However, activity of MITF is suppressed in quiescent mast cells, in part through its interaction with Hint-1 repressor. Transcriptional activation requires the disruption of MITF:Hint-1 interaction (Nechushtan and Razin, 2002; Razin et al., 1999). Release of Hint-1 is specifically driven by antigen-IgE induced production of a signaling molecule, diadenosine tetraphosphate (AppppA or Ap4A) and consequent binding of Hint-1 to Ap4A. Simply increasing Ap4A (either by adding Ap4A to the cell medium or by knocking down in vivo Ap4A hydrolase) releases Hint-1 from MITF and activates transcription of MITF-targeted genes, establishing Ap4A as an important second messenger for transcriptional activation (Carmi-Levy et al., 2008; Lee et al., 2004).
LysRS has a special role in MITF-dependent transcriptional control, as it is the major resource for Ap4A production during mast cell activation and is responsible for expression of MITF-inducible genes (Carmi-Levy et al., 2008; Lee et al., 2004; Yannay-Cohen et al., 2009). More than half of the aaRS members, including LysRS, AlaRS and MetRS, are known to synthesize Ap4A in vitro as a side reaction that uses a second ATP to react with aminoacyl-adenylate, the intermediate product of aminoacylation (McLennan, 1992). However, LysRS is responsible for 70–80% of cellular Ap4A production and is the sole source for the increase of Ap4A in immunologically activated cells. Knockdown of LysRS expression in mast cells significantly decreases the transcription of MITF-inducible genes (Yannay-Cohen et al., 2009).
Phosphorylation is a critical event determining the transcriptional function of LysRS. Higher eukaryotes have evolved three new proteins (MSC p43, p38, and p18, also named AIMP1, AIMP2, and AIMP3) that function as scaffold proteins to interact with nine aaRSs to form the MSC with a huge molecular weight of ~1.5 million Da. In quiescent cells, LysRS is held in the cytoplasmic MSC via its interactions with p38/AIMP2 (Kim et al., 2002; Robinson et al., 2000). In immune-activated mast cells, allergens trigger IgE-FcεRI receptor-dependent activation of MAPK cascade, which phosphorylates LysRS at Ser207 (to give LysRSpS207), and stimulates its release from the MSC (Yannay-Cohen et al., 2009). Ser207 phosphorylation is also required for Ap4A production. Mutation of LysRSS207A, which cannot be phosphorylated, significantly reduces Ap4A production when replacing the endogenous LysRS. Forced expression of the phospho-mimetic mutant LysRSS207D is sufficient to provoke increases in the concentration of intracellular Ap4A and to activate transcription of MITF target genes in mast cells (Yannay-Cohen et al., 2009). Despite these findings, the molecular mechanisms by which how LysRS performs these distinct functions and how they are regulated by phosphorylation remain totally unknown.
Here we exploit structural and functional approaches to elucidate the mechanism underlying an unprecedented switch of LysRS from translation to transcription.
RESULTS
Crystal Structure of the Human LysRS:p38/AIMP2 Complex
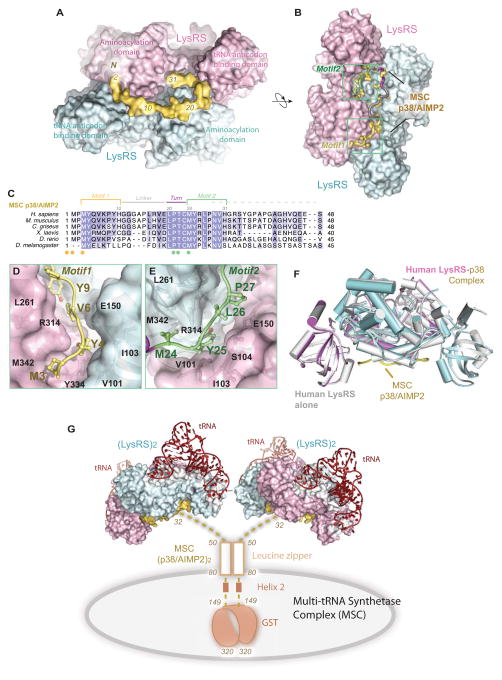
Investigations of MITF-dependent gene transcription reveal a close connection between phosphorylated LysRS and MITF activation. Upon antigen-IgE treatment, LysRS is specifically phosphorylated at Ser207 in immunologically activated mast cells, followed by a release of phosphorylated LysRS (LysRSpS207) from the cytoplasmic MSC to activate MITF in the nucleus (Yannay-Cohen et al., 2009). In quiescent mast cells, most LysRS is associated with the MSC (Halwani et al., 2004; Kyriacou and Deutscher, 2008; Yannay-Cohen et al., 2009), where it directly binds to the scaffold protein p38/AIMP2 (Kaminska et al., 2009; Quevillon et al., 1999). Although MSC is known as a critical reservoir of aaRSs, no crystal structure of the assembled MSC has been solved. The N-terminus of dimeric p38/AIMP2 binds LysRS with an affinity similar to that of full-length p38 (90 nM) (Robinson et al., 2000), and we have reconstituted a LysRS:p38 subcomplex by fusing the N-terminal 48 residues of p38 onto a carrier protein (Fang et al., 2011). To define the mechanism of LysRS in association with MSC and its release, we assembled the subcomplex of LysRS with the N-terminal 48 residues of p38/AIMP2 by co-purification in vitro and determined the crystal structure of this MSC assembly at 2.86 Å resolution (Figure 1A-B, Table S1). In the crystal structure, only the N-terminal 31 residues of p38 had electron density, indicating that p38 binds LysRS solely through its N-terminal end (1–31), and the rest of p38 was disordered in the solvent and did not bind to LysRS (Figure S1).
Figure 1. Structural basis for Reserving LysRS in the MSCdirect.
(A–B) Two orthogonal views of the human LysRS:p38/AIMP2 complex structure. Sequence of p38/AIMP2 is color-coded as in (C).
(C) Sequence alignment of the N-terminus of p38/AIMP2 from Drosophila to human. Motifs are colored as Motif 1 (yellow), Gly linker (gray), turn (red) and Motif 2 (green). Spheres represent two similar sets of residues (yellow/green) implicated in the LysRS:p38/AIMP2 interaction.
(D–E) The interface of the LysRS-p38/AIMP2 complex. The surface representation of LysRS is shown. D, Motif 1 of p38/AIMP2 and its interaction with the bottom groove of LysRS. E, Motif 2 of p38/AIMP2 and its interaction with the symmetric groove on the LysRS dimer.
