Abstract
Why do we need to sleep? What regulates when we sleep? And what dictates the number of hours we require? These are often viewed as three separate biological questions. Here, we propose they share molecular etiologies, whereby regulators of sleep schedules and sleep duration also govern the physiological purposes of sleep. To support our hypothesis, we review Mendelian human genetic variants sufficient to advance sleep-wake onset (PER2) and shorten sleep length (DEC2), and evaluate their emerging roles in immune responses that may rely on a sound night of slumber.
Keywords: circadian rhythms, sleep homeostasis, immune system, per2, dec2
Introduction
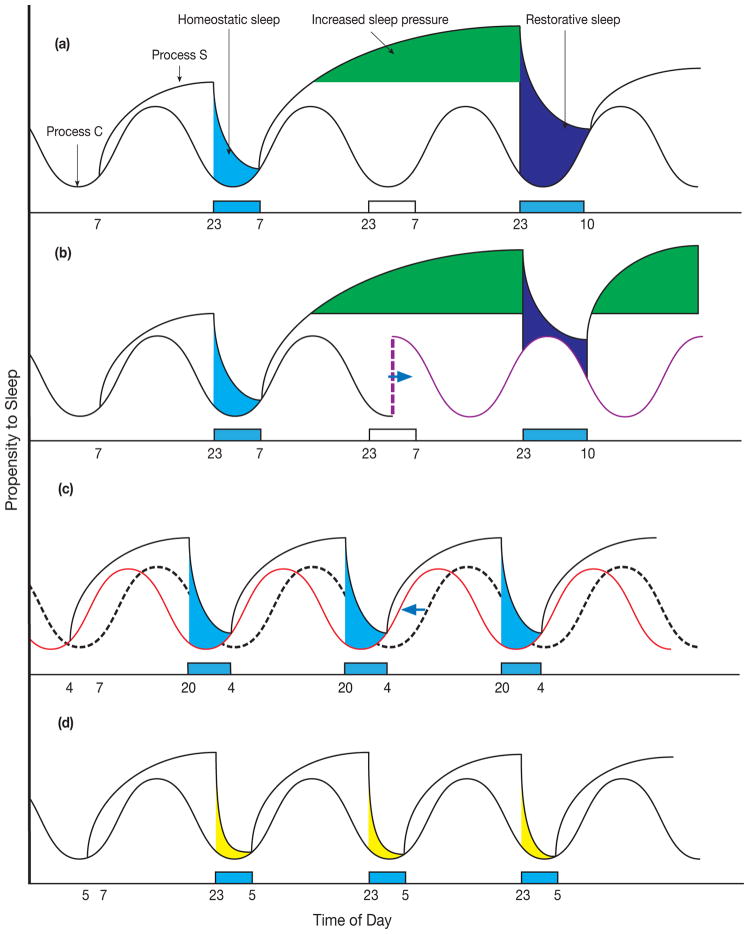
Sleep is an innate behavior that is evidently circadian for modern humans because it is usually a daily, consolidated event with predictable timing. Quality of sleep is of utmost importance, but it remains difficult to define because its purpose is highly debated in light of many intriguing possibilities [1,2]. Besides restorative biological processes, optimal sleep in conventional society also takes into account its timing (i.e. sleep schedules relative to the time of day, also known as Process C due to its association with circadian rhythmicity) and its duration (i.e. the number of hours that yield satiety, also known as sleep homeostasis or Process S) (Figure 1A) [3]. Sleep disorders such as insomnia or sleep deprivation distort the relationship between Process C and S and affect both (Figure 1B). Other variations in sleep patterns include those that specifically affect Process C such as advanced sleep phase (where affected individuals feel sleepy in the late afternoon and wake up before sunrise, though the total amount of sleep remains conventional) (Figure 1C), and those that perturb only Process S such as natural short sleep (Figure 1D) [4]. Therefore, understanding the biological underpinnings of Process C and S may lead to targeted treatment for sleep disorders.
Figure 1. The Integration of Process C and Process S.
