Abstract
Inflammatory bowel disease (IBD) is thought to result from a dysregulated immune response to intestinal microbial flora in individuals with genetic predisposition(s). Genome wide association studies (GWAS) in human IBD have identified more than 150 associated loci, some of which are key players in innate immunity and bacterial handling, reflecting the importance of the microbiota in disease pathogenesis. In fact, the presence of a microbial flora is not only crucial to the development of a normal murine immune system but also critical for the development of disease in the majority of animal models of IBD.
Although animal models do not perfectly recapitulate human IBD, they have led to the discovery of important concepts in IBD pathogenesis, such as the central role of microbiota in disease development and perpetuation. Many genetically susceptible models do not develop colitis when raised in a germ-free or Helicobacter-free environment. In fact, disease in most models can be attenuated or completely abolished with antibiotic treatment. Moreover, an interplay between intestinal microbiota and mucosal immune activation is suggested by the presence of serum antibodies against the Cbir1 flagellin, an immunodominant antigen that activates TLR5, in certain models of spontaneous colitis as well as in human patients. Furthermore, T cells reactive to Cbir1 are able to induce disease in recipient mice upon adoptive cell transfer, demonstrating the pro-inflammatory properties of certain bacterial products. In fact, it has been shown that transfer of certain intestinal bacteria from a specific genetically altered mouse model with spontaneous colitis can induce disease in wild-type mice upon co-housing or direct feeding. These observations demonstrate the pathogenic potential of intestinal microbiota in IBD.
However, intestinal bacteria are not always maladaptive in mucosal homeostasis. Both Bacteroides fragilis and Clostridium species promote the number and function of a certain regulatory T cell subset in the colon leading to protection against murine colitis. In fact, normal development of regulatory cells and epithelial cell integrity are abolished in the absence of an intestinal flora, suggestive of the need for certain microbial components to induce beneficial anti-inflammatory mechanisms.
All in all, altered immune responses to microbes play a crucial role in IBD pathogenesis. However, certain components of the microbiota are also likely critical for normal development of regulatory mechanisms that contribute to mucosal homeostasis. Findings in animal models highlight the concept that IBD is a disease that results from the interplay of genetics and microbial/environmental factors.
Inflammatory bowel disease (IBD), which encompasses Crohn’s disease and ulcerative colitis, affects 1 million individuals in the United States with often devastating impact on patient quality of life. The last two decades of discovery have underscored the complexity of IBD pathogenesis. Partial heritability of disease has long been recognized with a concordance rate in monozygotic twins of 10–15% in ulcerative colitis and 30–35% in Crohn’s disease [1] but underscores the potential contributors of additional host factors such as possible infectious triggers or environmental variations. More recently, genome-wide association studies (GWAS) in patients with IBD have identified more than 150 associated loci with many of the genetic variants pointing to the importance of barrier function, microbial defense, innate immunity regulation, autophagy, regulation of adaptive immunity, and metabolic pathways associated with cellular homeostasis [2–4]. However, despite rapid progress in identifying these various genetic susceptibility loci, they account for only <30% of the disease risk, which is evidence that IBD is a multi-factorial disease that encompasses not only genetic predisposition but also effects from the environment [5]. Thus, much attention has been directed to further elucidate the dynamic interaction between the mucosal immune response and the estimated 100 trillion microorganisms in the human gut [6]. Presently, inflammatory bowel disease is thought to result from a loss of homeostasis between the intestinal immune system and the intestinal microbial milieu in patients with specific genetic predispositions.
Investigations into the effects of the microbial flora and intestinal mucosal homeostasis has been made plausible with the use of various animal models of IBD, which were first identified in the early 1990’s starting with three genetically altered mice—T-cell receptor alpha (TCRα) [7], interleukin (IL)-2 [8], and IL-10 knockout (KO) mice [9], all noted to spontaneously develop chronic colitis [10]. Paralleling the complexity of the genetics implicated in IBD risk, more than 60 experimental models of IBD have been developed, each with varying strengths and limitations (ease and length of experiments, acute vs chronic inflammation, spontaneous vs chemically induced onset, subtle versus severe phenotype, susceptibility to colitis-induced cancer) [11]. The experimental models can be best characterized by the mechanism driving mucosal inflammation, the most common being genetic alterations resulting in universal or conditional knockout of a gene involved in maintenance of epithelial barrier or mucosal homeostasis. Additional mechanisms of colitis initiation include spontaneous disease development in certain inbred mice, chemical induction of disease (e.g. 2,4,6-trinitrobenzenesulfonic acid [TNBS], dextran sodium sulfate [DSS], and oxazolone), or adoptive transfer of naive CD4+ T cells into immunodeficient mice [12].