(F) The functional LysRS-p38/AIMP2 complex. Side view of free human LysRS structure (pdb: 3bju, gray) superimposed onto human LysRS:p38/AIMP2 complex, showing the close similarity of LysRS structures with and without binding to p38/AIMP2.
(G) Model of the human multi-tRNA synthetase complex. Dimerization of p38/AIMP2 is through the two helical regions and a C-terminal GST-domain outside of the LysRS binding site. The N-terminal linker (residues 32–48) is disordered as found in the crystal structure. p38/AIMP2 binds to at least 8 of the 11 members of the MSC and forms the core of the MSC. This model represents the canonical functional state of LysRS in the MSC for charging tRNA.
See also Figure S1
The LysRS:p38/AIMP2 structure showed that two related motifs (motif 1 and 2) in the N-terminus of p38 bind to the LysRS dimer (Figure 1B). These two motifs are linked by a natural Gly linker and a sequence that makes a 180º turn (Figures 1B-C). Interactions between p38/AIMP2 and LysRS are mainly hydrophobic, where the p38-binding grooves are formed between the N-terminal tRNA anticodon-binding domain of one LysRS subunit and the C-terminal aminoacylation domain of the other subunit. Side chains of Met3, Tyr4, and Val6 in p38 motif 1, and of Met24, Tyr25, and Leu27 in motif 2, form direct contacts with LysRS residues V101, Y103 from the N-domain, and with R314, Y334 and M342 from the C-domain, respectively. These residues comprise two symmetric pockets in LysRS dimer (Figures 1D-E). A similar assembly of LysRS into the MSC is likely present in all higher eukaryotes, because the N-terminal 31 residues, including the two motifs and the turn sequence, are conserved in p38/AIMP2 proteins (Figure 1C).
This complex structure answers several poorly understood functional characteristics of the MSC. First, aminoacylation activities of mammalian LysRS are similar whether bound in the MSC or free of the complex (e.g., after high salt treatment) (Dang et al., 1985; Mirande et al., 1983), indicating that the architecture of the MSC is compatible with aminoacylation activities. Accordingly, the reconstituted LysRS-p38/AIMP2 sub-complexes are fully functional for aminoacylation (Fang et al., 2011). In the complex, p38 is located underneath the LysRS dimer, leaving the top of the complex available for binding and charging tRNAs (Figure 1G). In support of this idea, LysRS binding to p38/AIMP2 does not involve a conformational change, with only a 0.619 Å (root mean square deviation for a total of 6374 atoms) difference between its free versus its p38/AIMP2-bound form (Fig. 1F).
Second, among all MSC components, LysRS has the highest stoichiometry, in which four LysRS are present per MSC--a unique feature that has been observed in the purified MSC for more than 20 years (Mirande et al., 1985). Previous SAXS analysis of the human LysRS:p38/AIMP2 subcomplex showed an α2β1:β1α2 organization in which a dimeric p38 scaffold (β:β) holds two LysRS α2 dimers in a parallel configuration (Fang et al., 2011). This crystal structure demonstrates the α2β1 architecture, in which one p38 polypeptide binds to one LysRS dimer, which represents one of the two copies of the LysRS:p38/AIMP2 subcomplex in the MSC.
Finally, as the core of the MSC, p38/AIMP2 directly interacts with most of the MSC components, including LysRS, GlnRS, AspRS, GluProRS, and IleRS as well as scaffold protein p43 and p18 (Kaminska et al., 2009; Kim et al., 2002; Robinson et al., 2000; Wolfe et al., 2005). p38 depletion leads to the complete disruption of MSC (Han et al., 2006; Kim et al., 2002). Modeling the LysRS:p38/AIMP2 complex into the MSC shows the unique structural framework of MSC, in which two dimeric LysRS molecules occupy only the N-terminal end of p38/AIMP2 with the rest of the p38/AIMP2 (residues 32–320) available for interactions with other MSC components (Figure 1G). This result also suggests when even one dimeric LysRS is released for its ancillary functions the other dimer can be maintained in the MSC bound to p38 to sustain protein synthesis.
Phosphorylation of Ser207 Triggers a Structural Opening of LysRS
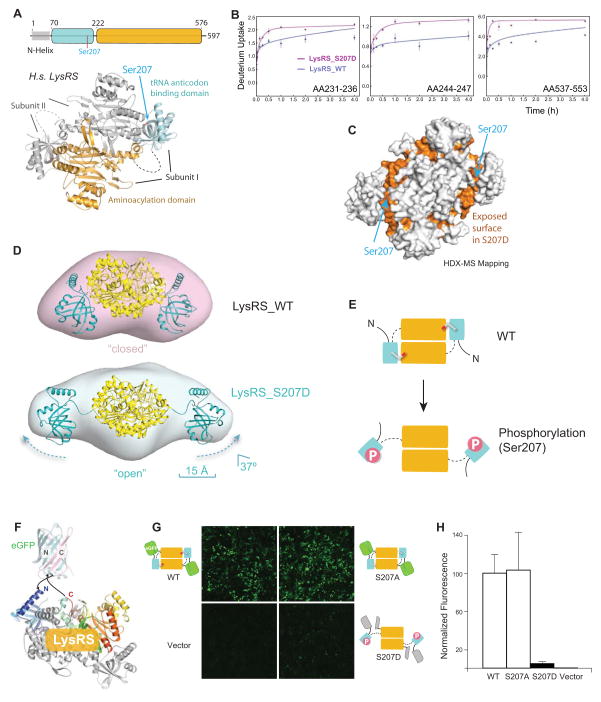
In both structures of the LysRS:p38/AIMP2 complex and of human LysRS alone (Guo et al., 2008), the dimerization of LysRS is mediated by the binding of an N-terminal domain of one subunit with the C-terminal aminoacylation domain of the other subunit (Figure 2A). Notably, Ser207 is found at this domain interface, where the addition of a phosphate group on Ser207 would provoke structural changes in the LysRS dimer. To assess the potential structural changes caused by Ser207 phosphorylation, hydrogen/deuterium exchange coupled with mass spectrometry (HDX-MS) analyses were performed. Comparison of the deuterium uptake for LysRSWT versus that for LysRSS207D revealed that four segments exhibited increased solvent accessibility (> 30%) in LysRSS207D (Figures 2B, S2). Mapping those segments onto the structure of human LysRS showed that they lie at the inter-domain interface where Ser207 is located (Figure 2C), indicating that the inter-domain interaction is disrupted in LysRSS207D.
Figure 2. Phosphorylation of Ser207 triggers an open conformation of LysRS.
(A) Schematic diagram of the structure of the human LysRS dimer.
