Abstract
Arsenic enhances genotoxicity of other carcinogenic agents such as ultraviolet radiation and benzo[a]pyrene. Recent reports suggest that inhibition of DNA repair is an important aspect of arsenic co-carcinogenesis, and DNA repair proteins such as poly (ADP ribose) polymerase (PARP)-1 are direct molecular targets of arsenic. Although arsenic has been shown to generate reactive oxygen/nitrogen species (ROS/RNS), little is known about the role of arsenic-induced ROS/RNS in the mechanism underlying arsenic inhibition of DNA repair. We report herein that arsenite-generated ROS/RNS inhibits PARP-1 activity in cells. Cellular exposure to arsenite, as well as hydrogen peroxide and NONOate (nitric oxide donor), decreased PARP-1 zinc content, enzymatic activity, and PARP-1 DNA binding. Furthermore, the effects of arsenite on PARP-1 activity, DNA binding, and zinc content were partially reversed by the antioxidant ascorbic acid, catalase, and the NOS inhibitor, aminoguanidine. Most importantly, arsenite incubation with purified PARP-1 protein in vitro did not alter PARP-1 activity or DNA binding ability, whereas hydrogen peroxide or NONOate retained PARP-1 inhibitory activity. These results strongly suggest that cellular generation of ROS/RNS plays an important role in arsenite inhibition of PARP-1 activity, leading to the loss of PARP-1 DNA binding ability and enzymatic activity.
Keywords: arsenic, PARP-1, ROS, RNS
INTRODUCTION
The balance between DNA damage and repair maintains genomic stability and disruption of this balance leads to DNA damage accumulation and consequent tumorigenesis. Many different enzymes participate in DNA damage repair and among these enzymes, Poly (ADP-ribose) polymerase (PARP) 1 is an important contributor to the repair processes. The PARP family of proteins include 17 different members and PARP-1 activity accounts for about 90% of the total cellular poly(ADP-ribose) formation [1, 2]. Activation of PARP-1 occurs as an immediate cellular response to DNA strand breaks, which can be induced by ionizing radiation, alkylating agents, or oxidative stress [1, 3]. PARP-1 is comprised of three functional domains [4]. The amino-terminal DNA-binding domain contains two zinc fingers that are important for the binding of PARP-1 to single-strand and double-strand breaks [1, 4]. A third zinc finger was described and found to be dispensable for DNA binding, but is important for coupling damage-induced changes in the DNA-binding domain to the catalytic domain leading to alterations in PARP-1 activity [5].
Our previous studies demonstrated that arsenic reduces ultraviolet radiation (UVR)-induced PARP-1 activation, and we found that the activity of PARP-1 could be depressed by exposure to low concentrations of arsenic in human keratinocyte cells [6, 7]. There is abundant evidence that cellular exposure to arsenic exposure generates reactive oxygen species (ROS) and reactive nitrogen species (RNS). The production of O2·−, H2O2, and ·OH has been reported in arsenite-exposed keratinocytes [7–11], and RNS is capable of interacting with certain DNA repair enzymes, including hOGG-1 and PARP-1 [7, 10, 12]. In this report we examined the potential relationship between cellular generation of ROS/RNS by arsenic and inhibition of PARP-1. We hypothesized that ROS/RNS induced by arsenic in cells play a major role in arsenic inhibition of PARP-1 activity.
MATERIAL AND METHODS
Cell culture and treatment
Immortalized human keratinocyte cells (HaCat) were cultured in Dulbecco’s Modified Eagle’s Medium F:12 HAM (Sigma-Aldrich, St. Louis, MO), supplemented with 10% newborn calf serum (Invitrogen, Carlsbad, CA), four-fold final concentration of MEM amino acids, 2 mM l-glutamine, and antibiotics (penicillin, 100 U/ml and streptomycin, 100 μg/ml) (all from Sigma-Aldrich, St. Louis, MO). Normal Human Epidermal Keratinocytes neonatal (HEKn) cells (Lifeline Cell Technology, Frederick, MD) were cultured in DermaLife K Medium Complete Kit (Lifeline Cell Technology, Frederick, MD) according to manufacturer’s instruction. Cells were cultured at 37°C in 95% air/5% CO2 inside a humidified incubator. For HaCat cell-based studies, cells at 60–70% confluence were placed into 1% serum containing DMEM/F12 medium overnight, then treated with sodium arsenite (As) (Sigma-Aldrich, St. Louis, MO), hydrogen peroxide (H2O2) (Bio-Rad Laboratories, Hercules, CA), NO donor NONOate (Cayman Chemicals, Ann Arbor, MI), vitamin C (Sigma-Aldrich, St. Louis, MO), or NOS inhibitor aminoguanidine hydrochloride (AG) (Cayman Chemicals, Ann Arbor, MI). For HEKn cells, cells at 70% confluence were treated with sodium arsenite (As), hydrogen peroxide, NONOate or catalase (EMD Millipore, Billerica, MA). Following incubation for 48 hours, PARP-1 was isolated by immunoprecipitation with PARP-1 antibody (Cell Signaling Technology, Danvers, MA). Alternatively, for studies with isolated PARP-1 protein, the protein was isolated from untreated HaCat cells by immunoprecipitation.
Isolation of PARP-1 from cells by immunoprecipitation (IP)
Cells were rinsed with ice-cold PBS 3 times, followed by addition of 500 μL ice-cold RIPA lysis buffer (Cell Signaling Inc., Danvers, MA) (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1mM EGTA, 1% triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1mM sodium vanadate, 1 μg/ml leupeptin and 1 mM PMSF). Cells were incubated on ice for 5 min before sonication and preparation of lysate by centrifugation at 12,000 rpm for 10 min at 4°C. Two μl monoclonal PARP-1 antibody were added to 200 μl supernatant and the samples were incubated at 4°C overnight with gentle shaking. Twenty μl Protein A beads (Sigma-Aldrich, St. Louis, MO) were then added to each tube and further incubated for 5 hours at 4°C. PARP-1 immunoprecipitates were isolated by centrifugation at 12,000 rpm for 10 min at 4°C and the pellets were washed 5 times with ice-cold RIPA lysis buffer.
