Abstract
Mammary stem (MaSCs) and progenitor cells are important for mammary gland development and maintenance and may give rise to mammary cancer stem cells (MaCSCs). Yet there remains limited understanding of how these cells contribute to tumorigenesis. Here we show that conditional deletion of focal adhesion kinase (FAK) in embryonic mammary epithelial cells (MaECs) decreases luminal progenitors (LPs) and basal MaSCs, reducing their colony-forming and regenerative potentials in a cell autonomous manner. Loss of FAK kinase activity in MaECs specifically impaired LP proliferation and alveologenesis, whereas a kinase-independent activity of FAK supported ductal invasion and basal MaSC activity. Deficiency in LPs suppressed tumorigenesis and MaCSC formation in a mouse model of breast cancer. In contrast to the general inhibitory effect of FAK attenuation, inhibitors of FAK kinase preferentially inhibited proliferation and tumorsphere formation of LP-like, but not MaSC-like, human breast cancer cells. Our findings establish distinct kinase dependent and independent activities of FAK that differentially regulate LPs and basal MaSCs. We suggest that targeting these distinct functions may tailor therapeutic strategies to address breast cancer heterogeneity more effectively.
Introduction
The mammary epithelium, mainly composed of an inner layer of luminal mammary epithelial cells (MaECs) and an outer layer of basal MaECs, is organized in a hierarchical manner (1–5). A single multipotent mammary stem cell (MaSC) in the basal layer can reconstitute a functional mammary gland by generating lineage-restricted progenitor cells, as shown in transplantation studies (2, 3, 6). By contrast, recent lineage-tracing experiments have alternatively proposed that distinct unipotent MaSC populations, located in the luminal and basal compartments, contribute to mammary gland development and maintenance under physiological conditions (7). Presently, the signaling mechanisms regulating these MaSC/progenitor populations remain to be characterized.
Breast cancer is a heterogeneous disease with six distinct subtypes based on gene expression profiling (8–11), suggesting possible origins from different subsets of MaECs in the mammary epithelial hierarchy. Indeed, genome-wide transcriptome analyses of different subtypes of breast cancers, as well as MaEC subpopulations in human BRCA1 mutation carriers, suggest that basal-like breast tumor may originate from aberrant luminal progenitors (LPs) whereas claudin-low subtype is closely associated with the signature of basal MaSC-enriched subsets (5, 12). However, direct experiments involving the selective depletion of potential tumor-initiating cell populations have not been reported.
Focal adhesion kinase (FAK), which mediates signaling pathways initiated by integrins and other receptors to regulate diverse cellular functions via kinase –dependent and –independent mechanisms (13–15), has been implicated in the development and progression of breast and other cancers (16–22). Further, we found that loss of FAK decreased the content of mammary cancer stem cells (MaCSCs) and compromised their self-renewal and tumorigenicity (18), suggesting that FAK may serve as a potential target in MaCSCs. However, it is unknown whether and how distinct activities of FAK contribute to different breast cancer subtypes possibly from different cells of origin. In this study, we demonstrate that FAK regulates MaSCs/progenitor activities via both kinase -dependent and -independent mechanisms that, in turn, affect normal mammary gland development as well as tumorigenesis and the maintenance of MaCSCs in different breast cancer subtypes.
Materials and Methods
Mice and Genotyping
FAK Ctrl (FAKf/f), MFCKO (FAKf/f, MMTV-Cre) and MMTV-PyMT transgenic mice have been described previously (18, 23, 24). MFCKD mice were created mating the FAKKD/+ mice (25) with MFCKO mice. MFCKO and MFCKD mice were mated with GFP transgenic mice (Jackson Laboratory, Stock Number: 003516) to obtain MFCKO-GFP (FAKf/f, MMTVCre, GFP), MFCKD-GFP (FAKf/KD, MMTV-Cre, GFP) and corresponding Ctrl-GFP (FAKf/f, GFP; FAKf/+, MMTV-Cre, GFP or FAKf/KD, GFP) mice. They were also crossed with MMTV-PyMT mice to obtain 3 cohorts of MFCKO-MT (FAKf/f, MMTV-Cre, MMTV-PyMT), MFCKD-MT (FAKf/KD, MMTV-Cre, MMTV-PyMT) and Ctrl-MT (FAKf/+, MMTV-Cre, MMTV-PyMT; FAKf/KD, MMTV-PyMT or FAKf/f, MMTV-PyMT) mice. Monitoring of mammary tumor formation was described as previously (18). All procedures using mice were carried out following the guidelines of The Unit for Laboratory Animal Medicine (ULAM) at the University of Michigan. The genotyping is described in the Supplementary Methods.
Cell Culture and Lentiviral/Adenoviral Infection
Preparation and culture of mouse MaECs or tumor cells from the virgin glands or mammary tumors is described in the Supplementary Methods or as described previously (18). Normal human breast tissues were obtained from reduction mammoplasties of premenopausal woman patients at the University of Michigan health system according to approved IRB protocols for research in human subjects (UM IRBMED #2001-0344). They were used to prepare human MaECs as described in the Supplementary Methods. Breast cancer cell lines SUM159 and SUM149 obtained from Dr. Stephen Ethier have been extensively characterized (26). MDA-MB231 and HCC1954 cell lines were purchased from American Type Culture Collection and maintained in culture conditions according to supplier’s recommendation (see Supplementary Methods for detailed conditions).
Recombinant adenoviruses encoding FAK or its mutants and lentiviruses encoding FAK shRNA/GFP or scrambled sequence/GFP have been described previously (27, 28). The detailed procedure for the infection of human and mouse MaECs and cancer cell lines with the viruses is described in the Supplementary Methods.
Antibodies and Flow Cytometry
The detailed list of antibodies and chemicals is described in the Supplementary Methods. MaECs or mammary tumor cells were suspended, labeled and analyzed by flow cytometry as described in the Supplementary Methods.
In vitro analysis of MaECs and mammary tumor cells
Mammosphere culture of mouse MaECs, primary human MaECs and human breast cancer cells was performed as previously described (29) and see Supplementary Methods for details. Apoptosis and cell proliferation were examined using TMR red (Roche) or cleaved Caspase-3 labeling and Ki67 or PCNA immunofluorescence. The detailed procedures for these experiments and Western blotting are described in the Supplementary Methods.
Mouse unsorted, luminal or basal MaECs at serial dilutions were used for transplantation assays to determine mammary repopulating unit (MRU) as described in the Supplementary Methods.