(B–C) Deuterium uptake profiles for LysRSS207D and LysRSWT. Segments with an increased deuterium exchange rate are mapped in orange onto the 3-dimensional LysRS structure (C).
(D) Solution envelopes of LysRSWT versus LysRSS207D calculated from their small angle x-ray scattering curves. The 3-dimensional structure of LysRSS207D is modeled by docking the N-domains into its extended solution envelope.
(E) Cartoon depicting the relationship between the two conformations of LysRSWT versus LysRSS207D.
(F–H) Structural model of the eGFP fragment complementation construct that fuses the N-terminal half of eGFP (eGFPN) and C-terminal half of eGFP (eGFPC) to the ends of human LysRS for monitoring the conformational change in HEK293T cells (F). Confocal images were taken 24 h after HEK293T cells were transfected with eGFPN-LysRS-eGFPC (G). GFP fluorescence was quantified by the ratio of green fluorescence to DAPI nucleic acid stain (H).
See also Figures S2–3
To further visualize the conformational change of LysRS caused by phosphorylation of Ser207, the solution shapes of LysRSWT and LysRSS207D were determined by small angle x-ray scattering (SAXS) (Figures S3A-C). Although LysRSWT adopted the same compact conformation as in the crystal structure of native LysRS in its “closed” state, the SAXS envelope of LysRSS207D had extensions of about 15 Å at both ends (Figure S3D). These long extensions correspond to the N-terminal tRNA anticodon-binding domains being flipped out in LysRSS207D (Figures 2D-E, S3D-E). Further inspection of the structures shows that the N-terminal domain of LysRSS207D moves 15 Å away with 37x° rotation, relative to the central dimeric catalytic domain. Specifically, Ser207 in LysRSS207D moves from 3.0 Å to 20 Å away from the catalytic domain. The exposed surface in the SAXS-based structure model of LysRSS207D (vs. LysRSWT), largely overlaps with the exposed surface identified in the HDX-MS analysis (Figure S3F). This result further highlights the open conformation that is caused by the phosphorylation of Ser207. Thus, the phosphate group on Ser207 triggers radical conformational changes of LysRS that open the N-terminal domain.
To confirm that this open conformer in LysRSS207D is manifest in cells, we monitored the conformational change of LysRS by use of fragment complementation assays (Michnick et al., 2007). The eGFP protein was split into two halves and separately fused to the N- or C-terminus of LysRS, which can fold together when the N- and C-termini are located in close proximity as in human LysRSWT (Figure 2F). Transfected eGFP-LysRSWT fragments generated strong fluorescence in cells (Figure 2G). However, the phospho-mimetic mutation S207D reduced fluorescence by 95 percent (Figure 2H), indicating an open structure of LysRSS207D. This effect is specific, as mutation of S207A gave the same fluorescence intensity as that of LysRSWT. Therefore, by structural opening of its N-domains, Ser207 phosphorylation triggers an “open” conformer (LysRSpS207) not seen in any previous LysRS structure (Figure 2E).
The Open Conformer Releases LysRS from the MSC
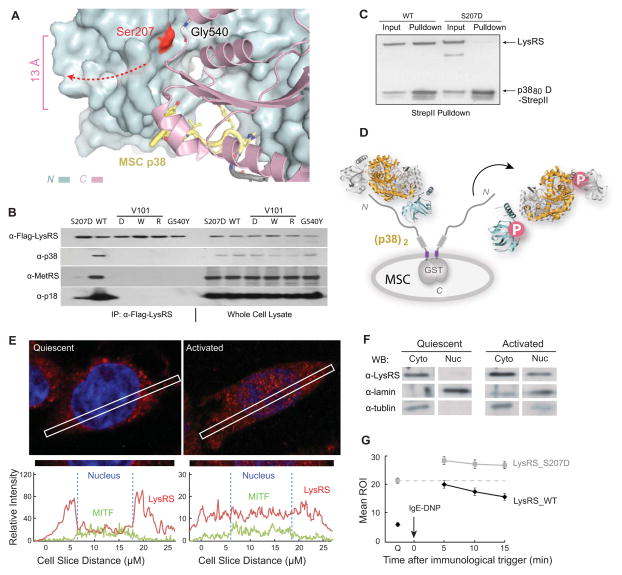
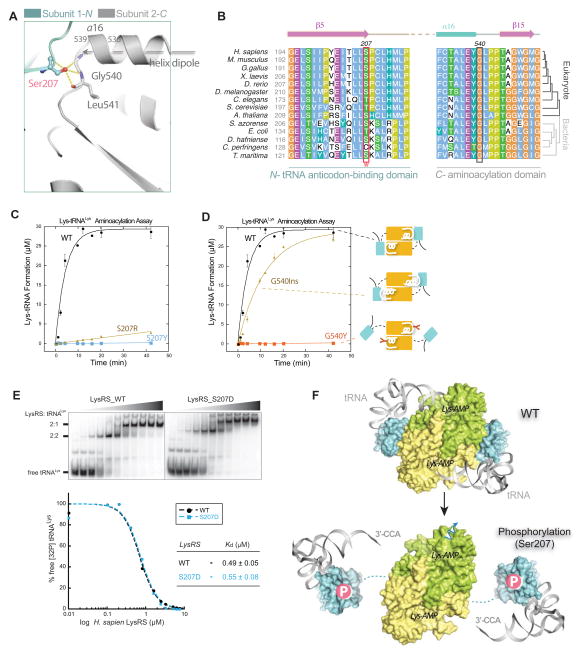
The LysRS that rapidly dissociates from the MSC following activation of mast cells is exclusively Ser207-phosphorylated (Yannay-Cohen et al., 2009). However, Ser207 is 13 Å away from the bound p38/AIMP2 in our crystal structure (Figure 3A). To understand the underlying mechanism, we first check to see if phosphorylation directly releases LysRS from the MSC, by assessing the binding of Flag-tagged LysRS to the MSC. In transfected cells, endogenous MSC bound to Flag-LysRSWT, whereas this interaction failed to occur in cells transfected with a plasmid encoding phospho-mimetic Flag-LysRSS207D (Figure 3B). In addition, expression of structure-guided LysRS mutants that have clashes within the p38/AIMP2-binding pocket (V101D/W/R, Figures 1D-E) showed that they failed to bind to any of the tested MSC component, including p38/AIMP2, p18/AIMP3, MetRS, GluProRS, and IleRS (Figure 3B). To further confirm that the release of Ser207-phosphorylated LysRS from the MSC is caused by its dissociation from p38/AIMP2, recombinant LysRS proteins were tested for their ability to bind directly to the N-terminal fragment of p38/AIMP2. Although LysRSWT bound tightly, the phospho-mimetic LysRSS207D completely lost its ability to bind to p38/AIMP2 (Figure 3C). These results indicate that Ser207-phosphorylation is the direct cause for release of LysRS from the MSC.
