Abstract
Systemic cancer therapy has traditionally exploited vulnerabilities in cancer cells, a strategy which has become more precise with the identification and targeting of driver oncogenes. However, persistent tumor growth due to primary (de novo) or secondary (acquired) resistance limits therapeutic efficacy for many patients. Alternative splicing is important for increasing the diversity of the cellular proteome, and is a process frequently de-regulated during cancer development and progression. In cancer cells, diverse splicing alterations have been identified that eliminate protein domains or enzymatic activities required for efficacy of cancer therapies, promote gain of novel signaling functions that circumvent cancer therapies, and uncouple signaling pathways from upstream regulatory points that are blocked by cancer therapies. The mechanisms underlying these splicing changes range from stable alterations in gene sequence/structure to de-regulation of splicing regulatory factors. In this review, the role of splice variants in cancer therapy resistance will be discussed, with examples of how mechanistic understanding of these processes has led to the development of novel strategies for therapy re-sensitization.
Keywords: splicing, cancer, therapy resistance, primary resistance, secondary resistance, targeted therapy, chemotherapy, radiation therapy
mRNA Splicing Variants and Cancer Therapy Resistance
The spliceosome is composed of small nuclear ribonucleoproteins (snRNPs: U1, U2, U4, U5, and U6) and more than 200 polypeptides. This macromolecular complex processes newly-transcribed precursor mRNAs (pre-mRNAs) in order to remove intronic sequences and join exons, giving rise to mature messenger RNAs (mRNAs). Intronic motifs that direct this process are the 5′ splice donor site, the 3′ splice acceptor site, and the branch site near the 3′ end of the intron, which form base pair contacts with the small nuclear RNA (snRNA) components of snRNPs. The U1 and U2 snRNPs recognize the splice donor and branch sites, respectively, which are key early steps in intron recognition. The recognition of these introns is regulated by diverse intronic and exonic splicing enhancer (ISE and ESE) and suppressor (ISS and ESS) elements. Alternative splicing is a process whereby introns are differentially identified and removed from pre-mRNAs, allowing multiple mRNA configurations of joined exons to arise from a single gene. This adds diversity to protein architectures and supports biological complexity. Indeed, most genes express multiple alternatively-spliced mRNAs simultaneously, although one isoform is usually dominant (1). A fundamental characteristic of cancer cells is the de-regulation of normal cellular processes, and de-regulation of splicing is no exception. This has been the topic of excellent reviews focused on de-regulation of the spliceosome and splicing plasticity in cancer cells (2, 3).
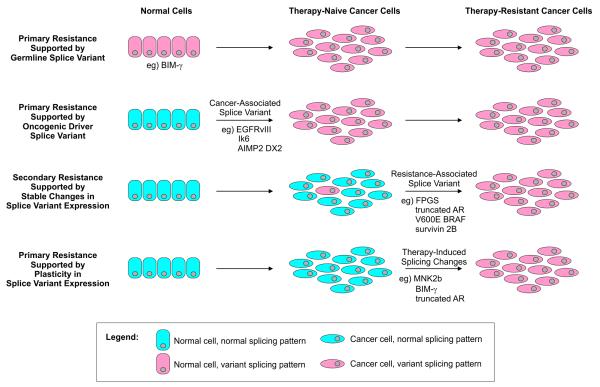
Many new targeted therapies have been approved for clinical use that inhibit specific de-regulated proteins or pathways, which has increased the precision of systemic cancer treatment compared with conventional chemotherapy. However, drug resistance is a major factor that limits effectiveness for both targeted and conventional therapies. Resistance can manifest as primary resistance, wherein cancer cells are intrinsically resistant prior to therapy, or secondary resistance, wherein resistance develops despite initial responses. The mechanisms underlying primary and secondary cancer therapy resistance are complex, including reduced intracellular drug accumulation, drug inactivation, alterations in the drug target, alterations in processing of drug-induced cellular damage, and evasion of apoptosis (4). Importantly, splicing alterations can be drivers of therapeutic resistance in cancer cells (5). In some instances, these resistance-associated splicing alterations are due to global defects in spliceosome regulation. However, mRNA splicing in cancer occurs against a backdrop of germline variability and genomic instability. In these cases, cancer cells can display stable patterns of “alternative” splicing due to sequence and/or structural alterations in the normal gene template. The goals of this review are to highlight select mechanisms of splicing alterations that underlie primary and secondary resistance to cancer therapy, and to assign points in the natural history of cancer development and progression where these splicing alterations can occur (Fig. 1).