Mammary Whole Mounts, Histology, and Immunofluorescence
The mammary ductal elongation and outgrowth after transplantation were examined by carmine staining of mammary whole mounts and observed under a dissecting microscope. In transplantation experiments using GFP-labeled donor cells, the whole mounts were observed under a fluorescent microscope. Mammary gland of virgin or pregnancy/lactation mice were also used for histology using H&E staining and immunofluorescence. The detailed procedures are described in the Supplementary Methods.
RNA isolation and qRT-PCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For qRT-PCR analyses, equal amounts of RNA were reverse-transcribed by SuperScript III first-strand synthesis system (Invitrogen) with oligo(dT) as a primer, and then the resulting cDNA templates were subjected to qRT–PCR using the SYBR Green PCR Core reagents system (Qiagen). Primer sequences are available upon request.
Statistical Analysis
Statistical significance was evaluated by paired student test, or two-way ANOVA using P < 0.05 as indicative of statistical significance. Kaplan-Meier tumor free survival data were compared using the log-rank test.
RESULTS
FAK plays an intrinsic role in the maintenance of MaSCs/progenitor cells
To define the roles of FAK in basal MaSCs and LPs, we analyzed MaECs from Ctrl and MFCKO mice (24) by flow cytometry using established markers (1–3). Three major subsets were obtained: CK8/18+ luminal cells (R6, Lin−CD24hiCD29lo), CK5+ basal/myoepithelial cells (R7, Lin−CD24loCD29mod-hi), and CK8/18−CK5− mammary stromal cells (lower, Lin−CD24−CD29lo)(Figs. 1A and S1A). Interestingly, in MFCKO mice, the MaSC-enriched fraction (Lin−CD24loCD29hi; R9 as a R7 subpopulation) was significantly reduced relative to that of Ctrl mice (Fig. 1B). We further verified that the R9 subpopulation contained a higher fraction of CK8+/CK5+ cells (characteristic of MaSCs) as opposed to the remainder of the R7 cells (Fig. S1B), supporting that it is enriched in MaSC activity (2, 3, 30). Further examination of the luminal population by CD61 (1) revealed that FAK deletion also reduced LPs (R10, Lin−CD24hiCD29loCD61+; Fig. 1C). Together, these results demonstrate that FAK deletion in MaECs decreases the content of both basal MaSCs and LPs.
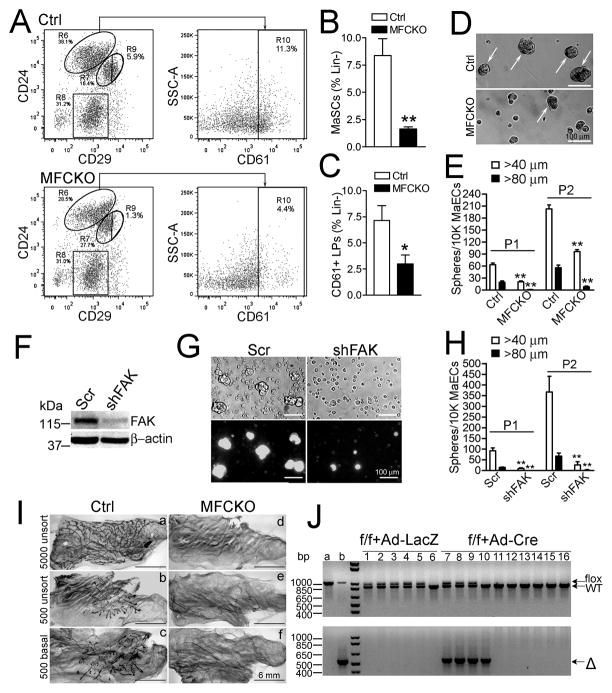
Fig. 1. FAK serves as an intrinsic determinant to maintain MaSC/progenitor activities.
(A–C) Flow cytometry of MaECs from Ctrl and MFCKO mice for LPs (R10, Lin−CD24hiCD29loCD61+) and basal MaSC-enriched subset (R9, Lin−CD24loCD29hi). Representative analyses (A) and Mean±SE (contents of MaSCs and LPs) from four independent experiments (B, C) are shown. (D, E) Primary (P1) and secondary (P2) mammosphere formation from Ctrl and MFCKO mice. Representative images (D, arrows mark spheres >40 μm) and Mean±SE from three independent experiments (E) are shown. (F–H) Human MaECs were infected with lentiviruses expressing FAK shRNA/GFP or control/GFP (Scr). Aliquots of cells were analyzed by Western blotting (F). Representative phase-contrast (top) and fluorescence (bottom) images of mammosphere (G) and Mean±SE of primary and secondary mammospheres from three independent experiments (H) are shown. (I) Representative mammary outgrowths from MaECs of Ctrl, but not MFCKO mice in cleared mammary fat pads of recipient mice. (J) FAKf/f MaECs infected with Ad-LacZ (a) or Ad-Cre (b) and outgrowths derived from 20,000 infected cells were examined by genotyping. Arrows mark WT (from recipient cells), flox and Δ (from donor cells) FAK alleles. *P< 0.05; **P<0.01.
We next examined mammosphere formation from Ctrl and MFCKO MaECs to assess their self-renewal potential (29). MaECs from Ctrl mice formed mammospheres (diameter >40 μm) whereas MaECs from MFCKO mice mostly remained as single or small clusters of cells (Fig. 1D). Quantitation of the mammospheres with diameters >40 μm and >80 μm verified a decrease in mammosphere formation by MFCKO MaECs compared to Ctrl MaECs (Fig. 1E). Similar results were obtained when MaECs were analyzed for spherical acini formation in 3D Matrigel culture (Fig. S2A). These results were not confined to murine MaECs, as knockdown of FAK by shRNA in human MaECs also greatly reduced mammosphere formation (Figs. 1F–1H). Together, these results support a model wherein FAK deletion impaired self-renewal of basal MaSCs and LPs, thereby reducing the content of these populations in MFCKO mice.