Figure 3. The Open Conformer Releases LysRS From MSC.
(A) View of the LysRS:p38/AIMP2 interface showing that the Ser207 phosphorylation site is at the inter-domain interface that forms the p38/AIMP2 binding groove. Ser207 is 13 Å away from the bound p38/AIMP2.
(B) Co-immunoprecipitation analyses showing that mutations of LysRS predicted to affect the p38/AIMP2 interaction (V101D, V101W, V101R), a mutation causing structural opening (G540Y) and the phospho-mimetic mutation (S207D), all dissociate LysRS from the endogenous MSC in HEK293T cells.
(C) Pull-down assay showing LysRSS207D loses the ability to bind a recombinant version of p38/AIMP2.
(D) Mechanism of Ser207 phosphorylation-directed release of LysRS from the MSC: The flipped-out anticodon-binding domain in LysRSpS207 opens the binding groove for p38/AIMP2 and prevents LysRS-p38/AIMP2 interaction.
(E) Confocal immunofluorescence microscopy showing the nuclear localization of the endogenous LysRS (red) and MITF (green) in quiescent RBL mast cells and following an IgE-DNP trigger. The blue signal is nuclear DAPI. The picture was taken by a confocal microscopy in Z-mode. The amount of LysRS and MITF signal in the cross section is quantified at the bottom.
(F) Nuclear localization of LysRS following RBL mast cell activation. Western blot analysis of LysRS in cytoplasmic versus nuclear fractions in quiescent mast cells versus in cells activated by the IgE-DNP trigger for 5 min.
(G) Confocal live imaging quantification of LysRS nuclear translocation. Means ROI (region of illuminated) of nuclear eGFP-LysRSwt versus eGFP-LysRSS207D at quiescent state (Q) and 5, 10, 15 min after immune activation. The means and standard errors of the means for 20 representative cells are shown (P<0.05).
See also Figure S4
Because the p38/AIMP2 binding pockets are formed by the inter-domain interface (Figure 3A) and p38 interacts with residues on both the N- and C- domain of LysRS (Figures 1D-E), the opening of the LysRS structure by Ser207-phosphorylation might itself disrupt the LysRS-p38 interaction. Importantly, Ser207 is nearly 20 Å away from the main dimer interface formed by the C-domain of LysRS, and phospho-mimetic LysRSS207D remains a dimer as indicated by the HDX-MS and SAXS results (Figure 2). To test if the open conformer of dimeric LysRS provoked by Ser207-phosphorylation is the basis for releasing LysRS from the MSC, the G540Y mutant was tested in the same assay. G540 is located at the opposite side of Ser207 across the inter-domain interface where G540Y will introduce a similar steric clash at Ser207 (Figures S4A-B). As predicted, LysRSG540Y did not bind to the endogenous MSC in transfected cells (Figure 3B). In addition, other mutations that either disrupts the LysRS-p38 interface (R314A) or the LysRS N-C domain-domain interface (G540R, G540R/S207D) also abolished interactions with the MSC (Figures S4C-D). Thus, through the open conformer of LysRSpS207 that disrupts the p38/AIMP2-binding pocket, Ser207-phosphorylation releases LysRS from p38/AIMP2 and the MSC (Figure 3D).
To test if the released LysRS translocates to the nucleus, RBL cells were activated by treatment with antigen-IgE for 5 min and LysRS and MITF localization was determined by confocal immunofluorescence analysis. Notably, a significant fraction of LysRS was present in the nuclei of activated mast cells and nuclear LysRS co-localized with MITF (Figure 3E and S4E). To confirm these findings, mast cell nuclei were separated from the cytosol and western blot analysis was performed with an anti-LysRS antibody. Again, the data showed that LysRS rapidly accumulated in the nucleus of activated mast cells (Figure 3F).
Finally, to test if phosphorylation of LysRS on Ser207 was also a prerequisite for its nuclear translocation, we assessed the localization of GFP-fusions of wild type LysRS (LysRSWT) or of the phospho-mimetic mutant LysRSS207D in transfected mast cells. GFP-LysRSWT accumulated in the nucleus within 5 min following immune activation, whereas GFP- LysRSS207D was detected in the nuclei even in quiescent mast cells (Figure 3G). These results demonstrate that Ser207-phosphorylation of LysRS induces an open conformer that disrupts its interaction with p38, releases it from the MSC, and directs nuclear translocation of the enzyme.
Phosphorylation of Ser207 Switches the Enzymatic Functions of LysRS
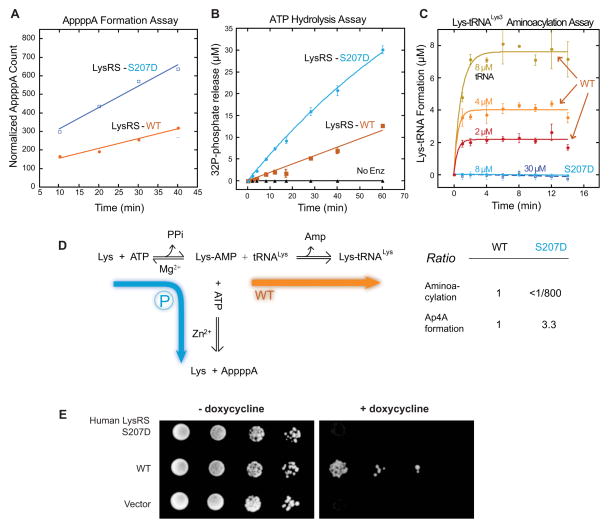
Upon antigen-IgE treatment, specific phosphorylation of LysRS at Ser207 is followed by a marked (3.5-fold) increase in the levels of Ap4A, the signaling molecule that specifically binds to the suppressor protein Hint-1 and dissociates Hint-1 from MITF (Lee et al., 2004; Yannay-Cohen et al., 2009). To test if phosphorylation of LysRS on Ser207 directs the catalysis of Ap4A, we assessed Ap4A synthesis of purified LysRSWT versus LysRSS207D in vitro. LysRS catalyzes Ap4A synthesis by conjugating the intermediate Lysyl-AMP with a second bound ATP, and then transfers the AMP moiety to form Ap4A, which can be detected by thin layer chromatography by use of α-[32P]-ATP as substrate. Notably, there was a 3-fold increase in Ap4A synthesis in LysRSS207D versus LysRSWT (Figure 4A). The formation of Ap4A involves the hydrolysis of one ATP and release of the β,γ-pyrophosphate. We therefore also measured ATP consumption by monitoring the released γ-[32P]-phosphate (Goerlich et al., 1982). Consistently, LysRSS207D also displayed a 3-fold increase in its relative activity for hydrolyzing ATP (Figure 4B). Thus, the phosphor-mimetic mutation LysRSS207D promotes its ability to hydrolyze ATP and catalyze Ap4A synthesis.