Measurement of PARP-1 activity by ELISA
PARP-1 activity was measured using the HT Colorimetric PARP/Apoptosis Assay kit (Trevigen Inc., Gaithersburg, MD) according to the manufacturer’s instructions. Activity was visualized by the addition of the TACS-Sapphire colorimetric substrate and the absorbance was read with a SpectraMax 340 plate reader at 450 nm wavelength. Activity of PARP-1 was calculated from the absorbance and the generated standard curve.
Western blot analysis of PARP-1
Western blots were performed to determine the levels of PARP-1 protein expression. Total protein lysate (4 μg) was resolved on an 8% SDS-polyacrylamide gel and transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA), then incubated for 1 h in TBST (10 mM Tris, pH 8.0, 150 mM NaCl and 0.1% Tween20) containing 5% non-fat milk at room temperature. The membrane was then incubated with the rabbit polyclonal anti-PARP-1 antibody (1:2000, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) overnight at 4°C. After washing with TBST, the membrane was incubated for 1 h with horseradish peroxidase conjugated secondary antibody (1:2000, Cell Signaling Technology, Danvers, MA), and the resulting signal was detected using the Super Signal West Pico chemiluminescent kit (Thermo Fisher Scientific Inc., Rockford, IL) on a Kodak Image Station 4000MM following the manufacturer’s instructions. To control sample loading and protein transfer, the membrane was stripped and re-probed to detect β-actin (1:2000, Cell Signaling Technology, Danvers, MA). The intensities were quantified by KODAK Molecular Imaging Software version 4.0. The PARP-1 protein levels were normalized to β-actin as compared with the untreated control.
PARP-1 DNA binding activity measurement with EMSA
Electrophoretic mobility shift assay (EMSA) was performed to measure PARP-1 binding to double strand DNA (20). The synthetic, 42-nucleotide double strand DNA poly-nucleotides (Integrated DNA Technologies, Coralville, IA) used as a probe in the studies had an upper strand nucleotide sequence of 5′-GAGTGTTGCATTCCTCTCTGGGCGCCGGGCAGGTACC-TGCTG-3′. The binding of PARP-1 with the double stranded DNA was measured using an EMSA kit (Molecular Probes, Eugene, OR) according to the vendor’s instructions. Briefly, the volume of the binding system is 10 μl (PARP-1 protein 2 μl, ds DNA probe 2 μl, 5×binding buffer 2 μl, deionized water 4 μl), mixed gently but thoroughly, and incubated during the reaction for 30 min at room temperature. At the end of the incubation period, 2 μl of 6×EMSA gel-loading solution was added for each 10 μl of reaction mixture, and mixed gently. The DNA-PARP-1 complexes were separated by electrophoresis using a native 6% polyacrylamide gel. After electrophoresis, the gels were stained for nucleic acids with SYBR Green EMSA Nucleic Acid Gel Stain. The results of the stained nucleic acids were visualized using 254 nm UV epi-illumination in a KODAK IS4000MM Image. The excitation wavelength is 465 nm and the Emission wavelength was 535 nm.
Detection of the zinc content of isolated PARP-1
Zinc content was measured by adding 10 μl of 1 mM 4,(2-pyridylazo)-resorcinol (Sigma-Aldrich, St. Louis, MO) to 100 μl of protein sample that was pre-treated with 10 mM H2O2 and scanning the spectra at 350–550 nm on a Beckman Coulter DU 800 spectrophotometer. The resorcinol indicator absorbance shifts from 411 to 493 nm in the presence of zinc and the peak at 493 nm is recorded and compared to a standard curve for zinc content [13]. The relative zinc content was normalized to protein concentration.
Data Analysis
Data was presented as the means ± S.D. Statistical analysis was carried out by analysis of variance (ANOVA) followed by multiple comparison tests (LSD). If the variance among groups was not homogenous, then a Kruscal-wallis H test was performed. A value of P < 0.05 was considered statistically significant and labeled by an asterisk (*).
RESULTS
Low concentrations of arsenite do not affect PARP-1 protein level
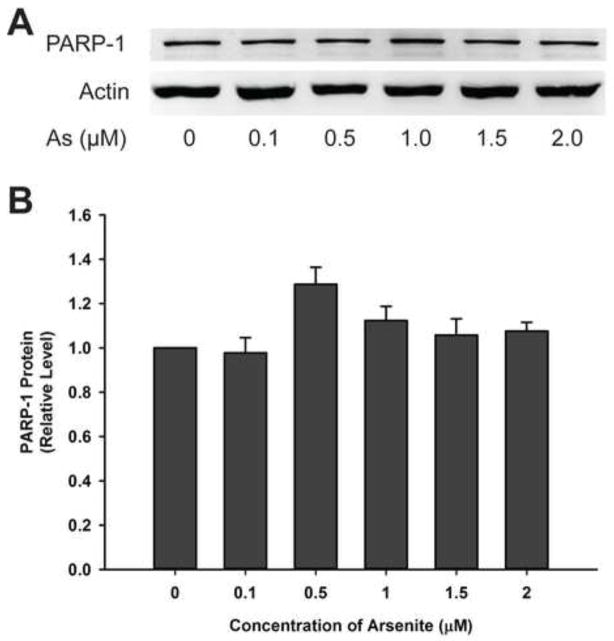
Enzyme activity in cells could be changed due to alteration of protein level. HaCat cells were incubated with low concentrations of arsenite (0–2 μM) for 48 hours and PARP-1 protein level was determined by western blotting (Fig. 1A). There is no significant difference in PARP-1 protein level with or without arsenite treatments (Fig. 1B) indicating that low concentrations of arsenite did not affect the total amount of the PARP-1 protein in HaCat cells under these conditions.
Fig. 1.