To determine directly the role of FAK in maintaining MaSCs in vivo, we used limiting dilution transplantation assays to assess the regenerative potential of MaSCs from Ctrl and MFCKO mice (2, 3). Analysis of unsorted MaECs and Lin−CD24loCD29mod-hi subpopulations enriched in basal MaSCs resulted in an estimated MRU (mammary repopulation unit) frequency of 1/634 and 1/407, respectively, in Ctrl mice, that was greatly diminished in MFCKO mice (<1/9514 and 1/11493) (Table 1 and Fig. 1I). Outgrowths were also generated from the Lin−CD24hiCD29lo luminal population of Ctrl (at low frequency and reduced size), but not MFCKO mice (Table 1). To ensure that the reduced MaSC/progenitor activity of MFCKO mice was caused by intrinsic defects in MaECs, we further examined MaECs from Ctrl (i.e. FAKf/f) mice following deletion of FAK by Cre recombinase in vitro. FAK ablation was verified in Ad-Cre infected Ctrl MaECs, and as predicted, these cells exhibited reduced mammosphere formation relative to control infected cells (Fig. S2B). Surprisingly, although lower than control infected cells (8/22 vs 7/12), a fraction of the Ad-Cre infected MaECs generated outgrowths in transplantation assays (Table S1). Genotyping of parts of outgrowths showed that those from Ad-LacZ infected cells retained the floxed FAK alleles (Fig. 1J, lanes 1–5; Fig. S3A, lane 1 & 2), while the recipient fat pads without outgrowth only contained the WT alleles (Fig. 1J, lane 6 and lane 11–16; Fig. S3A, lanes 3–6 and 11–18), as expected. Of the eight outgrowths generated from Ad-Cre infected cells, only two small outgrowths displayed the FAKΔ/Δ genotype (Fig. 1J, lane 10; Fig. S3A, lane 10), while the remainder exhibited a FAKf/Δ genotype (Fig. 1J, lanes 7–9; Fig. S3A, lane 7–9), suggesting that outgrowths derived from Ad-Cre infected cells were due to incomplete deletion of the floxed alleles. Together, these results demonstrate that FAK is required for the self-renewal and regenerative potential of MaSCs/progenitor cells in a cell-autonomous manner.
Table 1.
Outgrowth of unsorted, basal and luminal MaECs of Ctrl and MFCKO mice
| subsets | genotype | # of cells | take rate | percent fat pad filled | frequ. of MRU (95% CI) |
|---|---|---|---|---|---|
| unsort | f/f | 5000 500 200 |
5/5 3/5 1/5 |
|
1/634 (1/240–1/1681) |
| f/f, Cre | 5000 500 200 |
0/5 0/5 0/5 |
<1/9514 | ||
| basal | f/f | 1000 500 100 |
5/7 6/8 5/10 |

|
1/407 (1/226–1/732) |
| f/f, Cre | 1000 500 100 |
1/7 0/8 0/10 |

|
1/11493 (1/1635–1/80787) |
|
| luminal | f/f | 5000 1000 200 |
5/10 3/7 0/7 |

|
1/5302 (1/2545–1/11046) |
| f/f, Cre | 5000 1000 200 |
0/10 0/7 0/7 |
<1/19494 |
Basal and luminal MaECs from Ctrl (f/f) and MFCKO (f/f, Cre) mice at age of 2 months were sorted based on Lin−CD24loCD29mod-hi and Lin−CD24hiCD29lo signature respectively. Serial dilution transplantation of unsort, basal and luminal cells was performed in cleared mammary fat pads of 3-wk-old FVB recipient mice. Key:
 =100%,
=100%,
 =75%,
=75%,
 =50%,
=50%,
 =25%,
=25%,
 =<25%
=<25%
FAK kinase activity is required for the sphere/acini-forming activity of MaECs, but not their mammary regenerative potential
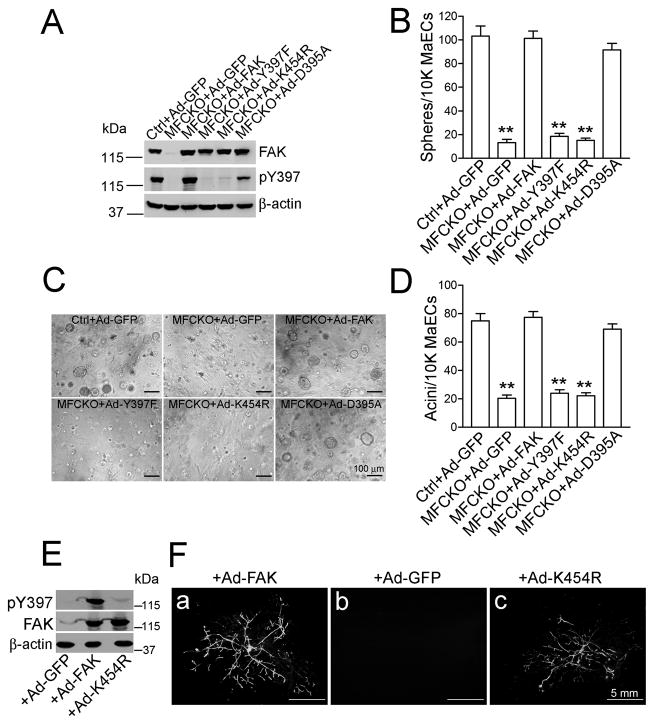
To investigate the mechanisms by which FAK regulates MaSCs/progenitor cells, a series of FAK mutants were analyzed for their ability to rescue the deficient sphere/acini-forming activity of FAK-null MaECs in vitro. As shown in Fig. 2A, both FAK and the D395A (defective for binding to p85 of PI3K) mutant were autophosphorylated at Y397, while no autophosphorylation was detected in cells transduced with the Y397F (autophosphorylation defective) or KD (K454 to R mutation, kinase-defective) mutants. Further, FAK or the D395A, but not the Y397F or KD mutant, rescued the impaired sphere-forming activity of FAK-null MaECs (Fig. 2B). Similar results were obtained when these cells were analyzed for acini formation (Figs. 2C and 2D). These results demonstrate that FAK kinase activity and its autophosphorylation at Y397, but not downstream PI3K activation, are important for the mammosphere/acini-forming activity of MaECs.
Fig. 2. Analysis of FAK and various mutants in FAK-null MaECs.
(A–D) MaECs from Ctrl or MFCKO mice were infected with low titer of various adenoviruses, as indicated. Aliquots of GFP+ cells were analyzed by Western blotting (A). Mean±SE of mammospheres (B) and acini (D) formed by GFP+ cells from three independent experiments are shown, and representative images of acini are shown in C. (E–F) MaECs of MFCKO-GFP mice infected by various adenoviruses were analyzed by Western blotting (E) or transplantation (F). **P<0.01.