Figure 4. Functional switch of LysRS by phospho-Ser207.
(A) Phospho-mimetic LysRSS207D has enhanced Ap4A synthetic activity.
(B) Phospho-mimetic LysRSS207D has enhanced ATP hydrolysis.
(C) Phospho-mimetic LysRSS207D loses aminoacylation activity for tRNALys. Four concentrations of tRNA were used for this assay. Error bars are standard deviations (SDs) from triplicates.
(D) Phosphorylation of Ser207 redirects the enzymatic flow of LysRS from aminoacylation to Ap4A synthesis to activate transcription.
(E) Functional replacement assays in yeast show that LysRSS207D is defective in essential translational functions in cells. The expression of endogenous yeast LysRS can be switched off by the addition of doxycycline to the yeast growth medium. Ten-fold serial dilutions of freshly grown yeast cells were spotted onto selective media SCM-HIS containing 2% raffinose with or without doxycycline and galactose.
We considered that the enhanced activity of LysRSS207D that hydrolyzes ATP might also increase its second step reaction that charges tRNALys. To assess if S207 phosphorylation affects the aminoacylation efficiency of LysRS, the aminoacylation activity of LysRSWT and LysRSS207D were compared by measuring the amount of [3H]-Lysine that is covalently attached to the saturating amount of tRNALys (Figure 4C). Surprisingly, this catalytic activity of LysRSS207D was reduced more than 800-fold relative to that of LysRSWT (Figure 4C). These data indicate that the Ser207-phosphorylation of LysRS promotes the Ap4A catalysis but specifically turns off the second step reaction on charging tRNALys, thus switching its enzymatic flow from aminoacylation to Ap4A production (Figure 4D).
The dramatic decrease in aminoacylation activity of LysRSS207D suggested that the Ser207-phosphorylated LysRS loses its essential functions in translation. To test this possibility in vivo, we substituted the endogenous LysRS in yeast (Saccharomyces cerevisiae) by galactose-induced human WT and mutant LysRSS207D. Human LysRSs can efficiently aminoacylate the yeast’s tRNALys (Francin and Mirande, 2006). Wild type LysRS could substitute for the yeast cytoplasmic LysRS (which is controlled by a tetracycline-induced promoter and can be suppressed by doxycycline) and sustain normal cell growth (Figure 4E). In contrast, yeast expressing the phospho-mimetic mutant LysRSS207D was completely non-viable (Figure 4E). Thus, by an undefined mechanism, the single phosphorylation of LysRS on Ser207 sufficiently switches from its canonical function in translation to becoming a producer of Ap4A.
Mechanistic Basis of the pSer207-Directed Functional Switch of LysRS
The ability of Ser207 phosphorylation to segregate within the same active site the two consecutive steps of aminoacylation, the synthesis of aminoacyl-AMP and the following transfer of the aminoacyl moiety to tRNA, is unusual. Ser207 is located on the last β–strand in the N-terminal anticodon-binding domain of LysRS and directly faces the catalytic domain of the other subunit of the LysRS dimer (Figure 5A). Further, three hydrogen bonds are formed across the interface between the hydroxyl group of Ser207 and the backbone of Gly540 and Leu541, and Ser207 is located at the C-terminal trajectory of a helix (α-16), where a positive charge is preferred because of the dipole moment of the helix (Figure 5A). Thus, the negative charge of the phosphate group on Ser207 would generate strong electrostatic repulsion and the combined effect of the bulk and charge of the phosphate group severs the interactions of the two domains. Finally, these interactions appear conserved from fruit fly to human, and Ser207 is substituted by similar residues (Thr or Cys) in lower eukaryotes and bacteria (Figure 5B).
Figure 5. Ser207 as a tipping point for turning off translational function of LysRS.
(A) Close-up view of the domain-domain interface in the human LysRS dimer.
(B) Sequence alignment of the LysRS anticodon-binding domain (N-) and the aminoacylation domain (C-) interface. Phosphorylation site Ser207 and the opposite side G540 mutations analyzed in (C, D) are highlighted in boxes.
(C–D) Aminoacylation assays show that mutations at the dimer interface affect tRNA charging. Mutations with positive charge (S207R) are less effective at inhibiting aminoacylation activity, suggesting that the negative charge of the phosphate group on LysRS_S207 is also important for introducing repulsion between the domain interfaces.
(E) Electrophoretic mobility shift assay shows that LysRSS207D binds tRNA with an affinity similar to that of LysRSWT. Increasing concentrations of the protein (0, 0.1, 0.2, 0.4, 0.8, 1.6, 2.4, 3.2, 5.0, 6.25, and 7.5 μM, respectively) were used.
(F) Mechanism that Ser207-phosphorylation switches off the aminoacylation function of LysRS. Top, a docking model of human LysRS in complex with tRNA by superimposition of the human LysRS structure (pdb3bju) with the yeast AspRS-tRNAAsp complex structure (pdb1asy) shows that the “closed” form of wild type LysRS places the 3′-CCA end of tRNA into the catalytic site. Bottom, the N-terminal tRNA anticodon-binding domain of human LysRSp207 is modeled on the SAXS envelope of LysRSS207D..
See also Figure S5
To investigate whether or not the structural opening of LysRSpS207 abolishes its aminoacylation functions, we created various mutations at S207 or its interacting residue G540, including S207Y/E/R and G540W/Y/D/E/R. Based on the LysRS structure, these mutations will introduce steric clashes and should mimic the effect of S207D to open up the domain-domain interface (Figures S5B). Indeed, the aminoacylation activity of all of these mutants was abolished or greatly diminished (e.g., S207R) (Figures 5C-D, S5A-C). These data support the notion that a pS207-directed changes in the structure of LysRS switches off its function in translation.
The N-terminal domain of LysRS connects to its C-terminal domain through a flexible linker that is disordered in the LysRS crystal structure. To test if flexibility of the N-terminal domain is necessary to switch off the function of LysRS in translation, an elongated helix in LysRS was constructed by duplicating the last turn of the helix α-16 before G540 (aa536-539) (Figure 5A). This insertion creates an extended structure in which this N-terminal domain is shifted ~5 Å away (the approximate size of a phosphate group) with the interface still fixed (mutant G540Ins, Figure 5D). Interestingly the G540Ins mutant appears to be in an intermediate structural state, because it is about 70% as active as LysRSWT. Thus, aminoacylation functions of LysRS are not abolished by the partial structural destabilization (~5 Å) of the domain interface. These findings further support the idea that a fully open structure of LysRSpS207 is critical to the functional switch.