Figure not drawn to scale to emphasize theoretical changes. (a) The hypothetical two process model of sleep as described by [3] integrates the daily (circadian) oscillation of sleep propensity (Process C) with the homeostatic sleep propensity accumulated in the awake state and relieved by restorative sleep (Process S). This model of integration assumes that Process C is unaltered in the setting of sleep deprivation. (b) A model of sleep integration whereby sleep deprivation also alters Process C (purple line) causing increased dys-synchrony between the two-processes and reconciliation of sleep need. (c) A model of integration where Process C is advanced (red line) over normal (dashed line) and Process S is unperturbed. (d) A model of integration where homeostatic sleep component of Process S is shortened and Process C is undisturbed. Blue bars, sleep. White bars, sleep deprivation.
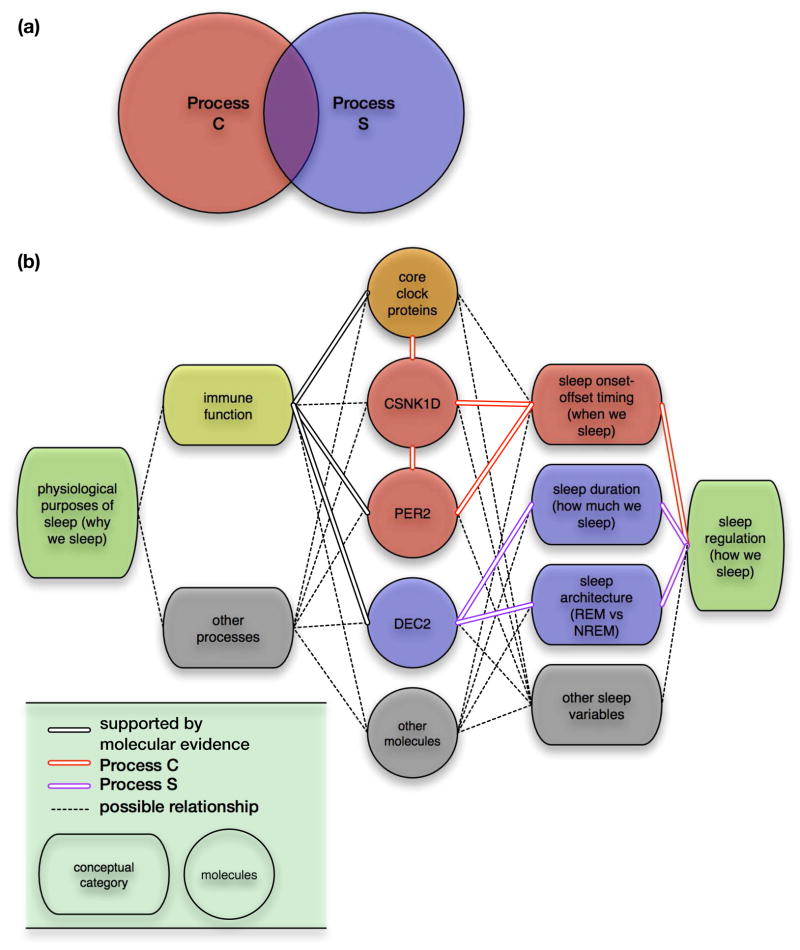
It is a common belief that there are separate molecular pathways for Processes C and S, and there may be coordinated mechanisms between them that together ensure “optimal” sleep quality. Therefore, the molecular basis of the two-process model is sometimes simplified as a Venn diagram with two partially intersecting circles (Figure 2A). Process C is better understood compared to Process S because it is associated with the “molecular core clock,” which defines a series of mechanisms that allow a cell to maintain circadian rhythmicity. The most defined aspects of the molecular core clock are a series of transcription-translation negative feedback loops that take approximately 24 hours to complete [5,6]. But despite a remarkable correlation between cellular and behavioral circadian periods (the time it takes to complete one cycle), [7] it is not clear how the molecular core clock regulates timing of sleep onset and offset. The molecular basis of Process S is even more nebulous because it is challenging to define and assay sleep homeostasis in vitro. Therefore, identifying molecular components sufficient to alter sleep timing and duration is of high research interest.
Figure 2. Molecular basis for the Two-Process Model of sleep regulation.
(a) The two-process model has been and remains instrumental for understanding the dynamics of sleep regulation, and recent research is focused on finding molecular correlates for these processes, with an implicit assumption that there are shared molecular mechanisms as well as separate pathways. (b) Our proposed model for understanding the purposes and regulation of sleep based on the Two-Process model [3]. Recent evidence suggests that PER2 and CSNK1D may be sufficient to alter sleep onset-offset timing, and DEC2 may be sufficient to reduce sleep duration and modify sleep architecture by reducing NREM sleep more than REM sleep. Emerging findings point to additional roles for PER2 and DEC2 in various aspects of immune function, which may contribute to the physiological purposes of sleep.