To further examine FAK signaling in mammary regenerative potential, MFCKO and Ctrl mice were crossed with GFP transgenic mice to generate FAKf/f;GFP (Ctrl-GFP); FAKf/+;MMTV-Cre;GFP (Cre Ctrl-GFP) and FAKf/f; MMTV-Cre;GFP (MFCKO-GFP) mice, thereby facilitating identification of donor cells in transplantation assays. As expected, limiting dilution transplantation of MaECs from Ctrl-GFP or Cre Ctrl-GFP, but not MFCKO-GFP mice, generated mammary outgrowths with strong GFP signals (Table S2). MaECs from MFCKO-GFP mice were then transduced with adenoviruses expressing FAK, KD, or GFP alone, and used in transplantation assays. Comparable expression levels of FAK and the KD mutant were observed, while Y397 phosphorylation was only detected in cells expressing FAK, but not the KD mutant (Fig. 2E). Re-expression of FAK but not GFP alone, rescued the mammary repopulating activity of FAK-null MaECs (Fig. 2F), generating mammary outgrowths in 6 out of 10 transplants. Surprisingly, re-expression of the KD mutant also restored mammary outgrowth in 8 out of 14 transplants, indicating that FAK kinase activity is disposable for the regenerative potential of MaSCs.
FAKKD mutant knockin preferentially affects luminal MaEC proliferation and alveologenesis
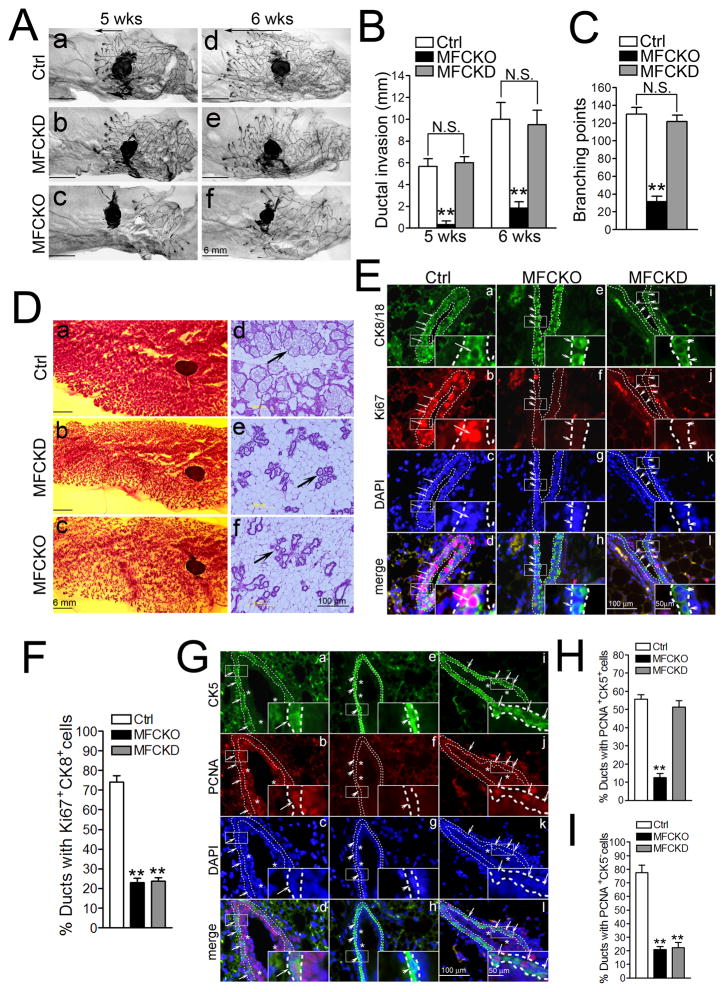
To better characterize the role of FAK kinase activity in MaSCs/progenitor cells in vivo, MFCKO mice were crossed with FAKKD/+ mice (25) to produce MaEC-specific KD knockin (FAKKD/f;MMTV-Cre; designed as MFCKD [mammary FAK conditional KD knockin]) mice. In virgin mice, comparable mammary ductal invasion and branch growth were observed in MFCKD and Ctrl mice, which is in contrast to MFCKO mice that displayed compromised ductal outgrowth (24)(Figs. 3A–3C). During pregnancy, however, both MFCKD and MFCKO mice display marked defects in milk production (data not shown). Whole mount staining revealed that despite normal ductal architecture, the lobulo-alveolar units in MFCKD and MFCKO mammary glands were smaller and more sparse relative to Ctrl mammary glands (Fig. 3D, left panels). Furthermore, while the Ctrl mammary glands were filled with large lobulo-alveolar units composed of multiple individual alveoli (arrow) lined by flattened epithelial cells, MFCKD and MFCKO mammary glands were dominated by adipose-rich stroma, and the parenchyma primarily consisted of dilated ductal networks and small clusters of alveoli (arrows) (Fig. 3D, right panels).
Fig. 3. Analyses of luminal and basal MaECs in MFCKD mice.
(A–C) Whole mount staining of ductal invasion (arrows mark distance between front edge and mammary lymph node) at 5 and 6 weeks. Representative images (A) and Mean±SE of ductal invasion (B) and branch points (C) are shown (n=8 for each). (D) Whole mount or H&E staining of mammary glands at the first day of lactation. (E–I) Mammary glands of virgin mice were analyzed by immunofluorescence using various antibodies, as indicated. Lines outline luminal (E) or basal (G) cells. Long and short arrows mark proliferating and non-proliferating luminal (E) or basal (G) cells, respectively. Asterisks in G mark luminal cells inside the basal/myoepithelial layer. Percentages (Mean±SE) of ducts with Ki67+CK8/18+ luminal cells (F), PCNA+CK5+ basal cells (H) or PCNA+CK5− luminal cells (I) are shown. **P<0.01 NS, not significant.
To explore the cellular basis of the preferential effect of KD knockin mutation on alveologenesis as opposed to ductal outgrowth, the luminal and basal compartments were examined for cell proliferation in virgin females. Double-labeling immunofluorescence revealed abundant proliferating luminal (Ki67+CK8+) cells in the ducts of Ctrl, but not MFCKO or MFCKD mice (Figs. 3E–3F). However, similar analysis showed that the number of proliferating basal (PCNA+CK5+) cells was significantly affected only in MFCKO, but not MFCKD, mice when compared to Ctrl mice (Figs. 3G–3H). This labeling also verified the decreased luminal cell proliferation (PCNA+CK5− cells) in the ducts of MFCKO and MFCKD mice (Figs. 3G and 3I). Together, these results suggest that FAK kinase activity plays a preferential role in controlling proliferation of luminal MaECs and alveologenesis, while the kinase-independent functions of FAK are sufficient to promote basal MaEC proliferation, ductal outgrowth and branching morphogenesis.