Lysine-specific tRNAs bind to LysRS by anchoring their anticodon stem-loops onto the N-terminal domain of LysRS (Cusack et al., 1996), where the 3′-acceptor end of the L-shaped tRNA is placed at the active site in the C-terminal domain of LysRS to allow the transfer of lysine from lysyl-AMP to tRNA (Figure 5F). tRNALys binds to the LysRSWT dimer with a Kd of 0.5 μM, where each of the subunits binds to one tRNALys (Figure 5E). The binding affinities of LysRSWT, and LysRSS207D, LysRSG540Y for tRNALys are similar (Figure 5E and S5D). However, structural modeling showed that when tRNALys anchors to the N-terminal anticodon-binding domain in LysRSS207D, the flipped anticodon-binding domain in the open form prevents tRNA from reaching the active site in the C-terminal catalytic domain of LysRS, abolishing transfer of activated lysyl-AMP to tRNA. Thus, phosphorylation of Ser207 causes a structural opening of LysRS that traps tRNA in an inactive state (Figure 5F).
The Open Conformer Drives the Ap4A production and LysRS-MITF interaction in Cells
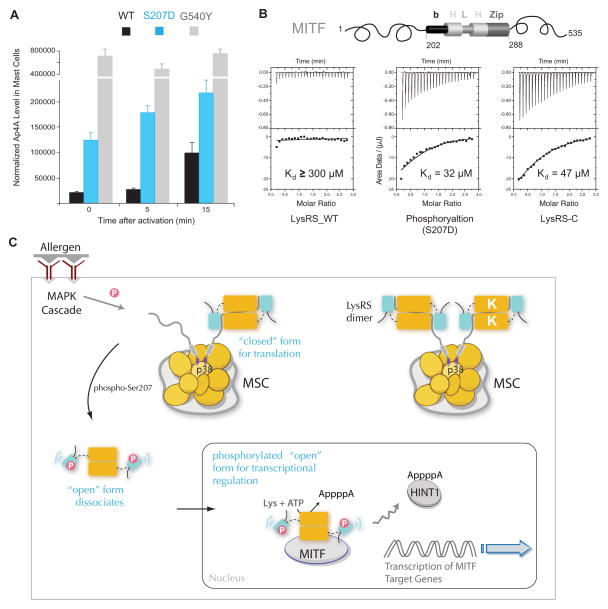
Ap4A synthesis occurs at the same active site at which Lys-AMP is transferred to tRNA. Thus, aminoacylation activity competes with Ap4A synthesis, which is blocked when free tRNA is available (Hilderman and Ortwerth, 1987). To determine whether or not the structural opening is the ultimate cause for the increased Ap4A production of LysRS in cells, the aminoacylation-dead mutant LysRSG540Y was introduced into RBL mast cells. SAXS analysis established that the G540Y mutation opened up the LysRS structure (Figures S4B). Since LysRSG540Y structure results solely from the introduced steric clash without changes at Ser207, it represents a pure open conformer. Significantly, like LysRSS207D, LysRSG540Y markedly increased Ap4A production even in quiescent mast cells (Figure 6A). In contrast, overexpression of recombinant LysRSWT augmented Ap4A production only in activated mast cells, whereas overexpression of LysRSS207D increased Ap4A levels in both quiescent and activated cells (Figure 6A). Further, expression of LysRSWT does not increase Ap4A levels when mast cells were treated with a MAPK kinase inhibitor to inhibit Ser207-phosphorylation of LysRS(Yannay-Cohen et al., 2009). Because S207D and G540Y open up the structure of the enzyme, these results indicate that the open conformer directs this new function of LysRS. Thus, by trapping tRNA in a nonfunctional state, LysRSpS207 switched off the translation function and allowed the production of Ap4A in activated mast cells (Figure S6A).
Figure 6. Structural opening directs LysRS to produce Ap4A and bind MITF in cells.
(A) In vivo Ap4A assay shows that the structural-opening mutant LysRSG540Y produced high level of Ap4A independent of mast cell activation. RBL cells were transfected with siLysRS and with plasmids expressing knockdown-resistant versions of LysRSWT, LysRSS207D or LysRSG540Y. After 24 h, cells were stimulated with the IgE-DNP trigger. The means and standard errors of the means for three experiments are shown. P<0.05.
(B) Isothermal titration calorimetry assays for binding of the MITF bHLH-Zip domain to LysRSWT, LysRSS207D or the C-terminal aminoacylation domain of LysRS.
(C) Model for the phosphorylation-dependent translation and transcription switch of LysRS driven by structural opening. In quiescent mast cells, LysRS is associated with p38 in a closed form and is retained in the cytoplasmic MSC. Antigen activation phosphorylates Ser207 and triggers an open form of LysRS. By opening up the structure, phosphorylated LysRS is released from the MSC, translocates from cytoplasm to the nucleus, binds to MITF and generates Ap4A to activate MITF transcription functions. Thus, by selecting two distinct conformers, phosphorylation could switch the functional of LysRS between translation and transcription.
See also Figure S6
Finally, interaction of LysRS and MITF has been previously shown in immunologically activated mast cells. To determine if phosphorylation of LysRS contributes to its association with MITF, endogenous LysRS and MITF were assayed by co-immunoprecipitation before and after the antigen-IgE trigger. Interaction of LysRS with MITF was observed only in activated mast cells (Fig. S6B), suggesting that it is the MSC-released Ser207-posphorylated LysRS that binds to MITF and that the Ser207-phosphorylation may be necessary for this interaction. The domain of MITF that binds to LysRS is the basic helix-loop-helix leucine zipper (bHLH-LZ) region (Lee et al., 2004). We quantified the LysRS-MITF interaction by use of isothermal titration calorimetry (ITC) assays with purified recombinant MITF bHLH-LZ and LysRS proteins. Notably, LysRSS207D bound with ~10-fold higher affinity to MITF (Kd≈30 μM) than to wild type LysRS (Figure 6B). Because the major effect of phosphorylation is to induce the opening of the N-terminal anticodon-binding domain of LysRS, this structural opening may promote the LysRS-MITF interaction. To test this hypothesis, a truncated LysRS mutant having only the C-terminal aminoacylation domain was generated (LysRS-C) and tested for binding to MITF. LysRS-C interacted with MITF with an affinity similar to that of LysRSS207D (Figure 6B). Moreover, binding studies with the N-terminal domain of LysRSWT or of LysRSS207D showed that the N-terminus of LysRS does not interact with MITF (Figures S6C-G). Thus, the binding site for MITF is auto-inhibited by the N-domain in LysRSWT, and the open conformer of LysRSpS207 promotes the interaction of LysRS and MITF. In conclusion, the open conformer of LysRSpS207 is necessary and sufficient to switch the functions of this aaRS from translation to binding transcription factor MITF.