Effect of arsenite (As) on PARP-1 protein level in HaCat cells. A) Western blotting analysis of PARP-1 protein level in cell lysates. HaCat cells were treated with indicated concentrations of sodium arsenite for 48 h. B) Quantification of western blot result with control group (As, 0 μM) normalized to 1.0. Data are presented as mean ± S.D., n=6.
Exposure to low concentrations of arsenite, hydrogen peroxide or NONOate reduces PARP-1 activity in HaCat cells
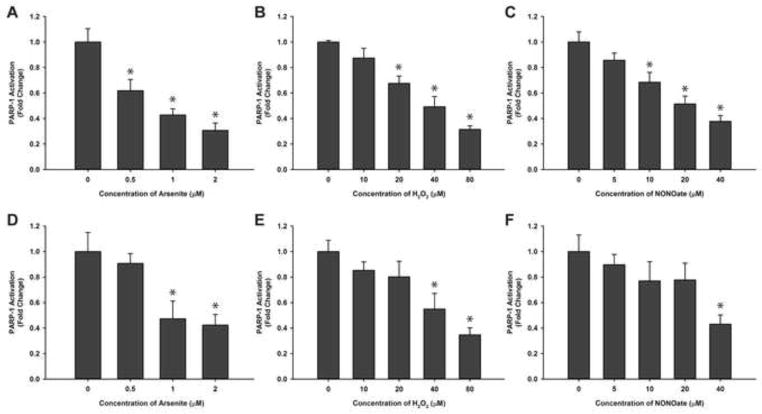
To investigate the effect of arsenite on PARP-1 activity, HaCat cells were exposed to low concentrations of arsenite for 48 hrs and the activity of immunoprecipitated PARP-1 protein was determined using the HT Colorimetric PARP/Apoptosis assay. The activity of PARP-1 decreased significantly after arsenite treatment in an arsenite concentration-dependent manner (Fig. 2A). This result is consistent with the results of other reports [6, 14]. Since arsenite exposure is known to generate ROS/RNS in human keratinocytes [8], we wanted to test whether ROS/RNS exposure has any impact on PARP-1 activity. HaCat cells were treated with hydrogen peroxide or NONOate, and the changes of PARP-1 activity were measured. As shown in Fig. 2B, the activity of PARP-1 decreased in HaCat cells in a hydrogen peroxide concentration-dependent manner. The reduction of PARP-1 activity is statistically significant at concentration of hydrogen peroxide at or above 20 μM. Similarly, we investigated the effect of NO on PARP-1 activity. NONOate is a NO donor which can liberate 2 mol of NO per mol of parent compound [15]. After HaCat cells were incubated with different concentrations of NONOate for 48 hrs, PARP-1 activity decreased in a NO concentration dependent manner (Fig. 2C). In order to demonstrate that the observed inhibitory effect on PARP-1 is not only restricted to HaCat cells, we also carried out the identical experiments using HEKn cells. Very similar results were obtained (Fig. 2D–F). These findings indicate that in cells, ROS/RNS can inhibit PARP-1 activity.
Fig. 2.
Effects of arsenite, hydrogen peroxide, and NONOate on PARP-1 activity in HaCat (A, B, C) and HEKn (D, E, F) cells. Cells were treated with sodium arsenite (A, D), hydrogen peroxide (B, E), or NONOate (C, F) separately for 48hours, and then PARP-1 protein was isolated by immunoprecipitation. PARP-1 activity was determined using a HT Colorimetric PARP/Apoptosis Assay kit. Data are presented as means ± S.D., * P<0.05 vs. 0 group, n=6 for HaCat (A, B, C), n=3 for HEKn (D, E, F).
Vitamin C, AG, and Catalase partially reverse arsenite inhibition of PARP-1 activity in HaCat cells
Vitamin C is a water soluble antioxidant that can remove ROS. To prevent lowering the pH value of the cell culture medium, we used the conjugate base, sodium L-ascorbate. AG is an inhibitor of NOS used to prevent cell production of NO. Catalase is an enzyme that catalyzes the decomposition of hydrogen peroxide to water and oxygen. In order to determine the role of ROS and RNS on PARP-1 activity, we blocked the ROS/RNS generation pathways induced by arsenite using vitamin C, AG or catalase, and then measured PARP-1 activity. HaCat cells were treated with Vitamin C, AG or catalase together with arsenite for 48 hours, and the activity of PARP-1 protein isolated from the cells was measured. As shown in Fig. 3, arsenite inhibition of PARP-1 activity was partially reversed by Vitamin C, AG or catalase treatment. These results suggest that arsenite-induced ROS/RNS generation plays an important role in arsenite inhibition of PARP-1 activity.
Fig. 3.
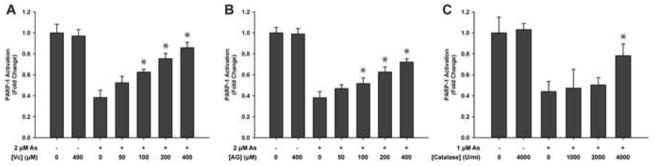
Effects of vitamin C, AG and catalase on the activity of PARP-1 in arsenite treated cells. HaCat cells were treated with arsenite as described in “Methods” except the indicated concentration of either vitamin C or AG was added for 30 min before arsenite addition. In catalase experiment, HEKn cells were treated with indicated concentrations of catalase for 30 min before arsenite addition. After incubation for 48 hours, PARP-1 was isolated by immunoprecipitation and the activity of PARP-1 was measured with a HT Colorimetric PARP/Apoptosis Assay kit. A) Effects of vitamin C on the activity of PARP-1. B) Effects of AG on the activity of PARP-1. C) Effects of catalase on the activity of PARP-1. Data are presented as means ± S.D. * P<0.05 vs. As alone group, n=6.
