Abstract
Arp2/3 complex mediates formation of complex cellular structures such as lamellapodia by nucleating branched actin filaments. Arp2/3 complex activity is precisely controlled by more than a dozen regulators, yet the structural mechanism by which regulators interact with the complex is unknown. GMF is a recently discovered regulator of Arp2/3 complex that can inhibit nucleation and dissemble branches. We solved the structure of the 240 kDa complex of Mus musculus GMF and Bos taurus Arp2/3 and found GMF binds to the barbed end of Arp2, overlapping with the proposed binding site of WASP family proteins. The structure suggests GMF can bind branch junctions like cofilin binds filament sides, consistent with a modified cofilin-like mechanism for debranching by GMF. The GMF-Arp2 interface reveals how the ADF-H actin-binding domain in GMF is exploited to specifically recognize Arp2/3 complex and not actin.
INTRODUCTION
Arp2/3 complex, a seven-subunit 224 kD ATPase, regulates the actin cytoskeleton by nucleating branched actin filaments in response to cellular signals. Branched actin networks created by Arp2/3 complex drive processes like endocytosis, lamellipodial protrusion, phagocytosis, and intracellular motility of bacterial pathogens1. Numerous cellular activators and inhibitors modulate the activity of the complex, providing tight control over the dynamics of branched actin networks in vivo. Nucleation promoting factors, or NPFs, bind directly to Arp2/3 complex and either actin monomers or filaments to switch on nucleation activity2. NPFs discovered to date include WASP (wiskott-aldrich syndrome protein) family proteins, which bind actin monomers and Arp2/3 complex, and cortactin, Abp1, and Pan1, which bind Arp2/3 complex and actin filaments, but not actin monomers2–4. In addition, several Arp2/3 regulators are known to directly or indirectly antagonize NPFs5–12. While some Arp2/3 complex regulators have been mutationally mapped to characterize their functionally relevant regions, little is known about surfaces of Arp2/3 that interact with regulators13. The limited structural information addressing how regulators bind to Arp2/3 complex has been an obstacle to our ability to understand how the activity of the complex is controlled.
GMF (Glial maturation factor) is a recently reported Arp2/3 complex regulator from the ADF-H (actin depolymerization factor homology) domain protein family12,14,15. Most ADF-H family members, including cofilin, twinfilin, Abp1 and drebrin, bind actin filaments or both monomers and filaments to directly regulate actin16. GMF, in contrast, does not bind actin but instead directly binds Arp2/3 complex to exert its influence on the actin cytoskeleton12,14. Both fission and budding yeast GMF have been shown to inhibit the nucleation activity of Arp2/3 complex in vitro12,14. Overexpression of GMF in yeast decreases the number of endocytic actin patches, actin networks which are nucleated by Arp2/3 complex17, supporting a function for GMF in down-regulating Arp2/3 complex activity in vivo. In addition to inhibiting the complex, budding yeast GMF was shown to disassemble branches nucleated by Arp2/3 complex12. Yeast treated with the actin depolyerizing drug latrunculin show decreased rates of actin patch disassembly when GMF is knocked out, supporting a role for GMF in turning over Arp2/3-nucleated actin networks in vivo14. Because the mode of interaction of GMF with Arp2/3 complex is not known, the mechanisms by which GMF inhibits the complex or causes debranching are unclear.
We set out to determine the structural bases for GMF function by solving the crystal structure of GMFγ (hereafter referred to as GMF) bound to Arp2/3 complex. The structure revealed that GMF binds to the end of Arp2 using a binding mode similar to the interaction of other ADF-H (actin depolymerization factor homology) domains with actin monomers. The structure showed how the ADF-H domain of GMF has evolved to bind Arp2 and not actin, providing the structural foundation for understanding how biochemical functions inherent to other ADF-H domain proteins, such as filament severing, could be co-opted to operate at branch junctions instead of filament sides. The structure also indicated that GMF may complete with the WASP C region for binding to Arp2, explaining how GMFs can inhibit nucleation by the complex. Finally, GMF binding caused ordering of subdomains 1 and 2 of Arp2, providing the new structural insights into how Arp2 senses the gamma phosphate of ATP to influence the stability of branch junctions.
RESULTS
Solution of the crystal structure of GMF bound to Arp2/3 complex
We co-crystallized bovine Arp2/3 complex with mouse GMFγ in the presence of ATP and calcium and collected x-ray diffraction data to 3.2 Å resolution. The data indexed as P65, with unit cell lengths of 231.5×231.5×109.7 Å. We used the structure of unliganded Arp2/3 complex as a starting model to solve the phases by molecular replacement (1K8K.pdb)18. The structure showed clear electron density for all seven subunits of Arp2/3 complex and one molecule of GMF, which contacts both the Arp2 and ARPC1 subunits (Fig. 1, Supplementary Fig.1). The refined model includes 2027 of 2117 total residues in the assembly, and side chains were located for all but 30 of the non-glycine residues. The final model had an Rwork of 21.6 % and an Rfree of 24.1 % (Table 1).
Figure 1.