Through the identification of Mendelian human sleep traits, genetic mutations that result in advanced sleep phase (PER2) and shortened sleep duration (DEC2) were found [8–10]. With regard to the two-process model, PER2 appear to participate in Process C and DEC2 in Process S. Interestingly, emerging evidence suggests an intimate relationship between Processes C and S, and clinical outcomes related to immune responses. Is it possible that instead of directly sharing molecular mechanisms, Process C and S may instead coordinate through participating in physiological reasons for sleep such as immune function? Here we posit that PER2 and DEC2 may function as regulators of sleep timing and duration respectively, yet both simultaneously impact the immune system via separate mechanisms (Figure 2B). Together, this hypothesis explains the observed correlation between sleep and immune responses, and also supports an alternative view of the two-process model. Finally, we discuss the challenges of untangling the molecular basis of sleep regulation (how much sleep and when), its physiological function (why we sleep), and the consequences (or feedback) of impaired sleep on molecular mechanisms underlying Processes C and S.
Human Genetics of Sleep Timing and Duration
The first Mendelian human circadian rhythm trait characterized was Familial Advanced Sleep Phase (FASP), a highly penetrant autosomal dominant trait [8,11]. Affected individuals awaken and go to bed exceptionally early to maintain a normal quantity and quality of sleep, and attempts at modifying circadian tendencies (such as the use of phototherapy) are usually unsuccessful [12,13]. This advancement in sleep phase is accompanied by a shortening of free-running activity period, which measures the endogenous behavioral sleep-wake cycle in the absence of environmental cues such as light, food timing, and social interaction [11,14].
Using human genetic methods, it was determined that PER2-S662G is associated with FASP in this pedigree. To demonstrate that PER2-S662G is sufficient to advance sleep timing, BAC transgenic mice carrying PER2-S662G and PER2-S662D were generated, with the latter mimicking phospho-serine at the same site because S662 was hypothesized to be a phosphorylation site. Remarkably, PER2-S662G mice recapitulate advanced activity onset and shorter free-running period observed in FASP individuals, whereas PER2-S662D mice demonstrate longer free-running periods [9]. Further supporting the importance of PER2 phosphorylation in sleep timing, an additional genetic variant identified for FASP is located in CSNK1D, which reduces enzymatic activity and therefore hypophosphorylates PER2 in vitro [15,16]. In addition, constitutive expression of PER2 reversibly disrupts circadian rhythms of activity [17].
Together, these findings suggest that the circadian oscillation of PER2 may be sufficient for regulating sleep timing [6]. However, FASP individuals do not exhibit overt changes in Process S according to EEG measures of sleep architecture [11]. In addition, Per2 knockout (KO) mice are reported to exhibit no significant differences in sleep homeostasis [18], suggesting that PER2 may be responsible for only Process C. But since PER2 and CSNK1D mutations appear to be sufficient for advancing sleep phase and transmit in a Mendelian manner, it was hypothesized that a rare genetic variant that changes sleep duration may also exist. Indeed, Familial Natural Short Sleepers (FNSS) were found to sleep 6–6.5 hours per night (~2 hours less than controls), and they do not report a sense of sleep deprivation. These FNSS individuals carry a mutation in DEC2/BHLHE41, which encodes a transcription repressor that belongs to the Hairy/Enhancer of Split subfamily [19]. The mutation replaces a proline at position 384 with arginine, and BAC transgenic mice carrying DEC2-P384R exhibit altered sleep homeostasis. Specifically, DEC2-P384R mice undergo shorter duration of rapid eye movement (REM) (~2%) and non-REM (NREM) (~6%) sleep, and recover more readily from sleep deprivation. Together, these findings suggest that DEC2-P384R is sufficient for reducing sleep length [10].