DISCUSSION
The findings presented herein show that, by adopting a new conformer, phosphorylated LysRS switches its function from translation to transcription. The data demonstrate that, in the absence of Ser207 phosphorylation, LysRS is strongly associated within the MSC in a “closed” form, which catalyzes aminoacylation for protein synthesis. However, phosphorylation of Ser207 triggers a distinct conformer that functions exclusively for transcription. By opening up the structure, phosphorylated LysRS is released from the MSC, translocates from cytoplasm to the nucleus, binds to MITF, and generates Ap4A to activate the transcription of MITF target genes (Figure 6C).
The mast cell is the principal effector for all allergic response. Following exposure to an allergen, the mast cells explode with histamine and other inflammatory mediators from pre-stored granules that cause the early-phase response (wheezing, sneezing, itchy eyes, etc.), and new cytokines and proteases are made for more severe and sustained reaction, such as later processes in tissue remodeling and acute inflammation (Church et al., 1997). MITF is a transcription factor in mast cells important for this process (Nechushtan and Razin, 2002). Our results establish a direct involvement of MSC-released, Ser207-phosphorylated LysRS that forms physical interaction with MITF and produces the signaling molecules Ap4A to activate MITF-dependent gene transcription. The co-crystal structure of LysRS:p38/AIMP2 complex and the pull-down results reveal the binary 2:1 interaction of LysRS with the N-terminal end of p38 scaffold protein that allows LysRS to be readily released upon phosphorylation. Moreover, through this binary interaction, LysRS is reserved in the MSC with a unique 2+2 stoichiometry that makes it the most abundant component in the MSC. This availability may allow LysRS to be easily mobilized from MSC as an efficient amplifier for induced synthesis of inflammatory mediators in the activated mast cells.
This “open conformer” mechanism establishes a structural framework for understanding the expanded functions of other housekeeping proteins (Guo et al., 2010; Park et al., 2008; Warner and McIntosh, 2009). Although structural details are not yet known, earlier work showed that, after stimulation with IFNγ, EPRS (GluProRS) is released from the MSC and becomes (together with ribosome subunit L13a) part of the IFNγ-activated inhibitor of translation (GAIT) complex (Mukhopadhyay et al., 2008; Sampath et al., 2004; Yao et al., 2012). Several other components of the ribosome also have ex-translational functions, related to the surveillance of ribosome biosynthesis, p53 regulation, and the DNA damage/repair response (McGowan et al., 2008; Mitrovich and Anderson, 2000; Takagi et al., 2005). These housekeeping multi-component machineries are regarded as depots for regulatory proteins (Ray et al., 2007), and phosphorylations of tRNA synthetases and of ribosomal subunits can promote their release from the MSC and ribosome, respectively (Arif et al., 2009; Carvalho et al., 2008; Jia et al., 2008). Recent studies showed that LeuRS can function as an amino acid sensor of the mTOR pathway, by binding and activating the RagBD GTPase in a leucine-dependent manner(Han et al., 2012). Our data for LysRS demonstrate a structural mechanism of protein metamorphosis, following an environmental stimulus (phosphorylation or ligand binding) that activates a new function of the translation apparatus.
METHODS SUMMARY
Human full-length LysRS, LysRS70–584 and their mutants were expressed in the bacterial strain BL21(DE3). To generate protein complexes, untagged pET20-LysRS was co-transformed with pET28-p3848-His into the BL21(DE3) strain. RBL-2H3 cells were maintained in RPMI 1640 medium as previously described (Lee et al., 2004).
Supplementary Material
Highlights.
Phosphorylation of Ser207 Triggers a Structural Opening of LysRS in Mast Cell
The Open Conformer Releases LysRS from the Multi tRNA-Synthetase Complex
It Traps tRNA in an Inactive State and Switches off the Canonical Function of LysRS
It Drives Ap4A Production and LysRS-MITF interaction for gene transcription in Cell
Acknowledgments
We thank John Cleveland for critical review and editing of the manuscript. Use of the Advanced Photon Source and Advanced Light Source are supported by the U. S. Department of Energy under Contract No. DE-AC02-06CH11357, DE-AC02-05CH11231. This work was supported in part by grants from NIH [GM088278 (XY), GM23562 (PS), GM78359 (AM), GM100136(MG)]; NSF Division of Materials Research through DMR-06-54118 (AM); by a fellowship from the National Foundation for Cancer Research; by the Global Frontier Project [NRF-M1AXA002-2011-0028417] and the WCU project [R31-2008-000-10103-0] of the Ministry of Education, Science and Technology, Korea (SK); by the Israel Science Foundation, the United States Binational Science Foundation, the National Research Foundation of Singapore (HUJ-CREATE), German-Israel Foundation for Scientific Research and Development, the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel’s Ministry of Science and Technology (MOST) (E.R.); and by funding from The State of Florida to Scripps Florida (MG).
Footnotes
Supplementary Information includes Extended Discussion, Extended Experimental Procedures, six figures, one table, and two movies and can be found with this article online.
Author Contributions E.R. and M.G. designed the experiments. Y. O-B., P.F., S.P.B., H-M.Z. J.W., I.R., R.S., J.S., A.D. and J.P. performed the experiments. Y. O-B., P.F., S.P.B., H-M.Z. H.N. and M.G. analyzed the data. Y. O-B., P.F., S.P.B., H-M.Z. S.K. A.G.M. X.-L.Y., P.S., E.R. and M.G. wrote the paper. All authors discussed the results and commented on the manuscript.
Author Information Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 4DPG for the LysRS:p38/AIMP21-48 complex.