Figure 1.
Diverse roles for alternative splicing in primary and secondary resistance to cancer therapy. Models for splice variant association with cancer therapy resistance, and the various points in the natural history of cancer progression where these splice variants have been shown to occur. In cases of primary resistance, the majority of tumor cells express resistance-associated splice variants, or can undergo rapid, acute changes in splicing dynamics in response to therapy. In cases of secondary resistance, rare cancer cells expressing splice variants that support survival may exist prior to therapy, or could arise during therapy, ultimately leading to therapy-resistant tumor cell populations. Detailed mechanisms by which the indicated splice variants can support primary or secondary resistance to therapy, and mechanisms underlying their synthesis are discussed in the text.
Primary Resistance Supported by Germline Splicing Variants
BIM-γ and Tyrosine Kinase Inhibitor Resistance
The tyrosine kinase inhibitor (TKI) imatinib is the first targeted cancer therapy developed for inhibition of the BCR-ABL fusion in CML. Up-regulation of BCL2-like 11 (BIM) is required for induction of apoptosis by TKIs, and BIM suppression is sufficient for TKI resistance (6). Multiple BIM splice variants have been described, one of which is BIM-γ, lacking the exon 4-encoded BH3 domain required for BIM pro-apoptotic function. Whole-genome sequencing of imatinib-resistant CML samples led to the identification of a recurrent 2,903 bp deletion in intron 2 of the BIM gene (7). This deletion, which favors BIM exon 3 splicing over exon 4, is a germline polymorphism occurring in 12.3% of the East Asian population that was found to be associated with an increased likelihood of CML resistance to imatinib and second-line TKIs (7). This BIM deletion polymorphism was also found to be associated with decreased progression free survival in lung cancer, a disease that can be treated with the epidermal growth factor receptor (EGFR) TKI, gefitinib (7). Based on its germline origin, this deletion polymorphism would be expected to drive primary resistance to TKIs. Indeed, cell line models of CML and non-small cell lung cancer with an engineered BIM deletion displayed resistance to imatinib and gefitinib, respectively. However, imatinib or gefitinib sensitivity could be restored therapeutically in deletion polymorphism-positive CML or lung cancer cells by treatment with a BH3 mimetic drug (7).
Primary Resistance Supported by Oncogenic Driver Splicing Variants
EGFRvIII and Radiation/Chemotherapy Resistance
Various EGFR splice variants have been described, the most frequent of which is EGFRvIII, an NH2-terminally truncated EGFR variant similar to the v-erb-B oncogene (8). EGFRvIII does not bind any known ligand, and displays constitutive, ligand-independent tyrosine kinase activity (8). EGFRvIII is expressed in 20-30% of glioblastoma multiforme (GBM), a disease which is highly resistant to radiation and chemotherapy. Radiation resistance of GBM is mediated by EGFRvIII activation of the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), which supports enhanced repair of DNA double strand breaks (9). This led to the discovery that EGFRvIII-expressing cells could be re-sensitized to radiation therapy via inhibition of DNA-PKcs kinase activity, which delayed DNA repair kinetics (9). Chemoresistance of GBM has also been linked to EGFRvIII through sustained activation of mammalian target of rapamycin (mTOR) complex 2 (mTORC2) (10). mTORC2 is activated frequently in GBM, leading to activation of NF-κB, and this EGFRvIII-mTORC2-NF-κB pathway was shown to drive resistance to cisplatin, temozolomide, and etoposide. This led to the discovery that blockade of the mTORC2 or NF-κB pathways with pharmacologic inhibitors re-sensitized GBM cells to these DNA damaging agents (10).
In GBM, expression of EGFRvIII is linked to complex rearranged and amplified EGFR gene structures (8). For example, there is a strong statistical correlation between EGFR intragenic losses encompassing EGFR exons 2-7 and expression of EGFRvIII in GBM (11). Similarly, EGFRvIII is expressed in ~10% of lung squamous cell carcinoma due to underlying intragenic deletions in the EGFR gene, and this is associated with TKI resistance (12). These studies demonstrate that oncogenic EGFRvIII expression in cancer is a stable phenomenon caused by somatic architectural changes in the EGFR gene, which drives primary radio- and chemo-resistance in various human cancers.