How does the DEC2-P384R mutation reduce sleep quantity? Early evidence suggests that DEC2 participates in the molecular core clock, but it is unclear whether circadian molecular mechanisms are responsible for its effects on sleep quantity [20–22]. P384R is located in a novel proline-rich domain with no known circadian function. In addition, unlike PER2-S662G, neither DEC2-P384R nor Dec2 KO mice demonstrate a change in free-running period of activity [10,23]. These findings suggest that at least on a behavioral level, sleep quantity and timing are separate processes. In addition, the evidence presented so far seems to suggest that PER2 regulates sleep timing through the molecular core clock, whereas DEC2 alters sleep duration through other pathways that require further investigation. Therefore, Process C and S may not directly share molecular mechanisms responsible for co-regulating sleep timing and duration. However, recent studies demonstrate a clinical correlation between sleep and immune responses. Perhaps PER2 and DEC2 both exert distinct effects on immune responses, resulting in co-regulation of a potentially important function of sleep? To address this possibility, we explore immune-related roles of PER2 and DEC2 in the next section.
Genes that make you tick can make you sick
While there are numerous studies aimed at understanding circadian effects on metabolism, cardiovascular function, and other physiological processes, similar research for immunological responses is just beginning to emerge [5,24]. Clinical observations suggest that the time of day influences susceptibility to disorders of human immunologic activity, which implies that circadian molecular mechanisms may regulate immune function. For example, the risk of mortality in human patients suffering from sepsis is increased between the hours of 2 am and 6 am [25]. Some rheumatic arthritis and asthma patients experience daily cyclical variations in the severity of symptoms [26–29]. Similarly, changes in sleep quantity can interfere with appropriate immunologic responses. For instance, adults with short sleep duration had lower secondary antibody responses to Hepatitis B vaccine, resulting in decreased predicted clinical protection [30]. In turn, this observation is associated with increased T lymphocyte activation and reduced natural killer (NK) cell activity [31]. Together, these findings suggest that sleep timing or duration and some aspects of immune function may be regulated by common molecular mechanisms.
PER2 and DEC2 appear to be sufficient for altering sleep timing and duration respectively, and these proteins are recently implicated in immune processes, potentially one of the many purposes for sleep. Indeed, Per2 KO mice demonstrate resistance to endotoxemic shock compared to wild-type mice after intraperitoneal injection with lipopolysaccharide (LPS) [32], and this finding is attributed to reduced oscillations and absolute quantities of IFN-γ and IL-1β cytokines. Furthermore, peritoneal macrophages of Per2 KO mice downregulate the expression of Toll-like receptor 9 (TLR9), a pattern recognition receptor that participates in both innate and adaptive immunity. When challenged with TLR9 agonist, peritoneal macrophages from Per2 KO animals had reduced IL6 and TNFα production in vitro. These effects likely involve the molecular core clock, because in vivo challenges of wild-type mice performed at the peak of TLR9 oscillation revealed increased morbidity and mortality [33].
Supporting the involvement of the molecular core clock in immune responses, other core clock components are also implicated to regulate inflammatory potential. When injected with LPS, both systemic Rev-Erbα KO and macrophage restricted Bmal1 KO mice had elevated cytokine production by abolishing the robust time-sensitive generation of IL-6 compared to controls [34]. Macrophages from Cry1/2 double KO mice have increased nuclear factor kappa B (NF-κB) activity, causing elevated baseline cytokine expression in vitro and generating greater inflammation when challenged with LPS in vivo [35]. In addition, in the absence of Clock, T cells fail to proliferate in a circadian manner [36]. As many immune relevant transcription factors, such as members of the signal transducer and activator of transcription family (STATs) and NF-κB, also fall in the domain of circadian regulation, it is likely that the molecular core clock drives downstream activities of immune responses, with PER2 responsible for a subset that remains to be fully elucidated [37].
Similar to PER2, DEC2 also exerts influences on immune function. Specifically, DEC2 is involved in the maturation of T helper type 2 (TH2) cell lineages associated with humoral (antibody mediated) immunity. TH2 cells highly express DEC2 compared to other T-cell lineages [38,39]. Dec2 KO mice demonstrate defective TH2 responses after repeated stimulation with OVA peptide, decreased alveolar infiltrate and reduced TH2 cytokine production after exposure to an in vivo model of allergic asthma [39]. DEC2 overexpression in undifferentiated T cells drives a TH2 cell polarization while in vivo allergic asthma challenges yield increased TH2 cytokine production and increased lung interstitial infiltrate compared to WT animals [38]. To our knowledge however, there are no current studies addressing the effect of Dec2 on TH2 cell populations or the larger immune system in the context of circadian timing or, more relevantly, distortion of sleep length. As DEC2 has been shown to affect pathways as diverse as cellular proliferation, differentiation, apoptosis and responses to hypoxia, DEC2 may exert its effects on the immune system and sleep duration outside of the molecular core clock. Further research is necessary for defining pathways downstream of DEC2, and it will be interesting to see whether DEC2-P384R confers beneficial or detrimental immune outcomes in addition to its role in reducing sleep duration.