As appreciated in human IBD, the importance of the intestinal flora in disease development in experimental models is being increasingly recognized. Even more than ten years ago, attenuated disease was noted in some genetically altered mice (including IL-10 and TCRα KO animals) when raised in germ-free conditions and varying degrees of severity of disease was observed in the IL-10−/− mice maintained in specific pathogen-free compared to conventional housing conditions [13, 14]. Early investigation into the role of microbial flora include observations of higher numbers of bacteria adherent to the epithelium in IL-10 KO compared to control mice despite similar total colonic bacterial counts between the two groups [15]. In addition, it was noted that antibiotics administered in various regimens, including continuous versus only during the first few weeks of life versus delayed initiation, resulted in attenuation of disease in these genetically altered mice [15]. Since then, antibiotics have been demonstrated to prevent disease in other models of colitis, such as mice with defective TGFβ and IL-10 (the two most crucial anti-inflammatory cytokines) signaling in which administration of antibiotics with activity against aerobic and anaerobic bacteria (ciprofloxacin and metronidazole) completely prevented disease and resulted in near-complete abrogation of pro-inflammatory cytokine production [16]. Garrett et al. also demonstrated complete resolution of colitis in Tbet/RAG-2 double KO (TRUC) mice with broad-spectrum antibiotic treatment with vancomycin, metronidazole, neomycin, and ampicillin [17]. In these studies, conventional culture of fecal pellets of antibiotic-treated compared to control mice demonstrated relatively unchanged aerobic colony counts (1010.11 versus 1010.16) but dramatically decreased anaerobic colony counts (109.74 versus <104.01; log10CFU/gram dry weight of stool) [17].
In an attempt to identify a particular member of the microbiome with colitogenic potential, Helicobacter species, a group of gram-negative, microaerophilic, helical-shaped organism, have been intensely investigated. Since its first identification in the early 1990s, Helicobacter hepaticus has been shown to be associated with chronic typhlocolitis in multiple immunodeficient but not immunocompetent mice [18–20]. In fact, many of genetically altered models of colitis do not develop disease when re-derived in an Helicobacter-free environment and re-introduction of one or several Helicobacter speciesleads to recapitulation of disease originally observed in normal specific pathogen-free conditions [21]. The pathogenicity of additional enterohepatic Helicobacter species, such as H. bilis and H. rodentium, has also been recognized [22]. However, it is not clear whether Helicobacter species themselves are directly pathogenic in genetically predisposed animals or whether they act through altering the intestinal microbial membership.
Additionally, infection with Helicobacter has been recognized to also drive the development of colitis-associated colorectal cancer. In RAG-2 KO mice, infection with H. hepaticus resulted in high-grade dysplasia and progression to adenocarcinoma arising from colitis-associated dysplastic epithelial foci [23]. Similarly, Maggio-Price et al. found dual-infection of mdr1a KO mice (which lack the membrane efflux pump p-glycoprotein) with H. bilis and H. hepaticus developed the most severe dysplasia relative to uninfected mice with a 5- to 12-fold increased expression of the oncogene c-myc relative to wild-type and uninfected mice by quantitative real-time polymerase chain reaction on colonic epithelial preparations [24]. Furthermore, colon cancer developed in Smad3 KO mice (which have defective TGF-β signaling) only in the setting of Helicobacter infection where dual-infection with H. bilis and H. hepaticus resulted in colitis and colorectal cancer in 50–60% of Smad3 KO mice [24, 25]. The mechanism underlying oncogenesis induced by Helicobacter is not clear, but it is known that adoptive transfer of CD4+CD25+CD45RBlo regulatory T cells (Tregs) into Rag KO mice significantly reduced H. hepaticus-induced inflammation and carcinogenesis in these mice [23].