Ribbon diagram of Bos taurus Arp (actin-related protein) 2/3 complex with bound Mus musculus GMFγ, ATP and calcium. Arp3, Arp2, and actin-related protein complex subunits 1–5 (ARPC1-5) are indicated. GMFγ (pink) binds to the barbed end of Arp2 (red) and also contacts ARPC1 (green). The barbed and pointed ends of Arp2 and Arp3 are labeled (BE) or (PE), respectively.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P65 |
| Cell dimensions | |
| a, b, c (Å) | 231.54, 231.54, 109.74 |
| α,β,γ (°) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50-3.21(3.34-3.21)* |
| Rsym | 0.16(0.81) |
| I / σI | 10.5(1.7) |
| Completeness (%) | 98.9(98.3) |
| Redundancy | 3.4(2.9) |
| Refinement | |
| Resolution (Å) | 36 - 3.2 |
| No. reflections | 57721 |
| Rwork / Rfree | 0.211/0.241 |
| No. atoms | |
| Protein | 16146 |
| ATP | 62 |
| Calcium | 2 |
| Water | 0 |
| B factors | |
| Protein | 61.0 |
| Ligand/ion | 57.0 |
| Water | 53.0 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.0064 |
| Bond angles (°) | 1.18 |
Values in parentheses are for highest-resolution shell.
GMF binds the Arp2 subunit
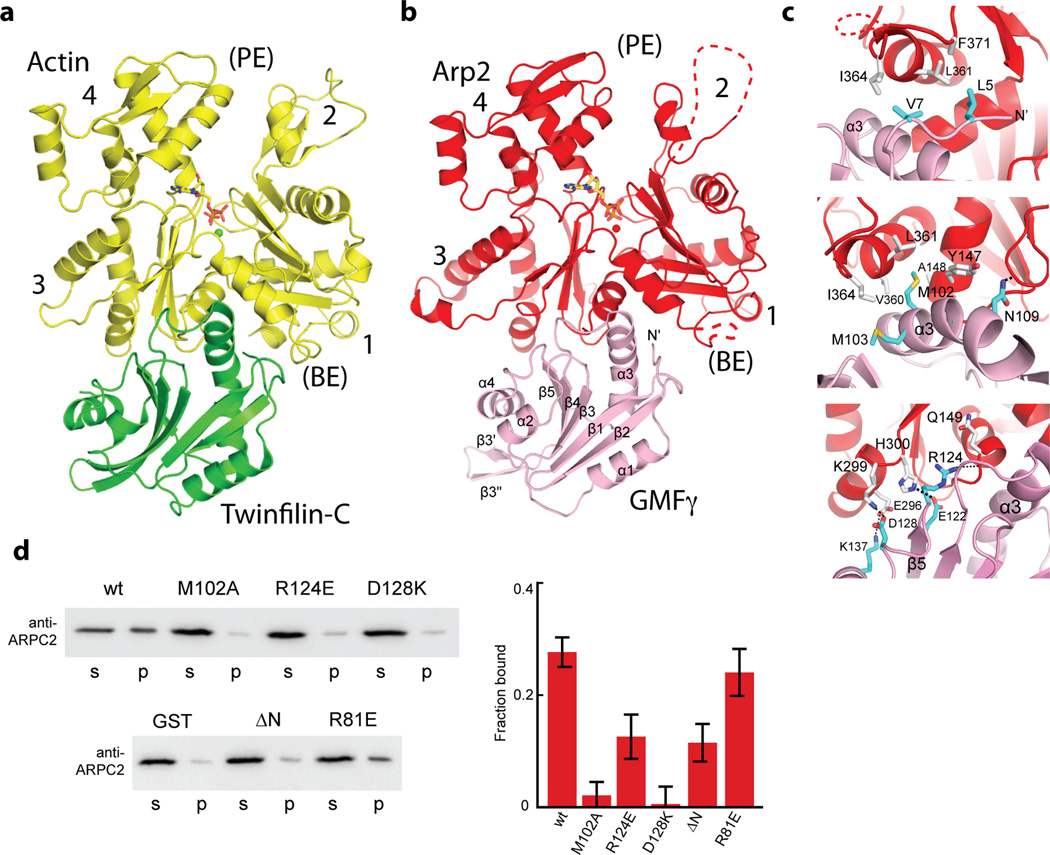
Arp2 provides the major contact surface between GMF and the complex, burying 980 Å2 of its solvent exposed surface area at the interface. This interaction occurs at the barbed end of Arp2 (Fig. 2). The mode of binding is similar to the interaction of ADF-H domain proteins twinfilin-C and cofilin with the barbed end of isolated actin monomers or with actin subunits in a filament, respectively19,20 (Fig. 2a,b). The similarities in these interfaces suggest that minor changes fine-tune the ADF-H domain of GMF to allow it to discriminate between Arp2/3 complex and actin (see below). As with other ADF-H domain proteins, the interaction with Arp2 can be broken into three regions of GMF; the N-terminus, the α3 helix, and the β5/α4 loop (Fig. 2c). The N-terminus adopts a different trajectory in the Arp2/3 bound GMF than free GMF21 (Supplementary Fig. 2), allowing it to form a hydrophobic interface with αL and the αL/αM loop on subdomain 1 of Arp2. This interface includes residues Val5 and Val7 in GMF and Leu351, Ile364 and Phe371 in Arp2. Hydrophobicity at the position of Val7 is maintained in other GMF sequences (Supplementary Fig. 3). Consistent with our structural observations, deletion of the first seven residues of GMF decreased binding to Arp2/3 complex in a GST pull-down assay (Fig. 2d, Supplementary Fig. 4). In cofilin, the N-terminus plays a critical role in mediating interactions with actin filaments, and deletion of the first five residues in budding yeast cofilin is lethal22. In addition, phosphorylation at Ser2 in cofilin regulates its activity by abolishing actin binding23. Previous reports have suggested that like cofilin, phosphorylation of GMFγ on a serine residue near the N-terminus (Ser 2) may regulate its activity15. Ser2 is disordered in the structure, and we did not find any obvious structural basis for an effect of phosphorylation of Ser2 on GMF activity.