Our hypothesis assumes that one of the main purposes of sleep is immune-related, and most findings that investigate the relationship between sleep and immune responses do so through disrupting sleep. Transcriptome based analysis of human subjects after sleep deprivation (disruption of Process C and S) reveal significant changes in multiple immune related pathways [40,41]. Supporting these findings, sleep deprivation in animal models decreases circulating lymphocyte populations, reduces the cellularity of the spleen and bone marrow [42,43], and acutely elevated inflammatory markers [44]. Simulated jet-lag models (which attempt to mimic disruptions of Process C) altered coordinated rhythmic expression of cytolytic factors and cytokines in NK cells, leading to deficient cytolytic function [46] and markedly decreased survival after LPS injection [45,46]. Interestingly, when compared to control, a group of genes that maintain circadian oscillation after sleep deprivation were explicitly immune related, hinting at the critical nature of circadian immune regulation since they are relatively preserved [41]. Together, these findings support a correlation between sleep and immune function, and we conclude with a further discussion of this association in relations to our hypothesis.
Conclusions
Although it is difficult to study both adaptive and innate immune responses simultaneously, it is plausible that Process C has a broader effect on immunity encompassing both adaptive and innate functions, whereas Process S may more specifically affect adaptive immunity and T cell polarization. Sleep deprivation with its effect on both Process C and S would then encompass the broadest range of immune alteration. However, even though these data allude to a hierarchical model for sleep and immunity (i.e. disruption of Processes C and S results in impaired immune function), a forced alteration in sleep timing/duration may not be the same as natural/habitual short sleep that satisfies homeostatic requirements. For instance, sleep deprivation may disrupt both the normal physiological purposes of sleep and affect molecular cues for initiating and enforcing sleep.
As an alternative viewpoint, here we propose that Processes C, Process S and certain immune responses (that may contribute to the physiological necessity of sleep) share molecular components such as PER2 and DEC2. As FASP and FNSS models exhibit stable and inherent alterations of sleep timing and duration, the characterization of their immune responses may address this hypothesis. Furthermore, identification of novel genes for FASP and FNSS may provide additional molecular correlates for sleep timing and duration beyond PER2 and DEC2. Ultimately, physiological reasons for sleep and regulation of sleep itself may require a delicate balance of shared molecular events. Therefore, tuning these pathways using both KO/haploinsufficient animals (loss-of-function) and transgenic mice carrying human mutations (gain-of-function) may reveal the answers to age-old questions of how, when, and why we sleep.
Highlights.
2-Process Model for sleep has distinct circadian (Process C) & homeostatic (Process S) components
Mendelian genes exist for advancing sleep (PER2) & shortening sleep duration (DEC2)
Some immunologic responses are associated with circadian timing & sleep
PER2 is part of a molecular clock that influences innate & adaptive immune function
DEC2 is implicated in immune function through T cell polarization
Acknowledgments
We would like to thank Jonathan F. Russell and Arun Prakash for critically reviewing the manuscript. Research conducted in the authors’ laboratories is supported by NIH GM079180, HL059596 and NS072360 to Y.-H.F. and L.J.P. and by the Sandler Neurogenetics Fund. L.J.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pace-Schott EF, Hobson JA. The neurobiology of sleep: genetics, cellular physiology and subcortical networks. Nat Rev Neurosci. 2002;3:591–605. doi: 10.1038/nrn895. [DOI] [PubMed] [Google Scholar]