A recently described innate immune-driven model of colitis has clearly demonstrated the colitogenic and oncogenic potential of certain members of the intestinal microbiota. Severe colitis was noted to occur by 4 weeks of age in T-bet/RAG-2 double KO (TRUC) mice, which lack both lymphocytes (due to RAG-2 deficiency) and T-bet, a member of T-box transcription factor family that regulates the differentiation and function of immune cells [17]. However, disease is completely abrogated by treatment with a combination of vancomycin, metronidazole, neomycin, and ampicillin with more than 100,000-fold decrease in cultureable fecal anaerobes but relatively unchanged aerobic colony counts, again suggesting the potential pathogenicity of anaerobic community [17]. Most interestingly, Garrett et al. questioned whether this population of potentially colitogenic anaerobic commensals could transfer disease either vertically to offsprings or horizontally to housemates. In cross-fostering experiments, wildtype and RAG-2−/− mice (which usually do not have any abnormal gross phenotype) were observed to develop colitis when fostered by TRUC mothers, confirming vertical transmission of colitis by pathogenic microbial members even to mice without genetic predisposition [17]. Additionally, horizontal transmission of colitis was observed to occur in adult wildtype and RAG-2 KO mice co-housed with adult TRUC mice for 8 weeks.
By phylogenetic analysis with 16S rRNA sequencing of stool, fifty-seven candidate colitogenic bacterial species were identified in TRUC mice [26]. Given that treatment with gentamicin and metronidazole, but not vancomycin alone, abrogated disease, the candidate species were narrowed to two gram-negative facultative organisms, Klebsiella pneumoniae and Proteus mirabilis. With successful treatment of disease using anti-tumor necrosis factor (TNF)-α antibody therapy, significant decreases in K. pneumoniae and P. mirabilis colonization was observed [26]. Additionally, K. pneumoniae and P. mirabilis were detected in the fecal samples of the cross-fostered RAG-2 KO and wild-type mice at levels comparable to age-matched TRUC-fostered TRUC mice but not regular wildtype and RAG-2 KO mice [26]. Infection with either K. pneumoniae or P. mirabilis led to disease in T-bet-sufficient mice housed in SPF conditions. However, co-colonization of germ-free mice with the two Enterobacteriaceae did not result in colitis, suggesting a still unknown interaction with the gut microbial community is required to cause disease [26].
These provocative studies continue to highlight the complexity of the interactions between the gut microbial community and the host immune system. Although the pathogenicity of Helicobacter, K. pneumoniae and P. mirabilis has been demonstrated, the specific mechanism underlying the interaction with the host immune system remains unknown.
On a molecular level, given the constant exposure of the host intestinal epithelial cells and the mucosal immune system to a large amount of microbial antigens, innate immune recognition of an expansive assortment of microbe-associated molecular patterns on pathogens and commensals by Toll-like receptors (TLRs) is important in controlling infections; however, this same process may sometimes elicit an inappropriate immune response [27]. The fact that Nod2, an intracellular protein that recognizes muramyl dipeptide (a peptidoglycan constituent of bacteria) is defective in a subset of patients with Crohn’s disease [28] underscores the association between microbial detection and IBD. Using a molecular technique called serological expression cloning on sera from colitic C3H/HeJBir mice (a substrain of mice with spontaneous colitis), Lodes et al. identified a family of related novel flagellins, which activate TLR5, as a class of immunodominant antigens [27]. These 15 novel flagellin clones were most closely related to flagellins from Butyrivibio, Roseburia, Thermotoga, and Clostridium within the Clostridium subphylum cluster XIVa of gram positive bacteria. In fact, antibodies to Cbir1, a particular commensal-derived flagellin, were also detected in other mouse models of colitis including mdr1a KO mice that have epithelial barrier dysfunction and B6.IL-10 KO mice that lack the down regulatory cytokine IL-10 [27]. Not only did the three mouse models of colitis examined have T cells reactive to Cbir1 but adoptive transfer of Cbir1-specific CD4+ T cells into immunodeficient scid mice induced colitis in all recipients. Strikingly, similar to the murine models, serologic testing of a large panel of Crohn’s disease patients and controls found a significantly higher level of anti-CBir1 antibody in the serum of Crohn’s disease patients than that of unaffected controls or patients with ulcerative colitis [27]. Expanding on the finding of CBir1 antibodies in colitic humans and animals, Hand et al. hypothesized that acute gastrointestinal infection could lead to the priming of the adaptive immune system with not only antigens derived from the specific pathogen but those from commensals as well [29]. With a model of acute gastrointestinal infection with Toxoplasma gondii, the authors observed an increased physical association between commensal bacteria and the intestinal epithelium with bacterial translocation and marked increased bacterial load in mesenteric lymph nodes, spleen, and liver [29]. In addition, more than half of the resulting IFN-γ-producing T cells were not reactive to T. gondii but presumably commensal bacteria [29]. When transferred to T. gondii-infected hosts, T cells from a TCR transgenic mouse with reactivity to Cbir1 were shown to proliferate whereas T cells transferred into uninfected hosts did not, suggesting that T cells specific to commensals could be induced to respond in the setting of acute infection. Furthermore, CBir1-specific T cells were shown to be activated in mice treated with DSS [29], again supporting the hypothesis that compromise of mucosal integrity can lead to loss of CD+4 T cell tolerance to commensal antigens.