Figure 2.
Interaction of GMF with the barbed end of Arp2. (a) Ribbon diagram of twinfilin C-terminal ADF-H domain (green) bound to ATP-Ca2+-loaded actin (yellow) generated from 3DAW. (b) Ribbon diagram of GMF (pink) bound to ATP-Ca2+-loaded Arp2 (red). The barbed and pointed ends of Arp2 or actin are labeled (BE) or (PE), respectively. Disordered regions are shown as dotted lines. (c) Ribbon diagrams showing interactions of GMF with Arp2 broken down into three regions. The top panel shows interactions between the N-terminus of GMF and Arp2. The middle panel shows the α3 helix of GMF, and the bottom panel shows the β5/α4 loop. (d) Anti-ARPC2 western blots of GST pull-down assay. GST-GMF (wild type or mutant) or GST negative control at 60 µM was bound to glutathione sepharose beads, incubated with 1 µM Arp2/3 complex and pelleted before analysis by SDS-PAGE and western blotting. The R81E mutation is not at the GMF-Arp2/3 complex interface and did not significantly influence binding. Error bars are standard errors of the mean for six separate binding reactions. P-values comparing percent wild-type versus mutant bound are as follows: M102A, 1.1×10−5; R124E, 0.008; D128K, 2.4×10−5; ΔN, 0.002; R81E, 0.48 and were determined using the student’s t-test. Uncropped versions of anti-ARPC2 western blots are show in Supplementary Figure 4.
The β5/α4 loop of GMF provides several polar contacts with Arp2. These include a hydrogen bond between Glu122 in GMF and His300 in Arp2, and salt bridges between Asp128 in GMF and Lys299 in Arp2 and K137 in GMF and Glu296 in Arp2. Consistent with the importance of these contacts, mutation of Asp128 in GMF to lysine significantly decreased binding to Arp2/3 complex (Fig. 2d). Arg124, a residue conserved in most GMF sequences (Supplementary Fig. 3), also inserts into the interface, making a hydrogen bond with the backbone of Gln149. This interaction is also important for binding Arp2/3 complex (Fig. 2d), and may be critical for specifying recognition of Arp2 over actin, as discussed below.
The long helix α3 in GMF forms the closest contact surface with Arp2, inserting into the front half of the hydrophobic groove between subdomains 1 and 3 in Arp2, referred to as the barbed end groove. Met102, a residue conserved in GMF and other ADF-H domain proteins sequences, projects into the groove, contacting a hydrophobic face formed by I364, V360, L361, A148 and Y147. These contacts are important for the interaction, as mutation of Met102 to alanine significantly decreased binding (Fig. 2d). Compared to its position in free GMF, the α3 helix rotates slightly to position Met102 into the groove (Supplementary Fig. 2). The barbed end groove in actin is a hotspot for interactions with regulatory proteins, and several proteins, including tropomyosin, N-WASp, twinfilin, profilin, and gelsolin insert a hydrophobic face of a helix into the groove to bind to the barbed end of actin24. That GMF uses the same mechanism for interaction with Arp2 indicates that the barbed end groove of Arp2 may also be a hotspot for interaction of regulatory proteins.
Contacts between GMF and ARPC1
GMFγ binding buries 380 Å2 of accessible surface area on ARPC1, contacting the outside (D) β-strand in β-propeller blade 3 (Fig. 3). Residues from the α2/β3 loop and the β4/α3 loop in GMF contribute to the interaction, with Glu63 and Gln65 from GMF forming hydrogen bonds with Lys135 and Glu126 in ARPC1. Van der Waals interactions occur between Val133 and Trp131 in ARPC1 and the aliphatic portion of Arg64 and Lys97 in GMF. Trp131 also packs against the backbone of residues 95–97 in the β4/α3 loop of GMF. Comparisons to unbound Arp2/3 complex reveal that Trp131 changes rotamers when GMF binds, allowing the favorable interactions described above and preventing a steric clash between Trp131 and Lys97 in GMF. A comparison of ARPC1 sequences from diverse species revealed that residues in the β3D strand are well conserved. In contrast, most of the residues in GMF that contact ARPC1 are not conserved (Supplementary Fig. 3). Structural differences at the ARPC1-GMF interface may underlie potential differences in the influence of GMFs from different species on Arp2/3 complex.
Figure 3.
Interaction of GMF with the ARPC1 subunit. (a) Ribbon diagram showing GMF (pink) contacts β-propeller 3 in ARPC1 (green). (b) Ribbon diagram showing close up of interactions between GMF and ARPC1.