Ik6 and Imatinib Resistance
In BCR-ABL positive acute lymphoblastic leukemia (ALL), primary imatinib resistance has been linked to a loss-of-function Ikaros splicing variant, Ik6. Ikaros is important for development of lymphocytes and other hematopoietic lineages. The Ik6 splice variant resulting from skipping of exons 3-6 of the Ikaros gene (IKZF1) lacks all NH2-terminal zinc finger DNA binding motifs and displays localization to the cytoplasm as opposed to nucleus (13). Mechanistically, Ik6 synthesis is due to intragenic deletions of IKZF1 exons 3-6, which appears to be a driver event during lymphoid blast crisis when CML progresses to ALL (14). In a cohort of 47 BCR-ABL positive ALL patients, Ik6 expression was detected in 43 patients, and in 23 of these cases Ik6 was the predominant isoform (13). The level of Ik6 expression in these clinical specimens was proportional to the percentage of blast cells, indicating that Ik6 expression is restricted to the blast cell population. Importantly, ectopic expression of Ik6 in a BCR-ABL positive ALL cell line was sufficient to drive resistance to imatinib (13).
AIMP2-DX2 and Doxorubicin Resistance
Aminoacyl-tRNA synthetase-interacting multifunctional protein 2 (AIMP2) is a haploinsufficient tumor suppressor gene important for controlling cell fate. A splice variant of AIMP2 lacking exon 2, termed AIMP2-DX2, occurs in lung, and perhaps other cancers (15). Following treatment with etoposide or doxorubicin, wild-type AIMP2 mediates the apoptotic DNA damage response by binding p53 and preventing Mdm2-mediated degradation. The AIMP2-DX2 splice variant also binds p53, but does not inhibit Mdm2-mediated degradation. AIMP2-DX2 expression in lung cancer specimens is associated with lung cancer stage and shorter overall survival (15). In a carcinogen-induced mutation model, normal lung cells surviving carcinogen treatment showed increased expression of AIMP2 DX2, concomitant with several mutations in the AIMP2 gene. In a minigene reporter assay, modeling one of the mutations in AIMP2 exon 2 showed that this mutation disrupted binding of the spliceosome assembly factor serine/arginine rich splicing factor (SRSF)1 to an exon 2 ESE, leading to exon 2 skipping (15). These findings implicate a role for AIMP2 exon 2 ESE mutations and AIMP2 DX2 expression in oncogenesis as well as primary resistance to chemotherapy in lung cancer, which is consistent with p53 suppressive function.
Secondary Resistance Supported by Stable Changes in Splice Variant Expression
FPGS Splicing Alterations and Antifolate Resistance
Upon cellular uptake, folates and antifolates such as methotrexate undergo FPGS-mediated polyglutamylation, which prevents cellular efflux. In methotrexate-resistant ALL cell lines and clinical ALL specimens, diverse FPGS splicing aberrations occur, including intron retention and exon exclusion (16). FPGS splicing defects did not occur in methotrexate-sensitive parental lines, indicating that these events support secondary resistance. Indeed, a FPGS splice variant resulting from exon 10 skipping was unable to support FPGS activity in stably-transfected cells (16). Interestingly, altered FPGS splicing in methotrexate-resistant ALL cell lines was associated with altered splicing patterns for various additional genes, indicating a stable, global defect in splicing regulation. It will be important for future studies to identify component(s) of the spliceosome that may be responsible for these global splicing alterations in methotrexate-resistant ALL, as this may reveal opportunities for restoring therapeutic sensitivity.