Differential role of kinase-independent and -dependent functions of FAK in basal MaSCs and LPs
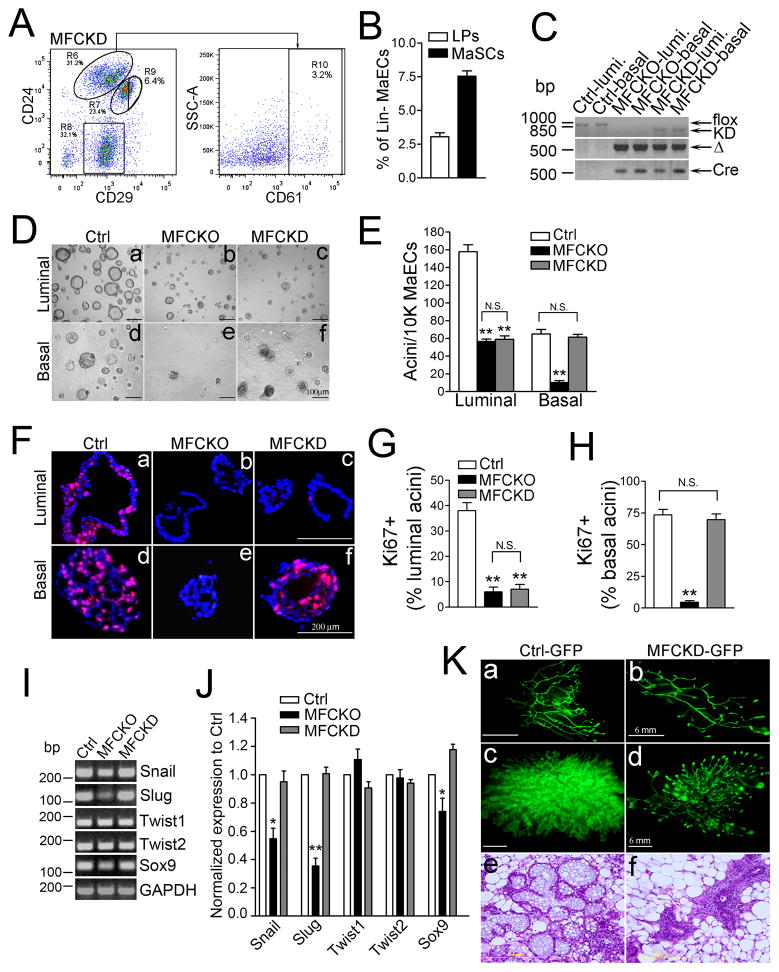
The differential requirement of FAK kinase activity to promote proliferation of luminal, but not basal, MaECs raised the interesting possibility that basal MaSCs and LPs are maintained through distinct functions of FAK. To investigate this possibility, we first examined MaSC and LP contents in MFCKD mice using phenotypic markers (Fig. 4A). Relative to Ctrl and MFCKO mice (see Figs. 1A–1C), decreased content of LPs, but not basal MaSCs, was found in MFCKD mice (Fig. 4B). Genotyping of luminal and basal MaECs verified that FAK was effectively ablated in both compartments of MFCKO and MFCKD mice, while the KD allele was not excised in either compartment of MFCKD mice (Fig. 4C). Interestingly, while luminal MaECs from MFCKO and MFCKD mice both showed reduced acini forming activity compared to those from Ctrl mice, basal MaECs from MFCKD, but not MFCKO, mice generated acini at levels comparable to Ctrl mice (Figs. 4D–4E). Ki67 staining revealed abundant proliferating cells in Ctrl luminal acini as well as both Ctrl and MFCKD basal acini, but few were detected in MFCKO and MFCKD luminal acini or MFCKO basal acini (Figs. F-4H). TUNEL staining did not reveal any apoptotic cells in the luminal cells in any of the genotypes (except those detached cells in the hollow space; arrows), but modest increases in apoptosis were found in the basal acinar cells from MFCKO, but not Ctrl or MFCKD mice (Fig. S3B). Examination in basal MaECs for the expression of several transcription factors implicated in regulating MaSC generation (31, 32) showed that, while the expression levels of Snail, Slug and Sox9 were all reduced in basal MaECs from MFCKO mice, normal levels were maintained in MFCKD basal cells (Figs. 4I–4J). Together, these results suggest that FAK kinase activity is required to promote cell proliferation to maintain colony-forming activity of LPs, but FAK kinase-independent functions are sufficient to support basal MaSC activity through regulation of both survival and proliferation and possibly by maintaining the expression of key transcription factors such as Snail, Slug and/or Sox9.
Fig. 4. MaEC-specific FAKKD knockin specifically impairs LP-, but not basal MaSC activity.
(A, B) Flow cytometry of MaECs from MFCKD mice for LPs (R10, Lin−CD24hiCD29loCD61+) and basal MaSC-enriched subset (R9, Lin−CD24loCD29hi). Representative analyses (A) and Mean±SE (contents of LPs and MaSCs) from four independent experiments (B) are shown. (C) Genotyping of sorted luminal (R6) and basal (R7) MaECs from Ctrl, MFCKO and MFCKD mice to detect different FAK (flox, KD and Δ) and Cre alleles. (D, E) Acini formation from luminal and basal MaECs of Ctrl, MFCKO and MFCKD mice (D) and data drawn from three independent experiments are shown in (E). (F–H) Cryosections of acinar colonies from luminal and basal MaECs of Ctrl, MFCKO, and MFCKD mice were stained with Ki67 antibodies (red) and DAPI (blue) (F), and the ratios of Ki67+ acini from the luminal (G) and basal (H) MaECs of each genotype were counted from three independent experiments. (I, J) qRT–PCR of different transcription factors in basal MaECs of Ctrl, MFCKO and MFCKD mice. Representative analysis (I) and Mean±SE of relative expression from 3 independent experiments (J) are shown. (K) Representative mammary outgrowths from basal cells of Ctrl-GFP and MFCKD-GFP mice before (a–b) and during late pregnancy (c–f). *P< 0.05; **P<0.01; NS, not significant.
We next performed transplantation assays to evaluate the role of FAK kinase-dependent and -independent activity in maintaining the regenerative potential of the basal and luminal MaECs. As expected, MRU activity in the luminal compartment of MFCKD-GFP mice was decreased to levels similar to those found in MFCKO mice (Table S3). Interestingly, while very low MRU activity was detected in basal MaECs of MFCKO-GFP mice (<1/12818), the MRU activity found in basal MaECs of MFCKD-GFP mice (1/530) was comparable to that detected in Ctrl-GFP mice (1/616, Table S3; Fig. 4K), thereby confirming the kinase-independent function of FAK in maintaining the regenerative potential of basal MaSCs. However, in contrast to the expanded mammary outgrowth and large lobulo-alveolar units observed in Ctrl-GFP transplants during pregnancy, the recipient fat pads transplanted with basal MaECs from MFCKD-GFP mice displayed little expansion of mammary outgrowths, with only small numbers of lobulo-alveolar units having been formed (Fig. 4K). Hence, basal MaSCs harboring FAK KD mutation are unable to support pregnancy-induced alveologenesis, despite their ability to generate complete mammary outgrowths.