GMF binding causes the ordering of subdomains 1 and 2 of Arp2
Comparisons to previously solved crystal structures of Arp2/3 complex revealed that GMF binding did not change the overall position of the individual subunits in the complex. However, GMF binding caused subdomains 1 and 2 of Arp2 to become ordered, whereas in all previously solved crystal structures of Arp2/3 complex, Arp2 is either partially or completely disordered25. We were able to build the entire Arp2 subunit, except for residues 36–52 and 366–368, which remained disordered (Fig. 4a). Subunits 1 and 2 of Arp2 are structurally very similar to the same subdomains in actin (Fig. 2), and overlay with an overall RMSD of 0.79 for alpha carbon atoms.
Figure 4.
Binding of GMF causes ordering of subdomains 1 and 2 of Arp2. (a) Electron density map contoured at 3.0 σ showing Fo-Fc electron density map calculated without contributions from subdomains 1 and 2 of Arp2. (b) Stereo figure comparing the nucleotide binding clefts (NBC) of ATP bound Arp2 and actin. Important structural features of the NBC are indicated by labeling representative residues in Arp2; P1 loop (Thr15), sensor loop (Asn76), and P2 loop (Val164). Note that the conformation of the sensor loop in Arp2 is identical to the sensor loop of ATP-bound actin.
ATP and calcium are bound to the Arp2 cleft, and the P1 and P2 loops are closed around the phosphates of ATP, reminiscent of ATP-bound actin structures25. Previous biochemical experiments showed that hydrolysis of ATP by Arp2 occurs after branch formation and serves as a timer to regulate the disassembly of Arp2/3 complex nucleated branches26,27. The lack of a crystal structure of the entire Arp2 subunit precluded a structural understanding of how the nucleotide state could control branch stability. In actin, the gamma phosphate is sensed through conformational changes in the P1 loop, which are then amplified by a backbone carbonyl flip in the nearby sensor loop28. Interactions with the gamma phosphate and the P1 loop keep the sensor loop flipped up, while dissociation of the phosphate causes the loop to flip down. In ATP-bound Arp2, the gamma phosphate hydrogen bonds to Thr15 in the P1 loop, and the sensor loop is switched to the up position (Fig. 4b). These interactions are identical to interactions observed in ATP-bound actin, and indicate that the sensor loop is structurally poised to sense loss of the gamma phosphate, as occurs in actin. These observations suggest that Arp2 and actin use a similar conformational relay mechanism to sense the gamma phosphate. While it is currently unknown how the flip of the sensor loop could destabilize branches, recent experiments show that small molecule inhibitors that change the conformation of the sensor loop in Arp3 may disrupt the lateral interaction between the Arp2 and Arp3 in the active state29. Changes in the position of the Arp2 sensor loop may use a similar mechanism to disrupt lateral interactions between Arp2 and the adjacent actin monomer in a daughter filament.
Conserved inserts in the actin core may modulate GMF-Arp3 interaction
Our structure suggests that GMF exerts its regulatory control over Arp2/3 complex through interactions with Arp2. Like Arp2 and actin, Arp3 has a hydrophobic barbed end groove that could potentially interact with GMF. To determine how GMF preferentially binds to Arp2, we overlaid Arp3 onto Arp2 in the GMF bound complex structure and examined the interface. While many of the residues that contact GMF in Arp2 are conserved in Arp3, Arp3 has two critical regions that differ from Arp2. The first is an insertion within its actin core, the αD/β9 insert, which lengthens the αD/β9 loop and extends the αD helix by one turn (Fig. 5a). The extended αD helix and the αD/β9 loop both clash with GMF in the model. In addition to this potential steric clash, two key interfacial residues in αD at the GMF-Arp2 interface, Tyr147 and Gly150, are Ala and Trp, respectively, in Arp3. The second critical difference is the C-terminus of Arp3, which is longer in than in Arp2 and actin, and contains a phenylalanine residue (Phe414) not present in Arp2 or actin. This residue pins the extension into the hydrophobic barbed end groove, where it would clash with α3 of GMF bound to the barbed end groove (Fig. 5b). While we cannot rule out the possibility that GMF binds to Arp330, binding would require conformational changes expected to weaken binding. Importantly, the αD/β9 insert and the C-terminal extension are present in all Arp3 sequences we examined (Supplementary Fig. 5), suggesting that Arp2 may provide the primary interaction surface for GMF from diverse species.
Figure 5.
Structural basis for the specificity of GMF for Arp2 over actin or Arp3. (a) Hypothetical structural model of Arp3 (orange) with bound GMF (pink) created by overlaying GMF bound Arp2 onto Arp3 from the GMF-Arp2/3 complex crystal structure. A close-up of the interface (right panel) shows the clash between the αD helix, the αD/β9 loop and GMF. (b) Close up of Arp3 barbed end from the GMF-Arp2/3 complex crystal structure with the α3 helix from GMF (pink) modeled into the barbed end groove as described in panel a. Arp3 (except for the C-terminal residues, shown as sticks) is shown in surface representation with hydrophobic residues colored green. (c) Stereo figure showing a comparison of the twinfilin-C:actin (3DAW) and GMF:Arp2 interfaces. Colors are as follows: GMF, pink; Arp2, red; Twinfilin-C, cyan; actin, blue. Key polar interactions are indicated with dotted lines. (d) Alternate view of interfaces described in panel (c). Approximate location of ARPC1 subunit relative to GMF is indicated.