Truncated Androgen Receptor Variants and Castration-Resistance
The androgen receptor (AR) is a steroid receptor transcription factor and lineage survival gene in cells of prostatic origin. Because of its lineage survival role, AR is a central therapeutic target in advanced prostate cancer. Multiple AR splice variants have been identified in castration-resistant prostate cancer cell lines, xenografts, and transgenic mouse models (17). These splice variants arise through cryptic exon inclusion or exon skipping, ultimately leading to synthesis of truncated AR variant proteins lacking the ligand-binding domain (LBD), which is the protein domain through which AR activity is targeted in prostate cancer (17). In various functional experiments, truncated AR variants have been shown to translocate to the nucleus and drive constitutive, ligand-independent activation of the AR transcriptional program in a manner insensitive to antiandrogens (18, 19). In castration-resistant prostate cancer cell lines and tissues, intragenic rearrangements in the AR gene have been shown to underlie high level expression of truncated AR variants (19-21). Moreover, in a cell-based model of prostate cancer progression, emergence of castration-resistant cells under conditions of AR-targeted therapy is associated with enrichment of cells harboring AR gene rearrangements and high-level expression of truncated AR variants (20). These studies indicate that AR gene rearrangements can drive secondary resistance to AR-targeted therapy by supporting expression of constitutively active truncated AR variants lacking the AR ligand binding domain.
Truncated V600E BRAF Splice Variants and Vemurafenib Resistance
Oncogenic V600E BRAF mutations are frequent in melanoma, and serve as the molecular target for the V600E BRAF kinase inhibitor vemurafinib. Vemurafinib inhibits constitutively active V600E BRAF monomers, but paradoxically activates wild-type BRAF dimers, which require active Ras signaling for dimerization and activation. In many vemurafinib resistant melanoma cell lines as well as tissue from patients displaying vemurafinib resistance, a V600E BRAF splice variant is expressed, which results from skipping of BRAF exons 4-8 (22). BRAF exons 4-8 encode the Ras-binding domain, and deletion of this domain allows the shorter splice variant of V600E BRAF to undergo constitutive dimerization in a Ras-independent manner. Despite vemurafinib binding to this truncated BRAF V600E splice variant, downstream activation of ERK persists (22), demonstrating that this is an important driver of BRAF V600E reactivation and secondary resistance to vemurafinib in melanoma. The mechanism for synthesis of this truncated BRAF variant is not clear, but skipping of exons 4-8 is restricted to mRNAs harboring the V600E mutation (22). This implies a mechanism of allele-specific alternative splicing, or underlying genomic alterations involving the BRAF allele harboring the V600E mutation.
Survivin 2B and Taxane Resistance
Increased survivin expression has been observed in taxane-resistant ovarian cancer. The survivin (BIRC-5) gene encodes 5 alternatively spliced isoforms. The development of isoform-specific RT-PCR primer sets led to the finding that the survivin 2B isoform is responsible for most of the increase in total survivin expression in isogenic sets of taxane-resistant vs. taxane-sensitive ovarian cancer cell lines (23), indicating that survivin 2B mediates secondary resistance to taxanes. Consistent with this finding, isoform-specific siRNA-mediated knockdown of survivin 2B increased apoptosis, inhibited cell cycle progression, and sensitized resistant ovarian cancer cell lines to docetaxel (23). This mechanistic finding was translated to a preclinical subcutaneous model of ovarian cancer, wherein liposome nanoparticles were used for systemic delivery of survivin 2B-targeted siRNA, resulting in reduced tumor growth and enhanced docetaxel sensitivity in vivo (23).
Resistance Supported by Plasticity in Splice Variant Expression
MNK2b and Gemcitabine Resistance
The mitogen activated protein kinase interacting kinase-2 (MNK2) gene gives rise to two splice variants differing only in composition of their COOH-termini due to alternative splicing of exon 13a (MNK2a isoform) or exon 13b (MNK2b). Exon 13a, but not Exon 13b, encodes a mitogen activated protein kinase (MAPK)-interacting domain. Therefore, the MNK2b splice isoform does not harbor a MAPK interacting domain, and displays constitutive, MAPK-independent kinase activity (24). Gemcitabine is one of the main treatment modalities for advanced pancreatic cancer, but primary resistance occurs frequently. Pancreatic cancer tissues display high levels of S209 phosphorylation of eIF4E, the core component of the translation initiation complex, and levels of eIF4E S209 phosphorylation are associated with worse overall survival (25). In pancreatic cancer cell lines, eIF4E S209 phosphorylation increases acutely in response to gemcitabine, cisplatin, and rapamycin, and these effects persist during treatment. In addition to increased eIF4E S209 phosphorylation, gemcitabine treatment also results in acute changes in MNK2 splicing favoring production of MNK2b (25). This plasticity in splicing favoring MNK2b was shown to be due to gemcitabine-mediated induction of SRSF1. Interestingly, although both MNK2a and MNK2b can mediate S209 phosphorylation of eIF4E, only MNK2a requires activation by upstream MAPK signaling. Indeed, ectopic expression of MNK2b but not MNK2a could drive gemcitabine resistance in pancreatic cancer cell lines. Importantly, an MNK chemical inhibitor or siRNA directed to MNK2 re-sensitized pancreatic cancer cells to gemcitabine. This effect was likely mediated through the MNK2b splice variant, as knock-down of SRSF1 led to an increased MNK2a:2b ratio, reduced eIF4E phosphorylation and gemcitabine re-sensitization (25).