Arsenic, hydrogen peroxide or NONOate inhibit PARP-1 DNA binding activity
DNA binding activity is critically important in the functionality of PARP-1 so we investigated the effect of arsenite, hydrogen peroxide, or NONOate on the ability of PARP-1 to bind DNA. After HaCat cells were treated with arsenite for 48 hours, PARP-1 was isolated by immunoprecipitation and PARP-1 DNA-binding was measured by EMSA. We found that exposure of cells to arsenite concentrations of 1.0 μM or above significantly decreased the DNA binding activity of PARP-1 (Fig. 4A, 4B). Similar results were obtained following cell treatment with hydrogen peroxide or NONOate (Fig. 4A, 4C and 4D). These results suggest that arsenite-induced ROS/RNS generation could be a mechanism for arsenite inhibition of PARP-1 DNA binding activity. In order to test this hypothesis, we treated HaCat cells with 2 μM arsenite together with 400 μM vitamin C or 400 μM AG. EMSA results revealed that treatment with vitamin C or AG partially restored PARP-1 DNA-binding ability (Fig. 4E). These findings demonstrate that blocking ROS/RNS during arsenite exposure can rescue the reduced PARP-1 DNA binding activity caused by arsenite.
Fig. 4.
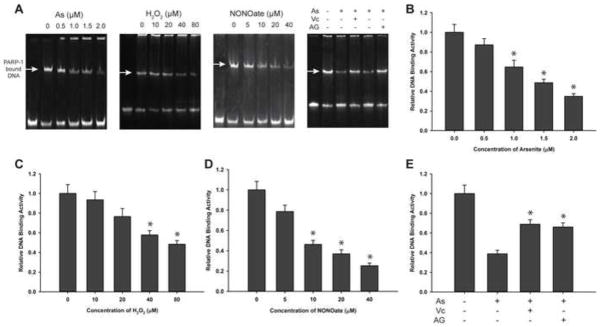
Effects of arsenite, hydrogen peroxide, or NONOate on PARP-1 DNA binding. Cells were treated with indicated concentrations of sodium arsenite, hydrogen peroxide, or NONOate for 48 hrs. PARP-1 was isolated by immunoprecipitation and PARP-1 binding to DNA was assayed by EMSA. A) Images of EMSA results after electrophoresis, visualization of nucleic acids with SYBR green. B) Effects of sodium arsenite on the DNA-binding of PARP-1. C) Effects of hydrogen peroxide on the DNA-binding of PARP-1. D) Effects of NONOate on the DNA-binding of PARP-1. E) Effects of vitamin C (400 μM) and AG (400 μM) on the DNA-binding of PARP-1 with arsenite (2 μM). Data are presented as means± S.D. * P<0.05 vs. concentration at 0 group, n=4.
Treatment of arsenite, hydrogen peroxide, or NONOate reduces zinc content in PARP-1 protein isolated from exposed cells
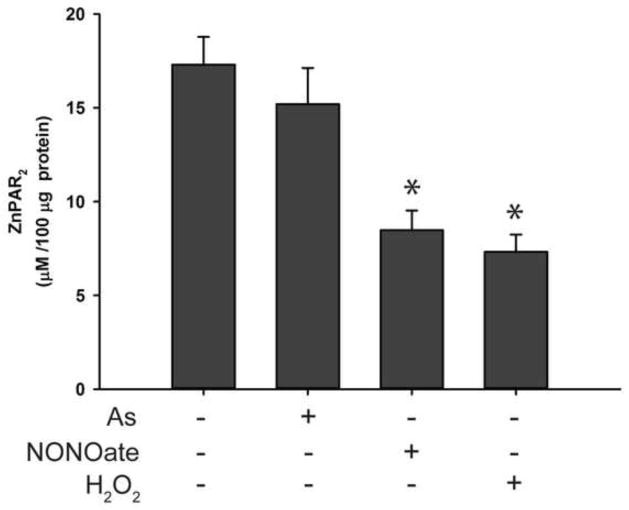
PARP-1 zinc finger domains are essential for DNA binding [1, 4]. Others have reported that the zinc finger domain is vulnerable to oxidative stress, causing zinc release [16, 17]. It is possible that ROS/RNS, either authentic or generated by arsenite, could interact with the zinc finger domain of PARP-1, resulting in zinc loss and decreased PARP-1 activity. To test this hypothesis, we measured zinc content in PARP-1 protein immunoprecipitated from cells exposed to arsenite, hydrogen peroxide, or NONOate for 48 hrs. Under our experimental conditions, these treatments had no significant effect on cell viability (data not shown). As shown in Fig. 5A, arsenite exposure reduced the amount of zinc associated with the PARP-1 in an arsenite concentration-dependent manner. The effects of hydrogen peroxide and NONOate on the zinc content of PARP-1 were similar (Fig. 5B and 5C). These results suggest that the decreased PARP-1 enzymatic activity (Fig. 2) and DNA binding (Fig. 4) due to the exposures were likely caused by the loss of zinc from the protein. In order to test whether the zinc loss induced by arsenite was related to ROS/RNS generation, HaCat cells were pretreated with vitamin C or AG before exposure to 2 μM arsenite for 48 hrs. We found that both vitamin C and AG partially prevented zinc loss from PARP-1 protein (Fig. 5D), suggesting that arsenite induced zinc loss in PARP-1 is through arsenite-generated ROS/RNS.
Fig. 5.