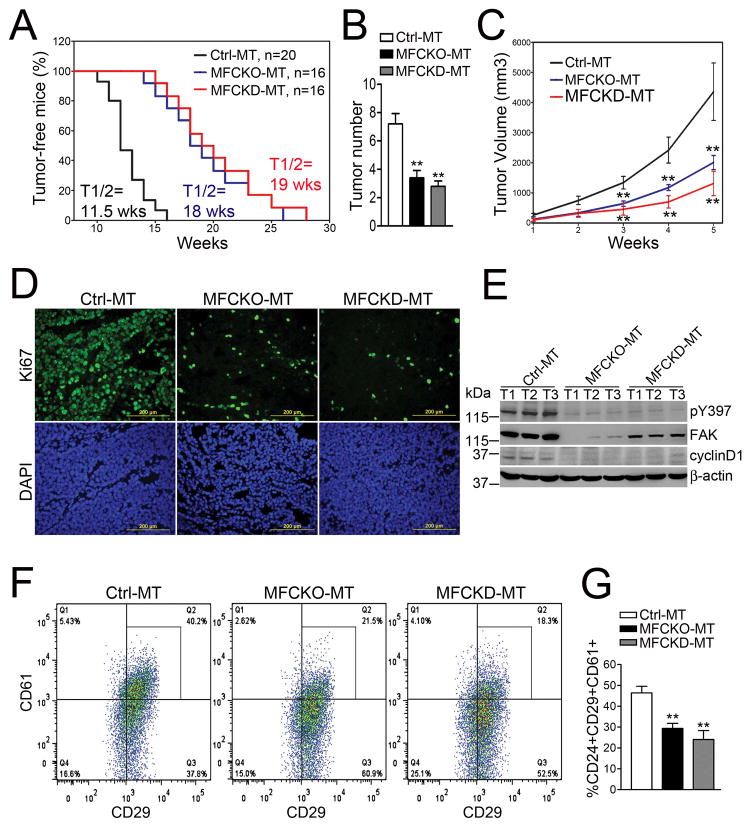
MaEC-specific FAKKD mutation suppresses MMTV-PyMT-induced tumorigenesis and maintenance of MaCSCs
The finding that FAK kinase activity is specifically required to maintain LPs, but not basal MaSCs, prompted us to examine whether this particular FAK function is important for tumorigenesis and maintenance of MaCSCs in tumors arising from LPs. Thus, we generated MaEC-specific FAKKD knockin mutation in the MMTV-PyMT breast cancer model that specifically targets LPs (33). Three cohorts of female mice with the genotype FAKf/KD;MMTV-Cre; MMTV-PyMT (designated MFCKD-MT mice), FAKf/KD;MMTV-PyMT, FAKf/f;MMTV-PyMT or FAKf/+;MMTV-Cre;MMTV-PyMT (collectively designated Ctrl-MT mice), and FAKf/f;MMTV-Cre;MMTV-PyMT (designated MFCKO-MT mice) were established and examined for mammary tumorigenesis. Interestingly, similar to MFCKO-MT mice (18), MFCKD-MT mice displayed an increased tumor-free interval compared to Ctrl-MT mice (Fig. 5A). Furthermore, decreased numbers of tumors (Fig. 5B) and reduced tumor growth (Fig. 5C) were also found in both MFCKO-MT and MFCKD-MT mice relative to Ctrl-MT mice. Consistent with these results, tumor cell proliferation was markedly decreased in MFCKD-MT and MFCKO-MT mice when compared to Ctrl-MT mice (Fig. 5D). Western blot of lysates from primary tumors showed that FAK expression was abolished in MFCKO-MT tumors and also decreased in MFCKD-MT tumors (likely due to the remaining KD allele) relative to Ctrl-MT tumors (Fig. 5E). Moreover, activated FAK (as measured by pY397) was not detected in MFCKO-MT or MFCKD-MT tumors. As reported previously for MFCKO-MT tumors (18), cyclin D1 expression was also decreased in MFCKD-MT tumors. Together, these results demonstrate that loss of FAK kinase activity in MFCKD-MT mice suppressed MMTV-PyMT-induced mammary tumorigenesis and tumor growth.
Fig. 5. MaEC-specific FAKKD knockin suppressed MMTV-PyMT induced tumorigenesis and maintenance of MaCSCs.
(A) Kaplan-Meier analysis of mammary tumor development in Ctrl-MT (n=20), MFCKO-MT (n=16) and MFCKD (n=16) mice. Ctrl-MT vs MFCKO-MT or MFCKD-MT: P< 0.01 by log-rank test. (B, C) Mean±SD of tumor number at week 8 (B) and tumor volume at various weeks (C) after first tumor appearance. (D, E) Tumors from Ctrl-MT, MFCKO-MT and MFCKD-MT mice were stained with Ki67 antibodies and DAPI (D) or analyzed by immunoblotting using various antibodies as indicated (E). (F, G) Flow cytometry of tumor cells from Ctrl-MT, MFCKO-MT and MFCKD-MT mice to determine the content of MaCSCs (Lin−CD24+CD29+CD61+). Representative analyses (F) and Mean±SE from six independent experiments (G) are shown. **P< 0.01.
To explore if the loss of FAK kinase activity affects the MaCSCs maintenance, we first performed flow cytometry for tumor cells and corresponding normal MaECs from various mouse models. As expected (see Fig. 1A), the luminal and basal MaECs from Ctrl, MFCKO and MFCKD mice segregated in gate R5 (luminal) and R6 (basal), respectively (Fig. S4A, upper panels). Interestingly, using these gating criteria, tumor cells from Ctrl-MT, MFCKO-MT and MFCKD-MT mice were confined entirely to the luminal gate R5 (Fig. S4A, bottom panels), supporting the notion that tumors arising in the MMTV-PyMT model originate in the luminal compartment (33), thereby defects in LPs upon loss of FAK kinase activity reduced tumorigenesis in MFCKD-MT mice. We next assessed MaCSC content directly using the Lin−CD24+CD29+CD61+ signature as described previously for MMTV-PyMT model (27, 34). The fraction of Lin−CD24+CD29+CD61+ tumor cells in MFCKD-MT and MFCKO-MT mice was significantly lower than that found in Ctrl-MT mice (Figs. 5F–5G). Hence, loss of FAK kinase activity suppressed mammary tumorigenesis in association with a dramatic decrease in MaCSC content via depletion of LPs (i.e. the proposed cell of origin for MaCSCs) in the MMTV-PyMT breast cancer model.