Reprints and permissions information are available at www.nature.com/reprints.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arif A, Jia J, Mukhopadhyay R, Willard B, Kinter M, Fox PL. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol Cell. 2009;35:164–180. doi: 10.1016/j.molcel.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi-Levy I, Yannay-Cohen N, Kay G, Razin E, Nechushtan H. Diadenosine tetraphosphate hydrolase is part of the transcriptional regulation network in immunologically activated mast cells. Mol Cell Biol. 2008;28:5777–5784. doi: 10.1128/MCB.00106-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Santos AA, Pires SR, Rocha CS, Saraiva DI, Machado JP, Mattos EC, Fietto LG, Fontes EP. Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS pathogens. 2008;4:e1000247. doi: 10.1371/journal.ppat.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church M, Bradding P, Walls A, Okayama Y. Human mast cells and basophils. In: Kay A, editor. Allergy and Allergic Diseases. Oxford, UK: Blackwell Science Ltd; 1997. pp. 149–170. [Google Scholar]
- Cusack S, Yaremchuk A, Tukalo M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 1996;15:6321–6334. [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Ferguson B, Burke DJ, Garcia V, Yang DC. Interactions of aminoacyl-tRNA synthetases in high-molecular-weight multienzyme complexes from rat liver. Biochim Biophys Acta. 1985;829:319–326. doi: 10.1016/0167-4838(85)90239-0. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. The eucaryotic aminoacyl-tRNA synthetase complex: suggestions for its structure and function. J Cell Biol. 1984;99:373–377. doi: 10.1083/jcb.99.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Zhang HM, Shapiro R, Marshall AG, Schimmel P, Yang XL, Guo M. Structural context for mobilization of a human tRNA synthetase from its cytoplasmic complex. Proc Natl Acad Sci U S A. 2011;108:8239–8244. doi: 10.1073/pnas.1100224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francin M, Mirande M. Identity elements for specific aminoacylation of a tRNA by mammalian lysyl-tRNA synthetase bearing a nonspecific tRNA-interacting factor. Biochemistry. 2006;45:10153–10160. doi: 10.1021/bi0606905. [DOI] [PubMed] [Google Scholar]
- Goerlich O, Foeckler R, Holler E. Mechanism of synthesis of adenosine(5′)tetraphospho(5′)adenosine (AppppA) by aminoacyl-tRNA synthetases. Eur J Biochem. 1982;126:135–142. doi: 10.1111/j.1432-1033.1982.tb06757.x. [DOI] [PubMed] [Google Scholar]
- Guo M, Ignatov M, Musier-Forsyth K, Schimmel P, Yang XL. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: Implications for multisynthetase complex formation. Proc Natl Acad Sci U S A. 2008;105:2331–2336. doi: 10.1073/pnas.0712072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halwani R, Cen S, Javanbakht H, Saadatmand J, Kim S, Shiba K, Kleiman L. Cellular distribution of Lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J Virol. 2004;78:7553–7564. doi: 10.1128/JVI.78.14.7553-7564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell. 2012 doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Han JM, Lee MJ, Park SG, Lee SH, Razin E, Choi EC, Kim S. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem. 2006;281:38663–38667. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- Hilderman RH, Ortwerth BJ. A preferential role for lysyl-tRNA4 in the synthesis of diadenosine 5′,5′″-P1,P4-tetraphosphate by an arginyl-tRNA synthetase-lysyl-tRNA synthetase complex from rat liver. Biochemistry. 1987;26:1586–1591. doi: 10.1021/bi00380a015. [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska M, Havrylenko S, Decottignies P, Gillet S, Le Marechal P, Negrutskii B, Mirande M. Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J Biol Chem. 2009;284:6053–6060. doi: 10.1074/jbc.M809636200. [DOI] [PubMed] [Google Scholar]
- Kellermann O, Tonetti H, Brevet A, Mirande M, Pailliez JP, Waller JP. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. I Species specificity of the polypeptide composition. J Biol Chem. 1982;257:11041–11048. [PubMed] [Google Scholar]
- Kim JY, Kang YS, Lee JW, Kim HJ, Ahn YH, Park H, Ko YG, Kim S. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc Natl Acad Sci U S A. 2002;99:7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell. 2008;29:419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YN, Nechushtan H, Figov N, Razin E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–151. doi: 10.1016/s1074-7613(04)00020-2. [DOI] [PubMed] [Google Scholar]
- Lee YN, Razin E. Nonconventional involvement of LysRS in the molecular mechanism of USF2 transcriptional activity in FcepsilonRI-activated mast cells. Mol Cell Biol. 2005;25:8904–8912. doi: 10.1128/MCB.25.20.8904-8912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan AG. Ap4A and other dinucleoside polyphosphates. Boca Raton: CRC Press; 1992. [Google Scholar]
- Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nature reviews Drug discovery. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- Mirande M, Cirakoglu B, Waller JP. Seven mammalian aminoacyl-tRNA synthetases associated within the same complex are functionally independent. Eur J Biochem. 1983;131:163–170. doi: 10.1111/j.1432-1033.1983.tb07244.x. [DOI] [PubMed] [Google Scholar]
- Mirande M, Le Corre D, Waller JP. A complex from cultured Chinese hamster ovary cells containing nine aminoacyl-tRNA synthetases. Thermolabile leucyl-tRNA synthetase from the tsH1 mutant cell line is an integral component of this complex. Eur J Biochem. 1985;147:281–289. doi: 10.1111/j.1432-1033.1985.tb08748.x. [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, Fox PL. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol Cell. 2008;32:371–382. doi: 10.1016/j.molcel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtan H, Razin E. The function of MITF and associated proteins in mast cells. Mol Immunol. 2002;38:1177–1180. doi: 10.1016/s0161-5890(02)00059-7. [DOI] [PubMed] [Google Scholar]
- Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci U S A. 2008;105:11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Jensen TH. Translation by remote control. Cell. 2010;143:501–502. doi: 10.1016/j.cell.2010.10.039. [DOI] [PubMed] [Google Scholar]
- Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J Mol Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci. 2007;32:158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Razin E, Zhang ZC, Nechushtan H, Frenkel S, Lee YN, Arudchandran R, Rivera J. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J Biol Chem. 1999;274:34272–34276. doi: 10.1074/jbc.274.48.34272. [DOI] [PubMed] [Google Scholar]
- Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J Mol Biol. 2000;304:983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe CL, Warrington JA, Treadwell L, Norcum MT. A three-dimensional working model of the multienzyme complex of aminoacyl-tRNA synthetases based on electron microscopic placements of tRNA and proteins. J Biol Chem. 2005;280:38870–38878. doi: 10.1074/jbc.M502759200. [DOI] [PubMed] [Google Scholar]
- Yannay-Cohen N, Carmi-Levy I, Kay G, Yang CM, Han JM, Kemeny DM, Kim S, Nechushtan H, Razin E. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol Cell. 2009;34:603–611. doi: 10.1016/j.molcel.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL. Coding Region Polyadenylation Generates a Truncated tRNA Synthetase that Counters Translation Repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.