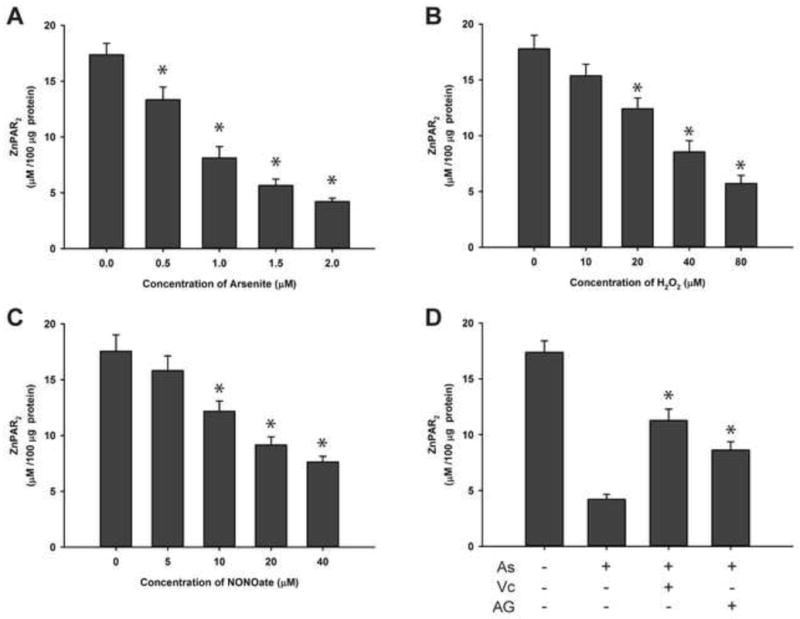
Effects of arsenite, hydrogen peroxide, NONOate on the zinc content of PARP-1 isolated from treated HaCat cells. Cells were treated with sodium arsenite, hydrogen peroxide, or NONOate for 48 hrs with or without pretreatment with vitamin C (400 μM) or AG (400 μM). PARP-1 was isolated from the treated cells by immunoprecipitation. The content of zinc in the isolated PARP-1 was detected by spectrophotometric method. Data were presented as means ± S.D., * P<0.05 vs. 0 group, n=6. A) Effects of arsenite; B) Effects of hydrogen peroxide; C) Effects of NONOate; D) Effects of arsenite (2 μM) with or without pretreatment with vitamin C (400 μM) or AG (400 μM).
Low concentrations of arsenite does not inhibit the activity of purified PARP-1
The above results demonstrate that arsenite inhibits PARP-1 activity in HaCat cells, and that the generation of ROS/RNS induced by arsenite in the cellular environment contributes to the response. To determine whether arsenite alone, in the absence of ROS/RNS, could inhibit PARP-1 activity, we performed PARP-1 activity assay using isolated protein from untreated HaCat cells. The PARP-1 immunoprecipitates were incubated with different concentrations of arsenite for 48 hrs before measurement of PARP-1 activity. As shown in Fig. 6A, arsenite did not significantly affect the PARP-1 activity until the concentration reached non-physiologic levels of 1,000 μM. This is in sharp contrast to the results observed in arsenite treated HaCat cells, where even 0.5 μM arsenite caused a significant reduction in PARP-1 activity (Fig. 2). These results suggest that arsenite alone is incapable of inhibiting PARP-1 activity and that arsenite inhibition of PARP-1 activity likely requires the generation of ROS/RNS within the cellular environment.
Fig. 6.
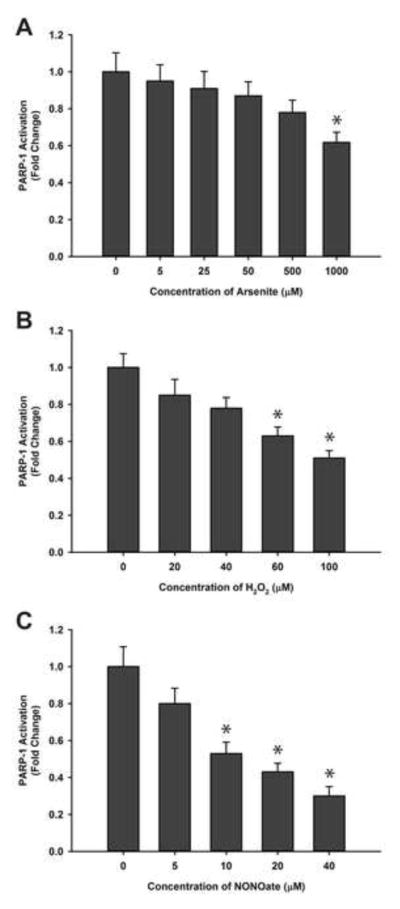
Effects of arsenite, hydrogen peroxide, or NONOate on the activity of PARP-1 isolated from untreated HaCat cells. HaCat cell lysates were prepared from untreated, subconfluent cells then PARP-1 was isolated by immunoprecipitation. The isolated PARP-1 was treated with the indicated concentrations of sodium arsenite, hydrogen peroxide, or NONOate at 4°C for 48 hrs before the activity of PARP-1 was determined using an HT Colorimetric PARP/Apoptosis Assay kit. Data were presented as means ± S.D., * P<0.05 vs. 0 group, n=6. A) Effects of arsenite. B) Effects of hydrogen peroxide. C) Effects of NONOate.
Hydrogen peroxide or NONOate reduces the activity of purified PARP-1 protein
Since arsenite does not inhibit the activity of purified PARP-1 (Fig. 6A), we next investigated whether authentic ROS/RNS is capable of inhibiting PARP-1 protein immunoprecipitated from untreated HaCat cells. PARP-1 was incubated with hydrogen peroxide or NONOate at 4°C for 48 hrs, and then the activity of PARP-1 was measured. Results shown in Fig. 6B and 6C demonstrate that unlike arsenite, both hydrogen peroxide and NONOate inhibit the activity of purified PARP-1 in a concentration dependent manner. These results show that hydrogen peroxide and NONOate inhibit PARP-1 activity, either in cellular environment or purified protein, suggesting that ROS and RNS have a direct effect on PARP-1.
Hydrogen peroxide and NONOate, but not arsenite, can reduce DNA binding activity of purified PARP-1
To further investigate the differential effect of arsenite versus hydrogen peroxide and NONOate on purified PARP-1 protein, we exposed purified PARP-1 with each of the three compounds at 4°C for 48 hrs, and PARP-1 DNA binding was measured by EMSA. Arsenite at concentrations up to 500 μM did not significantly alter the DNA binding ability of PARP-1 (Fig. 7A, 7B), which is in sharp contrast to the pronounced inhibitive effect of arsenite observed in cells at concentration as low as 1 μM (Fig. 4). Importantly, PARP-1 DNA binding activity decreased with the increasing concentration of hydrogen peroxide or NONOate (Fig. 7C & 7D), in a similar pattern as observed in cells (Fig. 4). These results demonstrate that while hydrogen peroxide and NONOate can inhibit PARP-1 DNA binding activity both in HaCat cells and in purified form, arsenite is only capable of doing so in cells, but not with purified protein.