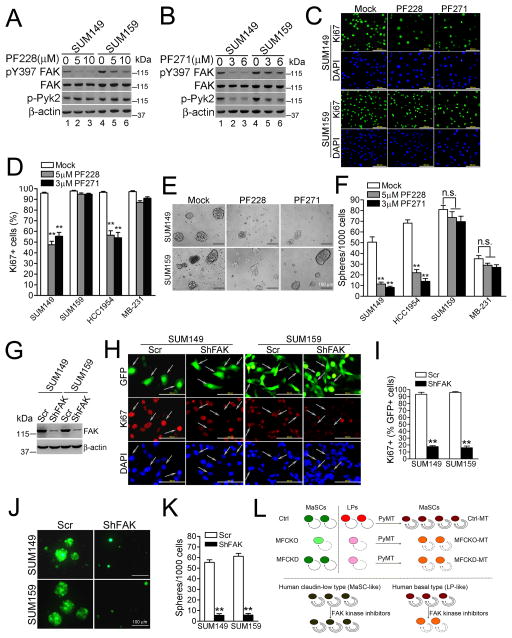
Differential requirement of FAK kinase activity is recapitulated in distinct subtypes of human breast cancer cells
Given our findings that FAK kinase activity is required for LPs, but not basal MaSCs, we next tested if FAK kinase inhibitors differentially affects human breast cancer cells that share gene signatures with LPs (LP-like, or basal subtype) and basal MaSCs (MaSC-like, or claudin-low subtype), respectively (5, 12). Western blotting revealed that MaSC-like SUM159 cells have higher level of FAK phosphorylation at Y397 than the LP-like, SUM149 cells (12, 35) (Fig. 6A). PF573228, a FAK specific inhibitor (27, 36), decreased FAK kinase activity as measured by its phosphorylation at Y397, but not affected Pyk2 phosphorylation (i.e. no Pyk2 compensation), in both SUM149 and SUM159 cells. PF562271, the dual FAK/Pyk2 inhibitor (27, 36), inhibited both FAK and Pyk2 phosphorylation in SUM149 and SUM159 cells (Fig. 6B). Treatment with either inhibitor did not significantly affect the survival of either cell line (Fig. S4B), but suppressed the proliferation of SUM149, but not SUM159, cells (Figs. 6C–6D). These inhibitors also decreased proliferation of HCC1954 (another LP-like breast cancer), but not MDA-MB-231 cells (another claudin-low subtype) (Fig. 6D). Similar to the differential effects observed on cell proliferation, tumorsphere formation by SUM149 and HCC1954 cells, but not SUM 159 and MDA-MB-231 cells, was significantly decreased in the presence of PF573228 or PF562271 (Figs. 6E–6F).
Fig. 6. Differential requirement of FAK kinase activity is recapitulated in human breast cancer cells sharing gene signatures of LPs and basal MaSCs.
(A–C) SUM149 and SUM159 cells were treated with different concentrations of PF228 or PF271 for 24h and then analyzed by immunoblotting with various antibodies as indicated (A, B) or Ki67 staining (C; 5 μm PF228 or 3 μm PF271). (D) Ratios of Ki67+ cells in SUM 149, SUM 159, HCC1954 and MDA-MB-231 cells following PF228 and PF271 treatment were determined in three independent experiments. (E, F) Representative images (E) and Mean±SE from three independent experiments (F) of tumorspheres (diameter > 40 μm) formed by various cancer cells in serum-free media containing DMSO (Mock), 5μM PF228 or 3μM PF271. (G–K) SUM149 and SUM159 cells were infected with lentiviruses expressing FAK shRNA/GFP or scrambled sequence/GFP (Scr) and analyzed for proliferation (H, I) and tumorsphere formation (J, K). Aliquot of cells were analyzed by Western blotting (G). Representative images (H, J) and Mean±SE of percentage of Ki67+ cells within GFP+ (arrows in H) cells (I) or the number of tumorspheres (K) from three independent experiments are shown. (L) A working model to show the roles of FAK kinase-dependent and independent actions in maintenance of LPs and basal MaSCs, as well as in PyMT-induced tumorigenesis and maintenance of MaCSCs in mouse models and human breast cancer cells of distinct subtypes. **P<0.01; NS. not significant.
Lastly, we examined the effect of FAK knockdown (i.e. blocking both kinase-dependent and –independent functions of FAK) on SUM149 and SUM159 cells. As shown in Fig. 6G, treatment of either SUM149 or SUM159 cells with FAK shRNA/GFP suppressed FAK expression relative to cells treated with control scrambled sequence/GFP. Analysis of the GFP-positive cells revealed that knockdown of FAK in either cell line blocked their proliferation (Figs. 6H–6I) and tumorsphere formation (Figs. 6J–6K). Together, these results suggest that while FAK kinase inhibitors are more effective in targeting LP-like, basal breast cancer cells, new approaches will be required to target the kinase-independent functions of FAK that drive the proliferation programs operative in the MaSC-like, claudin-low breast cancer subtype.
Discussion
Here, we have established distinct roles for FAK kinase-dependent and -independent functions in the regulation of LPs and basal MaSCs, and in promoting tumorigenesis and breast cancer heterogeneity. As summarized in Fig. 6L, in the normal mammary gland, FAK ablation in MFCKO mice decreases (broken lines) the colony forming activity of LPs (red to pink) and the self-renewal potential of basal MaSCs (green to light green). In MFCKD mice, loss of FAK kinase activity impairs the maintenance of LPs, but does not affect basal MaSCs, accounting for their normal ductal outgrowth. Both LPs and basal MaSCs might serve as targets for transformation by oncogenes such as PyMT to form MaCSCs (brown) with deregulated self-renewal (more lines). The decreased content of MaCSCs and their compromised tumorigenicity (broken lines; orange) in MFCKO-MT mice suggest either LPs or basal MaSCs (both depleted in MFCKO mice) could be the cells of origin in PyMT-induced mammary tumors. However, the reduced tumorigenesis, decreased MaCSC formation and self-renewal in MFCKD-MT mice, where the pool of LPs, but not basal MaSCs, was depleted after loss of FAK kinase activity, provides direct support that LPs (and not basal MaSCs) serve as the tumorigenic cell origin in the MMTV-PyMT mouse model and that FAK kinase activity is required for propagation of LP-like MaCSCs. Importantly, the differential role of FAK kinase-dependent and -independent functions in regulating LPs and basal MaSCs in mouse models are recapitulated in distinct human breast cancer cells (i.e., LP-like, basal subtype and basal MaSC-like, claudin-low subtype of breast cancer, respectively).