Also prompting an adaptive immune response, a particular member of the intestinal microbiota was recently found to be responsible for the induction of the helper T cell subset Th17, which produces interleukin (IL)-17, a cytokine that plays a crucial role in controlling bacterial and fungal infections [30]. Th17 cells were first noted a few years ago to be present in the intestinal lamina propria of wild-type B6 mice from Taconic Farms but notably absent in wild-type B6 mice from Jackson Laboratory [31], raising the speculation that differences in the gut microbial community was driving this difference. This hypothesis was proven by the induction of Th17 cells in the Jackson mice by transferring ileal or colonic bacteria from Taconic mice into Jackson mouse recipients [30]. Investigating the differences in bacterial composition of the small intestine contents of the Taconic and Jackson mice using 16S ribosomal RNA microarray revealed statistically significant differences of 479 taxa; yet, only two taxa were >25-fold more abundant in Taconic mice [30]. These two taxa were identified as members of Lactobacillaceae and Clostridiaceae families - Lactobacillusmurinus ASF361 and a segmented filamentous species of the candidate genus Arthromitus. Because Lactobacillaceae is known to be intentionally introduced into all Taconic mice strains and, in germ-free models, it has not been shown to induce Th17 cells, investigations were focus on the segmented filamentous species (SFB).
Segmented filamentous species, first discovered by Davis and Savage [32] in 1974, are yet to be cultured. They are commensal, gram-positive, anaerobic spore-forming bacteria with long filamentous morphology and well-defined septa often spanning the length of multiple villi. Segmented filamentous species are known to interact with the host immune system with colonization of germ-free animals leading to IgA production and recruitment of intraepithelial lymphocytes in the gut [33–35]. Using germ-free animals, Ivanov et al. demonstrated induction of Th17 cells after colonization with SFB, and ultimately, these Th17 cells were protective against experimental Citrobacter infection [30], suggesting a possible role in defense against intestinal pathogens. Further study is needed to understand the impact of Th17 induction as these cells have been implicated in the pathogenesis of autoimmune disease, including experimental autoimmune encephalomyelitis and colitis [36, 37].
Given the central role of Tregs in the maintenance of mucosal homeostasis, it was of high interest that two recent studies have implicated particular members of the commensal microbiota in induction of these regulatory cells. Given the temporal correlation between the timing of increasing Tregs in the intestinal lamina propria and the increasing exposure to commensals after birth, Atarashi et al. hypothesized a central role of a microbial community member in colonic Treg induction [38]. The use of antibiotics implicated an important role of gram-positive commensal bacteria in the accumulation of Tregs in the lamina propria as only animals treated with vancomycin showed a decreased presence of Tregs in the colon. Furthermore, animals inoculated with 3% chloroform-resistant fecal microorganisms (spore-forming fraction) were shown to have similar numbers of accumulated Tregs compared to untreated fecal inoculation, suggesting the critical organism to be spore-forming [38]. Within this subset of the commensal flora, Clostridium was hypothesized to be the particular organism given it is one of the most prominent organisms in the murine gastrointestinal tract and prior studies have implicated Clostridium clusters IV and XIVa (also known as Clostridiumseptum and coccoides groups, respectively) as potentially important in maintaining mucosal homeostasis in inflammatory bowel disease [39, 40]. Using germ-free mice, Atarashi et al. demonstrated colonization with 46 strains of Clostridium led to marked induction of IL-10-producing Tregs in the colonic lamina propria [38]. This Treg accumulation correlated with protection against colitis initiated with dextran sodium sulfate (DSS) or oxazolone administration [38]. This finding is in line with the fact that intestinal epithelial cells from mice colonized with Clostridium produce indoleamine 2,3-dioxygenase (IDO) [38], which is known to induce Tregs and limit disease severity in several colitis models [41, 42]. Ultimately, the authors also showed the impact of Clostridium inoculation to be most significant when exposure occurred early in life (at 2 weeks of age) [38], supporting the widely held hypothesis that early exposures have significant impact on adult gut microbial populations.