Fig. 7.
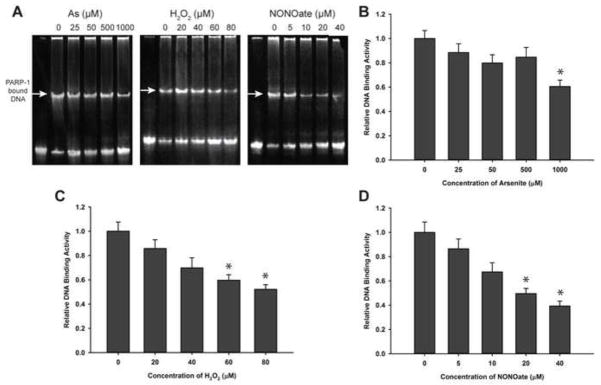
Effects of arsenite, hydrogen peroxide, and NONOate on DNA binding of purified PARP- 1. PARP-1 was isolated by immunoprecipitation from untreated HaCat cells, then the purified PARP-1 protein was treated with indicated concentrations of sodium arsenite, hydrogen peroxide, and NONOate for 48 hours at 4°C, and the DNA-binding ability was tested with EMSA as described in the legend to Fig. 4. Data are presented as means± S.D. * P<0.05 vs. concentration of 0 group, n=4. A) The images of EMSA. B) Effects of sodium arsenite. C) Effects of hydrogen peroxide. D) Effects of NONOate.
Hydrogen peroxide and NONOate, but not arsenite, deplete the zinc content of PARP-1 isolated from HaCat cells
The above findings demonstrate that arsenite does not affect the DNA-binding ability of purified PARP-1, in contrast to hydrogen peroxide and NONOate. In order to test whether these differential effects in DNA-binding are caused by disruption of the PARP-1 zinc finger as assessed by zinc loss from the protein, we measured the zinc content of PARP-1 following treatment with each of the three compounds at a selected concentration. As shown in Fig. 8, 50 μM hydrogen peroxide and 20 μM NONOate decreased the zinc content of the purified PARP-1, but 50 μM arsenite failed to affect the zinc content of purified PARP-1, which is in sharp contrast to the result in cells where 2 μM arsenite lead to the loss of more than 75% of zinc in the protein (Fig. 5A). These results suggest that the mechanism through which arsenite reduces the enzyme’s activity is likely through interaction with the zinc finger domains of PARP-1 via generation of ROS/RNS.
Fig 8.
Effects of arsenite, hydrogen peroxide, or NONOate on the zinc content of purified PARP-1. PARP-1 protein was isolated from untreated HaCat cells by immunoprecipitation, then the isolated PARP-1 was treated with sodium arsenite (50 μM), hydrogen peroxide (50 μM), and NONOate (20 μM) for 48 hrs at 4°C The zinc content in the isolated PARP-1 was detected spectrophotometrically (DU 800, Beckman Coulter). Data were presented as means ± S.D., * P<0.05 vs. control group; n=6.
DISCUSSION
Extensive evidence in the literature demonstrates that ROS and RNS generation pathways can be activated by arsenite. We have shown that treatment of keratinocytes with low micro-molar concentrations of arsenite lead to persistent generation of ROS [10] and NO [7]. The findings presented in this study demonstrate that low concentrations of arsenite (2 μM or below) can decrease the activity of PARP-1 in cells through the actions of ROS/RNS that are induced by arsenite exposure. Furthermore, the interaction of ROS/RNS with the zinc-finger domains of PARP-1 results in the loss of zinc from the protein, leading to the loss of the DNA binding ability of PARP-1. This finding was evident in cells or purified protein treated with H2O2 or NONOate, demonstrating that both ROS and RNS can directly interrupt PARP-1 function either in the cellular or in vitro environment.
Arsenite-induced ROS and RNS generation inhibits PARP-1 activity
Although mounting evidence has indicated that arsenic can inhibit the activity of PARP-1 in cells, little is known about the molecular mechanism of arsenic action. Arsenic has been well documented to stimulate ROS/RNS generation in many different cell types [18, 19]. We and other labs have reported that arsenite exposure generates ROS/RNS, such as O2·−, H2O2 and NO, in HaCat cells, leading to increased DNA damage [9, 10, 20, 21]. The findings presented in the current study show that low concentrations of arsenite could decrease the activity of PARP-1, having the similar effects as the treatment with H2O2 or NONOate, in both HaCat and HEKn cells (Fig. 2). These findings suggest that arsenite-induced ROS and RNS generation may inhibit PARP-1. To confirm this hypothesis, we treated the HaCat cells with arsenite in the presence of vitamin C, AG or catalase, which eliminate ROS or RNS, and we found that the activity of PARP-1 was partially restored (Fig. 3). These results provide evidence that arsenite-generated ROS/RNS is involved in the mechanism of arsenite inhibition of PARP-1 activity. Importantly, when arsenite was incubated with the purified protein, little inhibitory effect of arsenite on PARP-1 activity was observed, while hydrogen peroxide or NONOate remained effective (Fig. 6). The most logical explanation for the lack of effect by arsenite is that arsenite requires iNOS [7] and NADPH oxidase [10] in cells to generate ROS and RNS, respectively, and this aspect of arsenite action is lacking in the assays of purified PARP-1 protein.
Low concentrations of arsenite did not affect the transcription and/or translation of PARP-1 protein (Fig. 1), suggesting that arsenic inhibitory effect must be through other mechanisms. Numerous reports have shown that ROS/RNS may damage zinc finger structures through interaction with cysteine thiols, thereby releasing zinc [17, 22–24]. NO and nitroxyl (HNO) donors was found to inhibit activity of purified PARP-1 protein, indicating that PARP-1 could be modified by nitrosative and oxidative conditions [25]. However, it is not clear whether these nitrosative and oxidative modification would occur in cells, especially under conditions like arsenic exposure where relatively low level of ROS/RNS are generated. The current study was designed to answer this question.