While many factors are likely to regulate MaSC activities, recent work demonstrates that Slug and Sox9 act cooperatively to maintain the MaSC state (31). Interestingly, the expression levels of Slug and Sox9 as well as Snail were reduced in basal cells from MFCKO mice, but were rescued in MFCKD mice, suggesting that kinase-independent functions of FAK may support basal MaSC activity by maintaining the expression of these critical transcription factors. Additional studies will be required to clarify the specific roles of these potential targets and the mechanisms by which KD FAK regulates their expression.
Our studies revealed important differences in the regulation of normal MaSCs/LPs and MaCSCs by FAK signaling. We showed previously that the PI3K/Akt pathway, which is dependent on FAK kinase activity, was selectively activated in WT, but not FAK-null MaCSCs, and that the D395A mutant defective in PI3K binding failed to restore tumorsphere formation as well as tumorigenicity of FAK-null MaCSCs (27). Here, however, we found that the D395A mutant fully rescued sphere/acini-forming capacity of FAK-null MaECs, suggesting that PI3K pathways downstream of FAK are not required to maintain normal MaSC/LPs. Although more studies are needed to decipher the underlying mechanisms operative in this system, the differential requirement of PI3K pathway for MaCSCs, but not normal MaSCs/LPs, may be exploited to design specific strategies to target MaCSCs while sparing their normal counterparts.
The differential requirements for FAK kinase activity in distinct breast cancer subtypes has important implications for the development of therapies that take breast cancer heterogeneity under consideration. Recent studies in mouse models, as well as human patients with Brac1 mutations, provide strong support for the proposal that basal breast cancers originate from LPs (5, 37–39). Thus, our finding that FAK kinase activity plays a differential role in regulating LPs, as well as in tumorigenesis and the maintenance of MaCSCs in tumors derived from LPs, suggests that FAK kinase inhibitors could be an effective therapy for this breast cancer subtype. However, the recently identified claudin-low subtype of breast cancer exhibits mesenchymal characteristics (8), and is closely associated with the gene signature of basal MaSC-enriched cells (5, 12), suggesting that these cancers may be derived from basal MaSCs. Furthermore, following neoadjuvant chemotherapy or hormonal treatment, gene expression profiles of residual cancer cells (of both luminal or basal tumors) become more closely related to those of claudin-low subtype tumors (40). Our findings that kinase-independent functions of FAK support basal MaSCs and that FAK kinase inhibitors do not effectively target MaSC-like, human breast cancer cells, suggest that novel strategies may be required to inhibit the claudin-low breast cancer subtype and relapsed breast cancers that display similar molecular signatures.
Supplementary Material
Acknowledgments
We thank Dr. David Schlaepfer for FAK shRNA lentiviral construct, Drs. Alan Kraker and Donnie Owens for PF-573228 and PF-562271, Dr. Stephen Ethier for SUM149 and SUM159 cells, UMCCC Flow Cytometry core and Christine Bian for assistance, and Steve Weiss and our lab members for helpful comments. This research was supported by NIH grants to J.-L. Guan.
References
- 1.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 2.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 3.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 4.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 6.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 8.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 10.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 14.Schlaepfer DD, Mitra SK. Multiple connections link FAK to cell motility and invasion. Curr Opin Genet Dev. 2004;14:92–101. doi: 10.1016/j.gde.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 16.Golubovskaya VM, Cance WG. Focal adhesion kinase and p53 signaling in cancer cells. Int Rev Cytol. 2007;263:103–153. doi: 10.1016/S0074-7696(07)63003-4. [DOI] [PubMed] [Google Scholar]
- 17.McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat Rev Cancer. 2005;5:505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 18.Luo M, Fan H, Nagy T, Wei H, Wang C, Liu S, et al. Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Cancer Res. 2009;69:466–474. doi: 10.1158/0008-5472.CAN-08-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Provenzano PP, Inman DR, Eliceiri KW, Beggs HE, Keely PJ. Mammary epithelial-specific disruption of focal adhesion kinase retards tumor formation and metastasis in a transgenic mouse model of human breast cancer. Am J Pathol. 2008;173:1551–1565. doi: 10.2353/ajpath.2008.080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pylayeva Y, Gillen KM, Gerald W, Beggs HE, Reichardt LF, Giancotti FG. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009;119:252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC, et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci U S A. 2007;104:20302–20307. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahlou H, Sanguin-Gendreau V, Frame MC, Muller WJ. Focal adhesion kinase contributes to proliferative potential of ErbB2 mammary tumour cells but is dispensable for ErbB2 mammary tumour induction in vivo. Breast Cancer Res. 2012;14:R36. doi: 10.1186/bcr3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen TL, Park AY, Alcaraz A, Peng X, Jang I, Koni P, et al. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J Cell Biol. 2005;169:941–952. doi: 10.1083/jcb.200411155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy T, Wei H, Shen TL, Peng X, Liang CC, Gan B, et al. Mammary epithelial-specific deletion of the focal adhesion kinase gene leads to severe lobuloalveolar hypoplasia and secretory immaturity of the murine mammary gland. J Biol Chem. 2007;282:31766–31776. doi: 10.1074/jbc.M705403200. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Peng X, Sun S, Park AY, Guan JL. Role of kinase-independent and -dependent functions of FAK in endothelial cell survival and barrier function during embryonic development. J Cell Biol. 2010;189:955–965. doi: 10.1083/jcb.200912094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan H, Guan JL. Compensatory function of Pyk2 protein in the promotion of focal adhesion kinase (FAK)-null mammary cancer stem cell tumorigenicity and metastatic activity. J Biol Chem. 2011;286:18573–18582. doi: 10.1074/jbc.M110.200717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim Y, Lim ST, Tomar A, Gardel M, Bernard-Trifilo JA, Chen XL, et al. PyK2 and FAK connections to p190Rho guanine nucleotide exchange factor regulate RhoA activity, focal adhesion formation, and cell motility. J Cell Biol. 2008;180:187–203. doi: 10.1083/jcb.200708194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, et al. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K, Gjorevski N, Boghaert E, Radisky DC, Nelson CM. Snail1, Snail2, and E47 promote mammary epithelial branching morphogenesis. EMBO J. 2011;30:2662–2674. doi: 10.1038/emboj.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts WG, Ung E, Whalen P, Cooper B, Hulford C, Autry C, et al. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF- 562,271. Cancer Res. 2008;68:1935–1944. doi: 10.1158/0008-5472.CAN-07-5155. [DOI] [PubMed] [Google Scholar]
- 37.Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, et al. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci U S A. 2012;109:2772–2777. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.