Molecular determinants of GMF specificity for Arp2 over actin
GMF is the only one of five classes of ADF-H domain proteins (ADF/cofilins, twinfilins, Abp1/drebrins, coactosins and GMF) that does not bind actin16. To determine the structural basis for this molecular discrimination by GMF, we compared the GMF-Arp2 interface to the twinfilin-C/actin interface19. Twinfilin is unusual among the ADF-H domain proteins in that it contains tandem ADF-H domains. However, the C-terminal ADF-H domain, which is the only ADF-H domain crystallized with actin to date, binds both monomeric and filamentous actin, and thus provides a good model for understanding actin-ADFH interactions19. Comparing the structures of the two interfaces revealed that GMF specificity is achieved through matching of polar contacts at the interface and sliding of helix α3 in the barbed end groove in the GMF-Arp2 interface, which allows GMF to avoid clashing with ARPC1.
Matched polar contacts are evident in two key regions at the interface. Asp298 in the β5/α4 loop in twinfilin-C interacts with Arg147 in actin. GMF has an arginine (Arg124) in place of the Asp, creating the potential for steric clash and electrostatic repulsion in a hypothetical GMF-actin interaction (Fig. 5c). Residues at the N-terminus of α3 also appear to be critical for specificity. The basicity of Arg269 in twinfilin is conserved in twinfilins and cofilin, and forms a salt bridge with Glu334 in actin19. This interaction is not possible in a modeled GMF-actin interaction, because Arg269 in twinfilin-C is replaced by a glutamine (Gln101) in GMF, and Glu334 in actin is replaced with an arginine (Arg349) in Arp2. Together these interactions explain the specificity of GMF for Arp2 and also suggest how ADF-H domain proteins other than GMF can selectively bind actin over Arp2.
The proximity of ARPC1 to Arp2 in the assembled complex provides an additional level of specificity for the GMF-Arp2 interaction. When we overlaid actin from the twinfilin-C-actin structure with Arp2, twinfilin clashed with residues from β-strand 3D in ARPC1. To avoid this clash, GMF slides back in the barbed end groove, away from ARPC1 (Fig. 5d). Residues in the C-terminal end of the α3 helix specify this shifted binding register. In the twinfilin-C/actin interaction, a hydrogen bond between Lys276 (twinfilin-C) and Thr148 (actin) favors the twinfilin-actin register of the helix, which clashes with ARPC1. In Arp2, Thr148 is replaced by Leu152. The shift of GMF away from ARPC1 allows Leu152 in Arp2 to interact with the aliphatic portion of Lys108, and the amine moiety of Lys108 to form a salt bridge with Glu171 in Arp2. The sliding of GMF in the barbed end groove has important implications for understanding how GMF regulates Arp2/3 complex (see below)
GMF overlaps with the proposed C binding site on Arp2
GMF inhibits Arp2/3 complex activity stimulated by the VCA (Verprolin-homology, Central, Acidic) region of WASP and Scar family proteins12,14. A simple model for inhibition is that GMF blocks activation by competing with CA for binding to the complex. While neither of the two CA binding sites on the complex is well characterized, crosslinking showed that one site spans Arp2 and ARPC1, with C contacting Arp2 and the A region contacting mainly ARPC15,31. Mutational analysis suggested the C region forms an amphipathic helix, and sequence similarities comparing V and C lead to a model in which this helix fills the barbed end grooves of Arp2 and Arp3 using the same binding mode as V binding to the barbed end of actin32–34. To determine if GMF can directly disrupt CA-Arp2/3 complex interactions, we compared this model to the GMF binding site on Arp2 (Fig. 6). We found that the binding sites completely overlap, with α3 helix of GMF and the C-helix in CA superposing nearly perfectly in the barbed end groove. The helices have opposite polarity; the C-terminus of the α3 helix points away from ARPC1, while the C helix points toward it. Our analysis is consistent with biochemical experiments showing that VCA competes with GMF for binding to Arp2/3 complex14,30, and suggests a mechanism by which GMF may regulate Arp2/3 complex.
Figure 6.
GMF may block binding of C to barbed end of Arp2. (a) Model of the C region of VCA (green) binding to the barbed end of Arp2 (red). Model was constructed by overlaying structure of V bound actin (2A3Z) onto Arp2, and threading C sequence into V. GMF (pink) and Arp2 are shown as ribbons. ARPC1 is shown in surface representation with conserved residues colored either magenta or red for conservation scores of 8 (high) or 9 (very high) by analysis on the consurf server46. Residues that are basic and conserved (with scores of 8 or 9) are colored blue. Numbers indicate conserved basic or hydrophobic residues that may contact A region of VCA. The approximate position of the mother filament based on the branch junction EM model38 is indicated with a grey bar in left panel with an arrow in the direction of the barbed end. In right panel, barbed end (grey semicircle) is pointed out of the page. Regions of ARPC1 that contact ARPC4 or ARPC5 are shown with dotted lines.