- 2.Sehgal A, Mignot E. Genetics of Sleep and Sleep Disorders. Cell. 2011;146:194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borbély AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 4.Jones CR, Huang AL, Ptáček LJ, Fu Y-H. Genetic basis of human circadian rhythm disorders. Exp Neurol. 2013;243:28–33. doi: 10.1016/j.expneurol.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohawk JA, Green CB, Takahashi JS. Central and Peripheral Circadian Clocks in Mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong SYC, Ptáček LJ, Fu Y-H. Genetic insights on sleep schedules: this time, it’s PERsonal. Trends in Genetics. 2012;28:598–605. doi: 10.1016/j.tig.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown SA, Fleury-Olela F, Nagoshi E, Hauser C, Juge C, Meier CA, Chicheportiche R, Dayer J-M, Albrecht U, Schibler U. The Period Length of Fibroblast Circadian Gene Expression Varies Widely among Human Individuals. Plos Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toh KL. An hPer2 Phosphorylation Site Mutation in Familial Advanced Sleep Phase Syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptáček LJ. Modeling of a Human Circadian Mutation Yields Insights into Clock Regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Rossner MJ, Nishino S, Fu YH. The Transcriptional Repressor DEC2 Regulates Sleep Length in Mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones CR, Campbell SS, Zone SE, Cooper F, DeSano A, Murphy PJ, Jones B, Czajkowski L, Ptáček LJ. Familial advanced sleep-phase syndrome: A short- period circadian rhythm variant in humans. Nat Med. 1999;5:1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 12.Dodson ER, Zee PC. Therapeutics for Circadian Rhythm Sleep Disorders. Sleep Medicine Clinics. 2010;5:701–715. doi: 10.1016/j.jsmc.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahey CD, Zee PC. Circadian Rhythm Sleep Disorders and Phototherapy. Psychiatric Clinics of North America. 2006;29:989–1007. doi: 10.1016/j.psc.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Jones CR, Ptáček LJ, Fu Y-H. The Genetics of the Human Circadian Clock. Elsevier Inc; 2011. [Google Scholar]
- 15.Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. International Journal of Biochemistry and Cell Biology. 2011;43:465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptáček LJ, Fu Y-H. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 17.Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo S-H, Takahashi JS, Lee C. Rhythmic PER Abundance Defines a Critical Nodal Point for Negative Feedback within the Circadian Clock Mechanism. Molecular Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiromani PJ, Xu M, Winston EM, Shiromani SN, Gerashchenko D, Weaver DR. Sleep rhythmicity and homeostasis in mice with targeted disruption of mPeriod genes. Am J Physiol Regul Integr Comp Physiol. 2004;287:R47–57. doi: 10.1152/ajpregu.00138.2004. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto K, Shen M, Noshiro M, Matsubara K, Shingu S, Honda K, Yoshida E, Suardita K, Matsuda Y, Kato Y. Molecular Cloning and Characterization of DEC2, a New Member of Basic Helix-Loop-Helix Proteins. Biochemical and Biophysical Research Communications. 2001;280:164–171. doi: 10.1006/bbrc.2000.4133. [DOI] [PubMed] [Google Scholar]
- 20.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K-I. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 21.Hamaguchi H, Fujimoto K, Kawamoto T, Noshiro M, Maemura K, Takeda N, Nagai R, Furukawa M, Honma S, Honma K-I, et al. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem J. 2004;382:43–50. doi: 10.1042/BJ20031760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimoto K, Hamaguchi H, Hashiba T, Nakamura T, Kawamoto T, Sato F, Noshiro M, Bhawal UK, Suardita K, Kato Y. Transcriptional repression by the basic helix-loop-helix protein Dec2: multiple mechanisms through E-box elements. Int J Mol Med. 2007;19:925–932. [PubMed] [Google Scholar]
- 23.Rossner MJ, Oster H, Wichert SP, Reinecke L, Wehr MC, Reinecke J, Eichele G, Taneja R, Nave K-A. Disturbed Clockwork Resetting in Sharp-1 and Sharp-2 Single and Double Mutant Mice. PLoS ONE. 2008;3:e2762. doi: 10.1371/journal.pone.0002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hrushesky WJ, Wood PA. Circadian time structure of septic shock: timing is everything. J Infect Dis. 1997;175:1283–1284. [PubMed] [Google Scholar]