Induction of protective IL-10 Tregs has been additionally observed in association with Bacteriodes fragilis [43]. In germ-free mice, Tregs were found in similar percentages in the mesenteric lymph nodes and colon as compared to mice raised under conventional conditions, yet the production of IL-10 within the intestine was marked reduced in the germ-free mice. However, germ-free animals mono-associated with B. fragilis had a 2-fold increase in the percentage of IL-10-producing Foxp3+ Tregs, as well as resurgence of the expression of the anti-inflammatory cytokine TGF-β2. Previously, Mazmanian et al. had demonstrated that polysaccharide A (PSA), a molecule produced by B. fragilis, exerted a protective effect against a murine model of colitis by suppressing IL-17 production [44]. Here, the induction of IL-10-producing Tregs with B. fragilis monoassociation was found to be dependent on PSA through toll-like receptor 2 signaling. Similar to studies by Atarashi et al. [38], Treg induction with PSA was associated with attenuation of TNBS-induced colitis given either as pretreatment or 1–2 days after TNBS initiation [43].
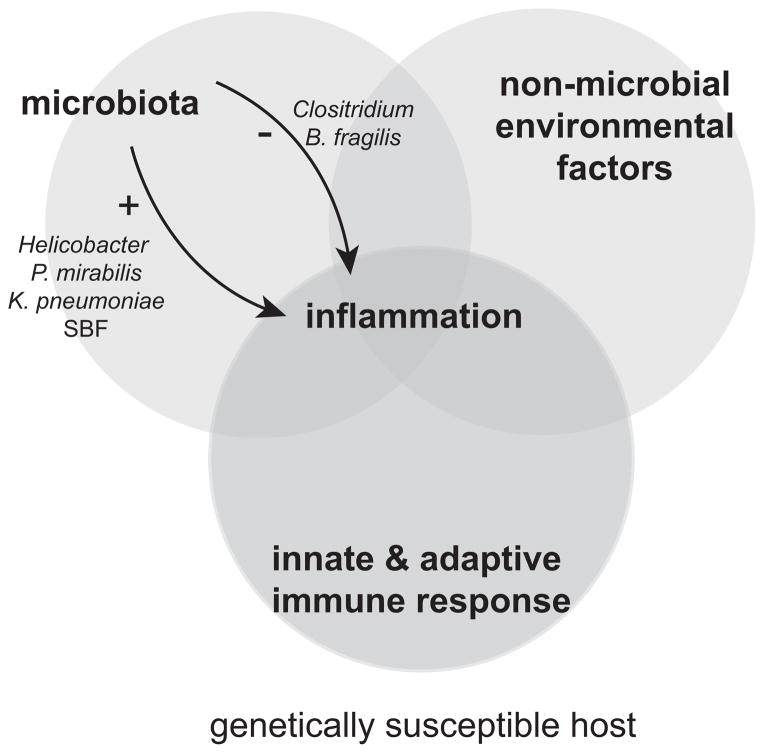
With more understanding, the importance of the interaction between the intestinal microbial milieu (especially the anaerobic community) and the host immune system in the development and potentiation of human disease has become increasingly apparent. In multiple examples, certain immune defects (e.g. TRUC mice) can create a niche for potentially colitogenic microbes, which may (e.g. Helicobacter) or may not be part of the commensal flora (e.g. K. pneumoniae and P. mirabilis). Additionally, some gut microbes (e.g. Helicobacter) can cause not only colitis but also colorectal cancer, while certain commensals play a critical role in the development of T cell subsets in the examples of SFB, Clostridium, and B. fragilis. Such findings in animal models highlight the interplay of genetics, microbial and environmental factors in the pathogenesis of inflammatory bowel disease (Fig. 1).
Figure 1. Complex interactions of host immunity, intestinal microbiota, and environmental factors lead to inflammation.