Because mutational data show that much of the binding affinity of CA for the complex is mediated through interactions with A35, we next asked if bound GMF overlaps with the A binding site on ARPC1. While the precise site of A on ARPC1 is not known, the surface area buried between ARPC1 and GMF is relatively small and leaves two major swaths of conserved surface residues on ARPC1 exposed (Fig. 6). Within one swath is a group of conserved basic residues that may interact with acidic residues in A. Some of these basic residues, either alone or in combination, have been shown to be important for viability of budding yeast, corroborating their functional relevance36. Also in the conserved swaths are candidate hydrophobic residues that could potentially interact with the conserved tryptophan in A, which is important for interaction of A with ARPC135. Therefore, our structural analyses suggest that A may partially or fully engage ARPC1 even if GMF is bound to the complex. Additional biochemical and structural information will be required to evaluate the importance of this potential contact in regulation of the complex by GMF.
Insights into debranching from a model of GMF at the branch junction
Budding yeast GMF has been reported to disassemble Arp2/3 nucleated branches12. Unlike cofilin, which stimulates debranching indirectly though interactions with the mother filament of actin37, GMF is thought to directly bind to Arp2/3 complex at filament junctions to dissemble branches12. We modeled GMF bound at a branch junction to investigate how debranching might occur38. Superposing Arp2 from the GMF-Arp2/3 co-crystal structure onto the EM reconstruction of a branch junction revealed that the GMF binding site is accessible in the assembled branch (Fig. 7). At the branch junction, GMF simultaneously contacts the barbed end of Arp2 and subdomains 1 and 2 of the adjacent actin monomer (subunit D2). The mode of binding is similar to interaction of cofilin with adjacent subunits in an actin filament20, and suggests that GMF may use a cofilin-like mechanism to sever the daughter filament at the branch junction. Consistent with this observation, recent mutational analysis of budding yeast GMF shows that residues in the “F-actin binding” region of GMF are important for debranching30.
Figure 7.
Model of GMF bound to a branch junction. (a) Model of GMF (spheres) placed into EM branch junction reconstruction by overlaying Arp2 from the GMF-Arp2/3 complex structure onto Arp2 from the EM branch junction model38. Spheres in GMF are colored according to conservation (blue=most variable, cyan= variable, pink=high conservation, magenta=highest conservation)46. Actin subunits in daughter filament are labeled D1 and D2. The approximate position of the mother filament is shaded grey, with barbed end pointed out of page. (b) Close up of hypothetical model of GMF at a branch junction. Regions of clash are indicated with red arrowheads. The direction of sliding of GMF in the barbed end groove is indicated with an arrow. Subdomains 1 and 2 of actin subunit D2 and beta propeller 3 of ARPC1 are indicated. (c) Same as (a), but rotated to show surface of GMF in contact with Arp2 and actin subunit D2. (d) Ribbon diagram of GFM showing most conserved residues in GMF sequences (magenta) and approximate regions of contact with Arp2 and subunit D2 (“F-actin binding region”). The orientation of GMF in this panel is the same as in (c).
However, key differences between cofilin and GMF point to potential mechanistic differences. First, sliding of GMF in the barbed end groove of Arp2, described above, moves GMF towards actin subunit D2, creating steric clash between residues in GMF with subdomains 1 and 2 of subunit D2 (Fig. 5d, 7). This suggests that binding of GMF might cause a change in the Arp2-D2 interface that could destabilize the junction. Because cofilin does not “slide back” in the groove of actin, this change is likely distinct from cofilin-induced changes to actin filaments20. Second, the region of GMF that contacts actin subunit D2, termed the “F-actin binding region” is structurally distinct in GMF. It includes a short antiparallel beta sheet (β3’ – β3”) between β3 and β4 not present in other ADF-H proteins (Fig. 7). This region harbors several conserved residues specific to GMF, including Asp79, Arg81, Ser83 and Pro85 (Supplementary Fig. 3). The analogous region in cofilin directly contacts actin filaments and has been shown to contribute to actin filament binding, but contains a distinct set of residues20,22 (Supplementary Fig. 3). GMF specific residues at the end of helix α1 and in the α1/β1 loop also contact subunit D2 in the branch model. While this segment has not been mutationally probed in cofilin, it makes close contacts with the filament in an EM reconstruction of cofilin bound filaments20. Together, these observations support a modified cofilin-like mechanism for GMF mediated disassembly of branch junctions.
GMF may block actin monomer recruitment during activation
Comparison of the GMF-bound complex to the EM reconstruction of a branch junction revealed a potential structural impediment to activation when GMF is bound38. As noted above, GMF at the branch junction clashes with the actin monomer (D2) bound to the barbed end of Arp2 (Fig. 7). This suggests that GMF may block longitudinal contacts with the actin monomer recruited by VCA to the barbed end of Arp2 during activation39, providing an additional level of regulation of the complex.
DISCUSSION
Diverse classes of Arp2/3 complex inhibitors target distinct steps in the branching nucleation pathway. Inhibitors like coronin5, tropomyosin8, caldesmon11 and EPLIN7 exploit the requirement of Arp2/3 complex to bind pre-existing filaments to downregulate the nucleation reaction. These proteins bind actin filaments to block Arp2/3 complex binding sites, thereby indirectly inhibiting the complex. Other inhibitors, such as the PDZ-BAR-domain protein Pick1, harbor acidic regions that mimic the A region of VCA, and directly compete with VCA for binding to the complex10. GMF may use a similar mechanism to block activator binding, targeting the C binding site instead of A. However, a simple competition mechanism cannot fully explain inhibition, because GMF binding is unlikely to block A binding sites on ARPC1. We hypothesize that in addition to competing with VCA for the C binding site, GMF may also block actin monomer recruitment through two mechanisms. First, displacement of C from Arp2 by GMF may prevent proper positioning of actin to the barbed end of Arp2. Second, GMF may directly block interactions between actin and the barbed end of Arp2, as mentioned above. Finally, we note that some Arp2/3 complex regulators, including small molecule inhibitors CK-666 and CK-869, have been shown to directly target the activating conformational change in Arp2/3 complex stimulated by VCA and actin monomers29. Averaged single particle EM images suggest GMF may block the movement of the complex into an activated conformation30. While our structure does not provide an obvious mechanism by which GMF could block this step, without higher resolution structures of the activated state we cannot rule out this possibility.