- 26.Cutolo M, Straub RH. Circadian rhythms in arthritis: Hormonal effects on the immune/inflammatory reaction. Autoimmunity Reviews. 2008;7:223–228. doi: 10.1016/j.autrev.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Straub RH, Cutolo M. Circadian rhythms in rheumatoid arthritis: Implications for pathophysiology and therapeutic management. Arthritis Rheum. 2007;56:399–408. doi: 10.1002/art.22368. [DOI] [PubMed] [Google Scholar]
- 28.Ferraz E, Borges MC, Terra-Filho J, Martinez JAB, Vianna EO. Comparison of 4 AM and 4 PM Bronchial Responsiveness to Hypertonic Saline in Asthma. Lung. 2006;184:341–346. doi: 10.1007/s00408-006-0017-0. [DOI] [PubMed] [Google Scholar]
- 29.Bechtold DA, Gibbs JE, Loudon ASI. Circadian dysfunction in disease. Trends in Pharmacological Sciences. 2010;31:191–198. doi: 10.1016/j.tips.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Prather AA, Hall M, Fury JM, Ross DC, Muldoon MF, Cohen S, Marsland AL. Sleep and antibody response to hepatitis B vaccination. Sleep. 2012;35:1063–1069. doi: 10.5665/sleep.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fondell E, Axelsson J, Franck K, Ploner A, Lekander M, Bälter K, Gaines H. Short natural sleep is associated with higher T cell and lower NK cell activities. Brain, Behavior, and Immunity. 2011;25:1367–1375. doi: 10.1016/j.bbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Mankani G, Shi X, Meyer M, Cunningham-Runddles S, Ma X, Sun ZS. The Circadian Clock Period 2 Gene Regulates Gamma Interferon Production of NK Cells in Host Response to Lipopolysaccharide-Induced Endotoxic Shock. Infection and Immunity. 2006;74:4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Silver AC, Arjona A, Walker WE, Fikrig E. The Circadian Clock Controls Toll-like Receptor 9-Mediated Innate and Adaptive Immunity. Immunity. 2012;36:251–261. doi: 10.1016/j.immuni.2011.12.017. TLR9 expression is under direct clock control and has a circadian expression pattern. Immune challenges initiated at maximal expression of TLR9 protein cause an increased of both severity of sepsis and lymphocyte proliferation. This is mediated through increased cytokine/chemokine expression, which is altered in Per2 deficient mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, et al. From the Cover: The nuclear receptor REVERB mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proceedings of the National Academy of Sciences. 2012;109:582–587. doi: 10.1073/pnas.1106750109. Relative increases in inflammation after LPS injection in mice is time-of-day dependent. Murine macrophages deficient for BMAL have reductions in relative increases in IL-6 after LPS exposure, a finding confirmed in REV-ERBα deficient macrophages. Isolated human macrophages exhibit similar time-dependent increases in IL-6 production that is inhibited by the REV-ERB ligand GSK4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **35.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:12662–12667. doi: 10.1073/pnas.1209965109. CRY1/2 double knockout mice have constitutively elevated cytokine expression in hypothalamus, fibroblasts and macrophages. CRY double knockout macrophages have increased expression of TNFα and IL-6 after LPS exposure, an effect that can be adoptively transferred. Absence of CRY leads ultimately causes increased NF-κB activity through a cAMP/PKA pathway mechanistically linking circadian proteins to chronic inflammatory states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Fortier EE, Rooney J, Dardente H, Hardy M-P, Labrecque N, Cermakian N. Circadian variation of the response of T cells to antigen. The Journal of Immunology. 2011;187:6291–6300. doi: 10.4049/jimmunol.1004030. Mouse lymph node cells express clock genes in a circadian manner. CD4+ and CD8+ T cells isolated from lymph nodes proliferate in a circadian manner after stimulation, which is abolished in CLOCK mutant mice. T cell activation to antigen in vivo is increased at specific times of day with a concomitant increase in CD44+INFg+ cells after in vitro restimulation. [DOI] [PubMed] [Google Scholar]