Expression plasticity of BIM-γ in breast cancer
SRSF1 has also been implicated in plasticity in BIM splicing in breast cancer, which highlights an additional mechanism for increased expression of the BIM-γ splice variant. In a model of breast cancer induced by overexpression of SRSF1, synthesis of the BIM-γ was observed (26). Overexpression of BIM-γ was able to drive increased acinar size and decreased apoptosis in mammary epithelial cells, indicating it may play an active oncogenic role (26). This contrasts with functional analysis of BIM-γ in CML harboring an underlying BIM deletion polymorphism indicated that this was a loss-of-function splice variant (7). More broadly, SRSF1 overexpression has been observed in various human cancers, and SRSF1 overexpression by itself is transforming in various cell types (26, 27). This indicates that SRSF1 may play a more global role in supporting splicing plasticity and therapeutic resistance in cancer cells, which can be manifest in part through expression BIM-γ and MNK2b splice variants.
Expression plasticity of truncated AR variants in prostate cancer
Plasticity in AR splicing has also been observed in prostate cancer, with expression of truncated AR variants lacking the AR ligand binding domain displaying acute expression increases in response to castration or treatment with enzalutamide, a next-generation AR antagonist (18). This indicates that another mechanism exists in addition to AR gene rearrangements as a basis for truncated AR variant expression. Interestingly, these acute increases in truncated AR variant expression were accompanied by increased expression of several genes involved in mitotic progression, indicating that truncated AR variants support a novel transcriptome that maintains prostate cancer cell mitosis during AR-targeted therapy (18).
Opportunities for Development of New Therapies
Several of the above examples have illustrated that mechanistic understanding of splice variants and their role in therapeutic resistance can lead to novel treatment ideas (eg. BH3 mimetics to enhance TKI sensitivity in BIM-γ driven cancers, TKIs to increase radiation sensitivity in EGFRvIII-driven GBM, nanoparticle delivery of siRNAs to selectively target the survivin 2B splice variant in ovarian cancer, etc.). However, all of these strategies are based on inhibiting expression of splice variants, or targeting pathways downstream of splice variant expression. More recently, strategies have been developed to target the splicesosome in cancer cells directly. For example, knock-down of SRSF1 can restore therapeutic sensitivity in cancers where resistance is driven by MNK2b or BIM-γ (25, 26). Similarly, shRNA-mediated knock-down of serine-arginine protein kinase (SRPK)-1, a kinase that phosphorylates SR domains in SRSF1 and other SR splicing factor proteins, leads to increased sensitivity of pancreas, breast, and colon cancer cell lines to gemcitabine and cisplatin (28). There are also several antitumor drugs such as spliceostatin A and pladienolides that target the splicing factor 3B (SF3B) component of the spliceosome U2 snRNP, and these are being tested in clinical trials (29). These SF3B inhibitors may be particularly useful in hematological neoplasms, where recurrent mutations in SF3B1 have been reported (29). Additionally, oligonucleotide-directed splice switching strategies have been developed to correct RNA mis-splicing or to prevent mutated exons from being incorporated into mature mRNAs (30). This splice modulating technology may be another approach to restoring or enhancing therapeutic responses in cancer cells where splice variants are drivers of resistacne..
Conslusions and Future Directions
The vignettes discussed in this review have demonstrated that the mechanisms leading to functional expression of alternatively spliced protein variants are diverse, ranging from germline or somatic alterations in gene sequence/structure to de-regulation of splicing regulatory factors. In order to obtain a more complete view of the contribution of these various mechanisms to splicing alterations, it will be important to design integrative analyses of genome sequence/structure and splicing patterns in therapy-resistant tissues. This knowledge is expected to reveal instances where there is plasticity in splicing-associated resistance mechanisms, or instances where these splicing alterations are stable. This may be important for designing optimal therapeutic re-sensitization strategies, which could include modulating the expression and/or activity of the splicing machinery, direct targeting of splice variant expression, or targeting downstream signaling pathways that are required for splice variants to drive therapeutic resistance.