Our data show that GMF overlaps with the proposed C site on Arp2, but does not bind to the proposed C site on Arp3. The ability of GMF to preferentially target Arp2 may play an important role in defining its influence on the complex, and is consistent with the conserved structural features on Arp3 that appear to block GMF binding. Another Arp2/3 regulator, cortactin, specifically binds Arp3 where it competes with VCA40. However, instead of inhibiting WASP-induced activation of the complex, cortactin synergizes with WASP to dramatically increase nucleation41. Determining precisely how the binding mode of a regulator influences its ability to modulate Arp2/3 activity will be critical for understanding not only the structural mechanism of activation, but the complex interplay between regulators in vitro and in vivo.
Arp2/3 nucleated branches dissociate on the timescale of minutes in vitro, but Arp2/3-medited networks turn over in seconds in vivo42. Debranching contributes to turnover in yeast actin patches and lamellipodia14,27, and may play a general role in remodeling dynamic actin networks. GMF, coronin1B and cofilin, three proteins reported to have debranching activity, each have a distinct mechanism that likely defines their function in a cellular context37,43. Our data suggests GMF targets branch junctions at the interface between Arp2 and the daughter filament, whereas cofilin and coronin1B bind actin filaments or both actin filaments and Arp2/3 complex, respectively. These distinctions will influence how effectively each debrancher competes with actin binding proteins that target and stabilize branch junctions, like cortactin, or filament sides, like tropomyosin44,45. Understanding how the debranching activity of GMF and other debranchers are influenced by the cellular milieu of actin binding proteins will be critical for understanding how actin filament networks are turned over in vivo.
ONLINE METHODS
Protein Expression and Purification
We purified Bos taurus Arp2/3 complex as previously described25. Mouse GMFγ was subcloned from a plasmid provided by Bruce Goode into the pGV6747, which tags the N-terminus with glutathione-S-transferase and a TEV protease tagging site. Point mutations and truncations of GMFγ were made in the context of the pGV67 plasmid. BL21(DE3)RIL E. coli transformed with GMF in pGV67 were grown to an OD600 of 0.6 before adding 0.4 mM IPTG and allowing expression at 22 °C overnight. Cells were harvested and lysed by sonication. Clarified lysate was loaded onto a GS4B glutathione affinity column, washed with binding buffer (20 mM Tris 8.0, 140 mM NaCl, 2 mM EDTA, 1mM DTT), and eluted with binding buffer containing 50 mM reduced glutathione. TEV protease was added to pooled fractions and the sample was dialyzed overnight against 10 mM CHES pH 9.5, 25 mM NaCl, 1 mM DTT. The dialyzed sample was then loaded onto a 6 mL resource Q column and eluted with a 25 mM to 500 mM gradient. Peak fractions were pooled, concentrated, and purified on a Superdex 75 gel filtration column in 20mM Tris 8.0, 100 mM NaCl, and 1 mM DTT.
GST pull-down assays
GST-GMF at 60 µM was bound to glutathione sepharose beads and incubated with 1 µM Bos taurus Arp2/3 complex in 50 mM KCl, 10 mM imidazole pH 7.0, 1 mM EGTA, 1 mM MgCl2, 0.2 mM adenosine diphosphate (ADP), and 1 mM dithiothreitol for 1 hr at 4 °C. Samples were spun, and both supernatant and washed pellet were loaded on SDS-PAGE gels. Arp2/3 complex in supernatant and pellet was visualized by blotting with an anti-ARPC2 antibody (Millipore #07-227) diluted 1:1000 and a donkey anti-rabbit IgG-HRP antibody (Santa Cruz, sc2313) diluted 1:10,000. The fraction bound was measured using Li-Cor imaging software.
Crystal growth, data collection and refinement
A solution containing 25 µM Arp2/3 complex, 25 µM GMFγ in 50 mM Tris pH 8.0, 500 µM ATP, 500 µM CaCl2, and 1 mM DTT was mixed 1:1 with 10.6 % polyethylene glycol 400 and allowed to equilibrate by vapor diffusion from a hanging drop at 4 °C. Crystals grew to ~50×50×100 microns in approximately 10 days. Crystals were cryoprotected by direct addition of a solution of 50 mM Tris pH 8.0, 50 % PEG 400, 500 µM ATP and 500 µM CaCl2 and flash frozen in liquid nitrogen. Data were collected at 100 K at a wavelength of 0.9793 Å at beamline 19-ID at Argonne National Laboratory and processed using HKL200048. A molecular replacement solution was found with Phaser49, using the ATP-bound-structure of BtArp2/3 complex as a search model (1TYQ). Refinement was initiated by rigid body minimization in ccp450, allowing each subunit of the complex, plus subdomains 1 and 2 of Arp3 to move independently. Minimization was continued in Refmac, using tight geometry constraints (weighting set at 0.002), jelly body refinement (sigma at 0.01) and TLS refinement. Density for GMF was clearly visible even in the first electron density maps, but was not added until the second round of refinement. Subdomains 1 and 2 of Arp2 were built piecemeal starting in the second round of refinement. The Ramachandran statistics for the final refined structure are: Most favored residues: 1581 (89.0%), additionally allowed regions: 195 (11.0%), generously allowed: 0 (0%), disallowed 0 (0%).