Zinc finger domain is the site of ROS/RNS interaction
PARP-1 protein contains three zinc finger domains, two of which are critically important for DNA binding [1, 4]. Our results presented here provide evidence for the interaction of arsenite-induced ROS and RNS with the zinc finger domains of PARP-1 and the subsequent zinc release from the protein (Figs. 5 & 8). Further studies are needed to investigate how exactly ROS and RNS interact with the zinc finger, and what specific structural modification (e.g., oxidation and nitrosation) occurs as a result of the interaction. It is important to note that our recent reports suggest that arsenic can also interact with the zinc finger domain directly [7, 26]. The fact that hydrogen peroxide or NONOate did not completely abolish PARP-1 activity (Fig. 2) and eliminating ROS/RNS did not completely restore PARP-1 function (Fig. 3&4) illustrates that the mechanism of arsenic inhibition of PARP-1 activity may be complex, perhaps involving multiple mechanisms. Clarifying these key issues will require additional studies. It is also worth noting that there are two seemingly “conflicting” processes going on at the same time when cells are exposed to ROS/RNS generated by arsenic: PARP-1 activation by ROS/RNS-induced DNA damage and any inhibition of PARP-1 catalytic activity by ROS/RNS. Depending on the experimental conditions and measured parameters, one could either find arsenic inhibiting the intrinsic activity of PARP-1 protein isolated from treated cells (such as [27]), or arsenic enhancing PARP-1 activity in cellular environment (such as [28]). Both findings are real and correct, and are not in conflict to each other.
Summary
Arsenic has been shown to enhance tumor development in animals either pretreated with other carcinogens [29, 30], or co-treated with ultraviolet radiation [31]. It is well known that DNA damage is an important contributing factor to the development of cancer. Arsenic could either directly induce the DNA damage itself [32] or inhibit the activity of DNA repair enzymes, such as PARP-1, thus enhancing carcinogenic effects of other agents. The novel findings presented here provide a better understanding of the underlying mechanism of arsenic co-carcinogenesis, highlighting the oxidation and/or nitrosation of the redox sensitive zinc finger proteins by arsenic-generated ROS and RNS as a critical deleterious molecular event following arsenic exposure.
Highlights.
Treatment with arsenite did not affect PARP-1 protein level in cells
Arsenite exposure of cells inhibited the activity of PARP-1
Effects of arsenite on cells were similar to H2O2 and NO in inhibiting PARP-1
Treatment with the antioxidant significantly blocked arsenite effect in cells
Arsenite did not inhibit the activity of purified PARP-1
Acknowledgments
Funding Sources
This work was supported in part by National Institutes of Health Grants R01ES15826 and R01ES021100. This work was also supported by University of New Mexico Cancer Center Grant NIH P30 CA118100 and by a pilot grant from the UNM HSC Environmental Health Signature Program.
Abbreviations
- ROS
Reactive oxygen species
- RNS
Reactive nitrogen species
- PARP-1
poly (ADP ribose) polymerase 1
- AG
Aminoguanidine
- IP
Immunoprecipitation
- NOS
Nitric oxide synthase
- ELISA
Enzyme linked immunosorbent assay
- EMSA
Electrophoretic mobility shift assay
- RIPA
Radioimmunoprecipitation assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nature reviews. Molecular cell biology. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 3.Burkle A, Diefenbach J, Brabeck C, Beneke S. Ageing and PARP. Pharmacological research: the official journal of the Italian Pharmacological Society. 2005;52:93–99. doi: 10.1016/j.phrs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Virag L. Structure and function of poly(ADP-ribose) polymerase-1: role in oxidative stress-related pathologies. Current vascular pharmacology. 2005;3:209–214. doi: 10.2174/1570161054368625. [DOI] [PubMed] [Google Scholar]
- 5.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. The Journal of biological chemistry. 2010;285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin XJ, Hudson LG, Liu W, Timmins GS, Liu KJ. Low concentration of arsenite exacerbates UVR-induced DNA strand breaks by inhibiting PARP-1 activity. Toxicology and applied pharmacology. 2008;232:41–50. doi: 10.1016/j.taap.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding W, Liu W, Cooper KL, Qin XJ, de Souza Bergo PL, Hudson LG, Liu KJ. Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. The Journal of biological chemistry. 2009;284:6809–6817. doi: 10.1074/jbc.M805566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H, Hudson LG, Ding W, Wang S, Cooper KL, Liu S, Chen Y, Shi X, Liu KJ. Arsenite causes DNA damage in keratinocytes via generation of hydroxyl radicals. Chemical research in toxicology. 2004;17:871–878. doi: 10.1021/tx049939e. [DOI] [PubMed] [Google Scholar]
- 9.Ding W, Hudson LG, Liu KJ. Inorganic arsenic compounds cause oxidative damage to DNA and protein by inducing ROS and RNS generation in human keratinocytes. Molecular and cellular biochemistry. 2005;279:105–112. doi: 10.1007/s11010-005-8227-y. [DOI] [PubMed] [Google Scholar]