Inflammatory bowel disease is thought to result from a loss of homeostasis between the host intestinal immune system and the intestinal microbial milieu (especially the anaerobic community) in genetically susceptible individuals. In animal models, several particular microbial members (e.g. Helicobacter, K. pneumoniae, and P. mirabilis) have been associated with colitis development while certain commensals (e.g. SFB, Clostridium, and B. fragilis) play a critical role in the development of T-cell subsets, some of which may protect against intestinal inflammation.
Highlights.
Animal models have uncovered a central role of the intestinal microbiota in colitis development
Some members of the intestinal microbiota are associated with induction of colitis
A subset of intestinal commensals has been shown to induce colitis-associated colon cancer
Certain commensals have been found to induce Th17 cells or regulatory T cells
Acknowledgments
Funding was from National Institutes of Health K08DK083430 awarded to DDN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spehlmann ME, Begun AZ, Burghardt J, Lepage P, Raedler A, Schreiber S. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14:968–76. doi: 10.1002/ibd.20380. [DOI] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–70. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mombaerts P, Mizoguchi E, Grusby MJ, Glimcher LH, Bhan AK, Tonegawa S. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:274–82. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- 8.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–61. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 10.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizoguchi A. Animal models of inflammatory bowel disease. Prog Mol Biol Transl Sci. 2012;105:263–320. doi: 10.1016/B978-0-12-394596-9.00009-3. [DOI] [PubMed] [Google Scholar]
- 12.Mizoguchi A, Mizoguchi E. Inflammatory bowel disease, past, present and future: lessons from animal models. J Gastroenterol. 2008;43:1–17. doi: 10.1007/s00535-007-2111-3. [DOI] [PubMed] [Google Scholar]
- 13.Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–20. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999;11:648–56. doi: 10.1016/s0952-7915(99)00032-1. [DOI] [PubMed] [Google Scholar]
- 15.Madsen KL, Doyle JS, Tavernini MM, Jewell LD, Rennie RP, Fedorak RN. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology. 2000;118:1094–105. doi: 10.1016/s0016-5085(00)70362-3. [DOI] [PubMed] [Google Scholar]
- 16.Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, et al. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foltz CJ, Fox JG, Cahill R, Murphy JC, Yan L, Shames B, et al. Spontaneous inflammatory bowel disease in multiple mutant mouse lines: association with colonization by Helicobacter hepaticus. Helicobacter. 1998;3:69–78. doi: 10.1046/j.1523-5378.1998.08006.x. [DOI] [PubMed] [Google Scholar]
- 19.Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–31. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward JM, Anver MR, Haines DC, Melhorn JM, Gorelick P, Yan L, et al. Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab Anim Sci. 1996;46:15–20. [PubMed] [Google Scholar]
- 21.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011;4:22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shomer NH, Dangler CA, Schrenzel MD, Fox JG. Helicobacter bilis-induced inflammatory bowel disease in scid mice with defined flora. Infect Immun. 1997;65:4858–64. doi: 10.1128/iai.65.11.4858-4864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erdman SE, Poutahidis T, Tomczak M, Rogers AB, Cormier K, Plank B, et al. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003;162:691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maggio-Price L, Bielefeldt-Ohmann H, Treuting P, Iritani BM, Zeng W, Nicks A, et al. Dual infection with Helicobacter bilis and Helicobacter hepaticus in p-glycoprotein-deficient mdr1a−/ − mice results in colitis that progresses to dysplasia. Am J Pathol. 2005;166:1793–806. doi: 10.1016/S0002-9440(10)62489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maggio-Price L, Treuting P, Zeng W, Tsang M, Bielefeldt-Ohmann H, Iritani BM. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 2006;66:828–38. doi: 10.1158/0008-5472.CAN-05-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–12. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 29.Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, et al. Acute Gastrointestinal Infection Induces Long-Lived Microbiota-Specific T Cell Responses. Science. 2012 doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov II, de Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–49. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun. 1974;10:948–56. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–11. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun. 1993;61:303–6. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 37.Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–67. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–9. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 41.Ciorba MA, Bettonville EE, McDonald KG, Metz R, Prendergast GC, Newberry RD, et al. Induction of IDO-1 by immunostimulatory DNA limits severity of experimental colitis. J Immunol. 2010;184:3907–16. doi: 10.4049/jimmunol.0900291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matteoli G, Mazzini E, Iliev ID, Mileti E, Fallarino F, Puccetti P, et al. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut. 2010;59:595–604. doi: 10.1136/gut.2009.185108. [DOI] [PubMed] [Google Scholar]
- 43.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]