Supplementary Material
ACKNOWLEDGEMENTS
Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, beamline 19ID. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank S. Ginell, J. Lazarz, B. Nocek and Y. Kim for assistance with remote data collection. We thank B. Goode (Brandeis University, Waltham, MA, USA) for sending mouse GMFγ expression plasmids, and K. Needham for help with cloning and protein purification. We would also like to acknowledge K. Prehoda for comments on the manuscript. This work was financially supported by a US National Institutes of Health Grant GM092917 (to B.J.N) and the Pew Scholars in the Biomedical Sciences program.
Footnotes
ACCESSION CODES
4JD2
AUTHOR CONTRIBUTIONS
Q.L. and B.J.N designed the research, Q.L. performed all experiments, Q.L. and B.J.N analyzed the data and wrote the paper.
REFERENCES
- 1.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2012;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 2.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 3.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 5.Liu SL, Needham KM, May JR, Nolen BJ. Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J Biol Chem. 2011;286:17039–17046. doi: 10.1074/jbc.M111.219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries CL, et al. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maul RS, et al. EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J Cell Biol. 2003;160:399–407. doi: 10.1083/jcb.200212057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 9.Maritzen T, et al. Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. Proc Natl Acad Sci U S A. 2012;109:10382–10387. doi: 10.1073/pnas.1206468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamakita Y, Oosawa F, Yamashiro S, Matsumura F. Caldesmon inhibits Arp2/3- mediated actin nucleation. J Biol Chem. 2003;278:17937–17944. doi: 10.1074/jbc.M208739200. [DOI] [PubMed] [Google Scholar]
- 12.Gandhi M, et al. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol. 2010;20:861–867. doi: 10.1016/j.cub.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci U S A. 2011;108:E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano K, Kuwayama H, Kawasaki M, Numata O, Takaine M. GMF is an evolutionarily developed Adf/cofilin-super family protein involved in the Arp2/3 complex-mediated organization of the actin cytoskeleton. Cytoskeleton (Hoboken) 2010;67:373–382. doi: 10.1002/cm.20451. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda K, et al. Glia maturation factor-gamma is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ Res. 2006;99:424–433. doi: 10.1161/01.RES.0000237662.23539.0b. [DOI] [PubMed] [Google Scholar]
- 16.Poukkula M, Kremneva E, Serlachius M, Lappalainen P. Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 2011;68:471–490. doi: 10.1002/cm.20530. [DOI] [PubMed] [Google Scholar]
- 17.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 18.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 19.Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. Structure of the actindepolymerizing factor homology domain in complex with actin. J Cell Biol. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci U S A. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goroncy AK, et al. NMR solution structures of actin depolymerizing factor homology domains. Protein Sci. 2009;18:2384–2392. doi: 10.1002/pro.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. Embo J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 24.Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- 27.Ingerman E, Hsiao JY, Mullins RD. Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J Cell Biol. 2013;200:619–633. doi: 10.1083/jcb.201211069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 29.Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small molecules CK-666 and CK-869 inhibit Arp2/3 complex by blocking an activating conformational change. Chem Biol. 2013 doi: 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ydenberg C, et al. GMF Severs Actin-Arp2/3 Complex Branch Junctions by a Cofilin-like Mechanism. Curr Biol. 2013;23:1–9. doi: 10.1016/j.cub.2013.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci U S A. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- 33.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irobi E, et al. Structural basis of actin sequestration by thymosin-beta4: implications for WH2 proteins. Embo J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 36.Balcer HI, Daugherty-Clarke K, Goode BL. The p40/ARPC1 subunit of Arp2/3 complex performs multiple essential roles in WASp-regulated actin nucleation. J Biol Chem. 2010;285:8481–8491. doi: 10.1074/jbc.M109.054957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boczkowska M, et al. X-Ray Scattering Study of Activated Arp2/3 Complex with Bound Actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver AM, et al. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- 41.Uruno T, Liu J, Li Y, Smith N, Zhan X. Sequential interaction of actin-related proteins 2 and 3 (Arp2/3) complex with neural Wiscott-Aldrich syndrome protein (N-WASP) and cortactin during branched actin filament network formation. J Biol Chem. 2003;278:26086–26093. doi: 10.1074/jbc.M301997200. [DOI] [PubMed] [Google Scholar]
- 42.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weaver AM, et al. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- 45.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the Factin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 46.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online methods
- 47.Nolen BJ, Pollard TD. Structure and biochemical properties of fission yeast ARP2/3 complex lacking the ARP2 subunit. J Biol Chem. 2008;283:26490–26498. doi: 10.1074/jbc.M802607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data dollected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.