- 37.Bozek K, Relógio A, Kielbasa SM, Heine M, Dame C, Kramer A, Herzel H. Regulation of clock-controlled genes in mammals. PLoS ONE. 2009;4:e4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **38.Liu Z, Li Z, Mao K, Zou J, Wang Y, Tao Z, Lin G, Tian L, Ji Y, Wu X, et al. Dec2 Promotes Th2 Cell Differentiation by Enhancing IL-2R Signaling. The Journal of Immunology. 2009;183:6320–6329. doi: 10.4049/jimmunol.0900975. Dec2 overexpression (Tg) drives TH2 differentiation through the induction of CD25, a process that is attenuated by the addition of Dec2 RNAi in vitro. Dec2 Tg naïve T cells showed increased TH2 polarization and appropriate cytokine production after restimulation ex-vivo. In vivo modeling with an OVA-induced asthma model demonstrates increased TH2 cytokine production and histological inflammatory response in lung tissue of Dec2 Tg mice. [DOI] [PubMed] [Google Scholar]
- **39.Yang XO, Angkasekwinai P, Zhu J, Peng J, Liu Z, Nurieva R, Liu X, Chung Y, Chang SH, Sun B, et al. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nature Immunology. 2009;10:1260–1266. doi: 10.1038/ni.1821. TH2 cells express Dec2 over other T cell subsets. Dec2 −/− caused less proliferation and cytokine expression of naïve CD4+ T cells in vitro and increased TH-17 polarization. In vivo, Dec2 −/− showed reduced bronchoalveolar lavage infiltrate in a model of allergic asthma. Early TH2 polarization processes are mediated by Dec2 regulation of transcription of Junb and Gata3, and that GATA-3 regulates Dec2 expression reinforcing the early polarization signaling paradigm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellegrino R, Sunaga DY, Guindalini C, Martins RCS, Mazzotti DR, Wei Z, Daye ZJ, Andersen ML, Tufik S. Whole blood genome-wide gene expression profile in males after prolonged wakefulness and sleep recovery. Physiological Genomics. 2012;44:1003–1012. doi: 10.1152/physiolgenomics.00058.2012. [DOI] [PubMed] [Google Scholar]
- **41.Möller-Levet CS, Archer SN, Bucca G, Laing EE, Slak A, Kabiljo R, Lo JCY, Santhi N, Schantz von M, Smith CP, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proceedings of the National Academy of Sciences. 2013;110:E1132–41. doi: 10.1073/pnas.1217154110. This is the first article to outline the changing circadian blood expression profiles of human subjects in the setting of adequate sleep followed by sleep deprivation versus restricted sleep followed by sleep deprivation. Clustered physiologic processes in some cases lost circadian rhythmicity while others gained oscillation, with clusters that remained circadian rhythmic showing more subtle alterations in expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guariniello LD, Vicari P, Lee KS, de Oliveira AC, Tufik S. Bone marrow and peripheral white blood cells number is affected by sleep deprivation in a murine experimental model. J Cell Physiol. 2011;227:361–366. doi: 10.1002/jcp.22743. [DOI] [PubMed] [Google Scholar]
- 43.Zager A, Ruiz FS, Tufik S, Andersen ML. Immune Outcomes of Paradoxical Sleep Deprivation on Cellular Distribution in Naive and Lipopolysaccharide-Stimulated Mice. Neuroimmunomodulation. 2012;19:79–87. doi: 10.1159/000329484. [DOI] [PubMed] [Google Scholar]
- 44.Yehuda S, Sredni B, Carasso RL, Kenigsbuch-Sredni D. REM Sleep Deprivation in Rats Results in Inflammation and Interleukin-17 Elevation. Journal of Interferon & Cytokine Research. 2009;29:393–398. doi: 10.1089/jir.2008.0080. [DOI] [PubMed] [Google Scholar]
- *45.Logan RW, Zhang C, Murugan S, O’Connell S, Levitt D, Rosenwasser AM, Sarkar DK. Chronic Shift-Lag Alters the Circadian Clock of NK Cells and Promotes Lung Cancer Growth in Rats. The Journal of Immunology. 2012;188:2583–2591. doi: 10.4049/jimmunol.1102715. The first study to show that chronic jet lag in rats alters circadian period and circadian gene expression, with subsequent disruption in the circadian cycling of cytokine and cytolytic factors involved in normal NK cell activity. Jet lagged rats have decreased NK cell cytotoxicity and increased lung tumor frequency and burden. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *46.Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, Besing RC, Menaker M, Gewirtz AT, Davidson AJ. Dysregulation of Inflammatory Responses by Chronic Circadian Disruption. The Journal of Immunology. 2010;185:5796–5805. doi: 10.4049/jimmunol.1001026. The first study linking chronic jet-lag to increased susceptibility to LPS endotoxemia in mice. Chronic jet lag alters levels of inflammatory cytokines after LPS injections, with a subsequent reduction in survival. Chronic jet-lag also changes expression of clock genes and circadian period in different tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]