Acknowledgments
Financial Support: American Cancer Society Research Scholar Grant (RSG-12-031-01), National Institutes of Health (R01CA174777), and Department of Defense Prostate Cancer Research Program New Investigator Award (W81XWH-10-1-0353). S.M.D. is a Masonic Scholar of the Masonic Cancer Center, University of Minnesota.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pal S, Gupta R, Davuluri RV. Alternative transcription and alternative splicing in cancer. Pharmacology & therapeutics. 2012;136:283–94. doi: 10.1016/j.pharmthera.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Wilson TR, Longley DB, Johnston PG. Chemoresistance in solid tumours. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2006;17(Suppl 10):x315–24. doi: 10.1093/annonc/mdl280. [DOI] [PubMed] [Google Scholar]
- 5.Eblen ST. Regulation of chemoresistance via alternative messenger RNA splicing. Biochemical pharmacology. 2012;83:1063–72. doi: 10.1016/j.bcp.2011.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DC, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci U S A. 2006;103:14907–12. doi: 10.1073/pnas.0606176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–8. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 8.Fenstermaker RA, Ciesielski MJ. EGFR Intron Recombination in Human Gliomas: Inappropriate Diversion of V(D)J Recombination? Current genomics. 2007;8:163–70. doi: 10.2174/138920207780833838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–9. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that promotes chemotherapy resistance. Cancer discovery. 2011;1:524–38. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arjona D, Bello MJ, Rey JA. EGFR intragenic loss and gene amplification in astrocytic gliomas. Cancer Genet Cytogenet. 2006;164:39–43. doi: 10.1016/j.cancergencyto.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006;103:7817–22. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iacobucci I, Lonetti A, Messa F, Cilloni D, Arruga F, Ottaviani E, et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood. 2008;112:3847–55. doi: 10.1182/blood-2007-09-112631. [DOI] [PubMed] [Google Scholar]
- 14.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 15.Choi JW, Kim DG, Lee AE, Kim HR, Lee JY, Kwon NH, et al. Cancer-associated splicing variant of tumor suppressor AIMP2/p38: pathological implication in tumorigenesis. PLoS Genet. 2011;7:e1001351. doi: 10.1371/journal.pgen.1001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark M, Wichman C, Avivi I, Assaraf YG. Aberrant splicing of folylpolyglutamate synthetase as a novel mechanism of antifolate resistance in leukemia. Blood. 2009;113:4362–9. doi: 10.1182/blood-2008-08-173799. [DOI] [PubMed] [Google Scholar]
- 17.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18:R183–96. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–62. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–9. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–17. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Hwang TH, Oseth L, Hauge A, Vessella RL, Schmechel SC, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–67. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivas-Mejia PE, Rodriguez-Aguayo C, Han HD, Shahzad MM, Valiyeva F, Shibayama M, et al. Silencing survivin splice variant 2B leads to antitumor activity in taxane--resistant ovarian cancer. Clin Cancer Res. 2011;17:3716–26. doi: 10.1158/1078-0432.CCR-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheper GC, Parra JL, Wilson M, Van Kollenburg B, Vertegaal AC, Han ZG, et al. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol Cell Biol. 2003;23:5692–705. doi: 10.1128/MCB.23.16.5692-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adesso L, Calabretta S, Barbagallo F, Capurso G, Pilozzi E, Geremia R, et al. Gemcitabine triggers a pro-survival response in pancreatic cancer cells through activation of the MNK2/eIF4E pathway. Oncogene. 2012 doi: 10.1038/onc.2012.306. [DOI] [PubMed] [Google Scholar]
- 26.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nature structural & molecular biology. 2012;19:220–8. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature structural & molecular biology. 2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–80. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- 29.Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nature reviews Drug discovery. 2012;11:847–59. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 30.Hammond SM, Wood MJ. Genetic therapies for RNA mis-splicing diseases. Trends in genetics : TIG. 2011;27:196–205. doi: 10.1016/j.tig.2011.02.004. [DOI] [PubMed] [Google Scholar]