- 10.Cooper KL, Liu KJ, Hudson LG. Enhanced ROS production and redox signaling with combined arsenite and UVA exposure: contribution of NADPH oxidase. Free radical biology & medicine. 2009;47:381–388. doi: 10.1016/j.freeradbiomed.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper KL, Liu KJ, Hudson LG. Contributions of reactive oxygen species and mitogen-activated protein kinase signaling in arsenite-stimulated hemeoxygenase-1 production. Toxicology and applied pharmacology. 2007;218:119–127. doi: 10.1016/j.taap.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Romero R, Canuelo A, Martinez-Lara E, Javier Oliver F, Cardenas S, Siles E. Poly(ADP-ribose) polymerase-1 modulation of in vivo response of brain hypoxia-inducible factor-1 to hypoxia/reoxygenation is mediated by nitric oxide and factor inhibiting HIF. Journal of neurochemistry. 2009;111:150–159. doi: 10.1111/j.1471-4159.2009.06307.x. [DOI] [PubMed] [Google Scholar]
- 13.Kopera E, Schwerdtle T, Hartwig A, Bal W. Co(II) and Cd(II) substitute for Zn(II) in the zinc finger derived from the DNA repair protein XPA, demonstrating a variety of potential mechanisms of toxicity. Chemical research in toxicology. 2004;17:1452–1458. doi: 10.1021/tx049842s. [DOI] [PubMed] [Google Scholar]
- 14.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Current medicinal chemistry. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 15.Keefer LK, Nims RW, Davies KM, Wink DA. “NONOates” (1-substituted diazen-1-ium-1,2-diolates) as nitric oxide donors: convenient nitric oxide dosage forms. Methods in enzymology. 1996;268:281–293. doi: 10.1016/s0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- 16.Smirnova J, Zhukova L, Witkiewicz-Kucharczyk A, Kopera E, Oledzki J, Wyslouch-Cieszynska A, Palumaa P, Hartwig A, Bal W. Quantitative electrospray ionization mass spectrometry of zinc finger oxidation: the reaction of XPA zinc finger with H(2)O(2) Analytical biochemistry. 2007;369:226–231. doi: 10.1016/j.ab.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Witkiewicz-Kucharczyk A, Bal W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicology letters. 2006;162:29–42. doi: 10.1016/j.toxlet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Kessel M, Liu SX, Xu A, Santella R, Hei TK. Arsenic induces oxidative DNA damage in mammalian cells. Molecular and cellular biochemistry. 2002;234–235:301–308. [PubMed] [Google Scholar]
- 19.Kitchin KT, Ahmad S. Oxidative stress as a possible mode of action for arsenic carcinogenesis. Toxicology letters. 2003;137:3–13. doi: 10.1016/s0378-4274(02)00376-4. [DOI] [PubMed] [Google Scholar]
- 20.Shi H, Hudson LG, Liu KJ. Oxidative stress and apoptosis in metal ion-induced carcinogenesis. Free radical biology & medicine. 2004;37:582–593. doi: 10.1016/j.freeradbiomed.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Qin XJ, Hudson LG, Liu W, Ding W, Cooper KL, Liu KJ. Dual actions involved in arsenite-induced oxidative DNA damage. Chemical research in toxicology. 2008;21:1806–1813. doi: 10.1021/tx8001548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Yoshida K, Kuroda K, Endo Y, Endo G. Effects of cysteine on the cytotoxicity of arsenic compounds. Archives of environmental contamination and toxicology. 2003;45:324–330. doi: 10.1007/s00244-002-0216-5. [DOI] [PubMed] [Google Scholar]
- 23.Schwerdtle T, Walter I, Hartwig A. Arsenite and its biomethylated metabolites interfere with the formation and repair of stable BPDE-induced DNA adducts in human cells and impair XPAzf and Fpg. DNA repair. 2003;2:1449–1463. doi: 10.1016/j.dnarep.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Jiang G, Gong Z, Li XF, Cullen WR, Le XC. Interaction of trivalent arsenicals with metallothionein. Chemical research in toxicology. 2003;16:873–880. doi: 10.1021/tx034053g. [DOI] [PubMed] [Google Scholar]
- 25.Sidorkina O, Espey MG, Miranda KM, Wink DA, Laval J. Inhibition of poly(ADP-RIBOSE) polymerase (PARP) by nitric oxide and reactive nitrogen oxide species. Free radical biology & medicine. 2003;35:1431–1438. doi: 10.1016/j.freeradbiomed.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, Hudson LG. Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. The Journal of biological chemistry. 2011;286:22855–22863. doi: 10.1074/jbc.M111.232926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter I, Schwerdtle T, Thuy C, Parsons JL, Dianov GL, Hartwig A. Impact of arsenite and its methylated metabolites on PARP-1 activity, PARP-1 gene expression and poly(ADP-ribosyl)ation in cultured human cells. DNA Repair. 2007;6:61–70. doi: 10.1016/j.dnarep.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Kang YH, Yi MJ, Kim MJ, Park MT, Bae S, Kang CM, Cho CK, Park IC, Park MJ, Rhee CH, Hong SI, Chung HY, Lee YS, Lee SJ. Caspase-independent cell death by arsenic trioxide in human cervical cancer cells: reactive oxygen species-mediated poly(ADP-ribose) polymerase-1 activation signals apoptosis-inducing factor release from mitochondria. Cancer Res. 2004;64:8960–7. doi: 10.1158/0008-5472.CAN-04-1830. [DOI] [PubMed] [Google Scholar]
- 29.Waalkes MP, Liu J, Ward JM, Diwan BA. Animal models for arsenic carcinogenesis: inorganic arsenic is a transplacental carcinogen in mice. Toxicology and applied pharmacology. 2004;198:377–384. doi: 10.1016/j.taap.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 30.Fischer JM, Robbins SB, Al-Zoughool M, Kannamkumarath SS, Stringer SL, Larson JS, Caruso JA, Talaska G, Stambrook PJ, Stringer JR. Co-mutagenic activity of arsenic and benzo [a]pyrene in mouse skin. Mutation research. 2005;588:35–46. doi: 10.1016/j.mrgentox.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Burns FJ, Uddin AN, Wu F, Nadas A, Rossman TG. Arsenic-induced enhancement of ultraviolet radiation carcinogenesis in mouse skin: a dose-response study. Environmental health perspectives. 2004;112:599–603. doi: 10.1289/ehp.6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong JT, Luo XM. Effects of arsenic on DNA damage and repair in human fetal lung fibroblasts. Mutation research. 1994;315:11–15. doi: 10.1016/0921-8777(94)90022-1. [DOI] [PubMed] [Google Scholar]