Abstract
Background
A key issue in otitis media (OM) is mucous cell metaplasia in the middle ear mucosa, a condition for hyperproduction of mucus in the middle ear mucosa and development of chronic OM. However, little is known about the driving force for the differentiation of mucous cells in OM.
Methods
Mouse middle ear epithelial cells (mMEEC) were used in this study to test whether Math1, a critical transcription factor for the development of mucous cells in the intestine, synergize with inflammatory cytokines (tumor necrotic factor alpha, TNFα) and other epithelial differentiation factors (retinoid acid, RA) to induce the differentiation of mMEEC into mucus-like cells in vitro. Simultaneously, Math1 was transduced into the middle ear mucosa and see whether it induces mucous cell hyperplasia in vivo.
Results
Math1 significantly increased the mucus cell numbers in the middle ear mucosa of mice. Math1, in the presence of TNFα and epithelial differentiation factor retinoic acid (RA), synergistically promoted the differentiation of mMEEC into mucus-like cells via upregulation of mucins and their chaperones:trefoil factors (TFFs) in vitro. RA treatment for 12 hours activated Math1 although RA alone had very limited effects on mucus-like cell differentiation.
Conclusion
Math1 plays a critical role in the pathogenesis of OM by induction of mucous cell differentiation, in the presence of TNFα and RA.
INTRODUCTION
Under normal conditions, few mucous cells exist in the middle ear mucosa of humans and rodents and they are mainly distributed in the orifice of Eustachian tube, promotary area, and inferior tympanium (so called “ciliated tract”). Under the pathological conditions, mucous cells are dramatically increased (1–7) with abundant production of mucins (2, 8) and mucin chaperones: trefoil factors (9). Upregulation of mucins and mucin chaperones is a biological basis for mucous cell metaplasia (MCM), a predisposition for chronic OM.
It is known that pro-inflammatory cytokines, such as TNFα, are involved in the development of MCM in animal models (10). However, little is known about the additional factors needed for the differentiation of mucous cells. Knockout of Math1 (Atoh1, atonal homolog 1) in mice developed no mucous cell lineage in the intestine, suggesting that Math1 is a candidate involved in the development of MCM in OM. Since the Math1 knock out mouse model is embryonic lethal (11) and not available for MCM, we thus sought to investigate this issue by using in vitro “knock-in” techniques in mMEEC.
A classic pathway for MCM is middle ear infection. Inflammatory cells produce cytokines and cytokines, in turn, stimulate the differentiation of mucous cells (2, 10). A number of cytokines generated from inflammatory cells upregulate the expression of mucins and induce MCM. These cytokines include TNFα (10, 12–14), and interleukin-4 (IL-4),(15) IL-10,(16) IL-8,(17) IL-9,(18) IL-13 (19–21). They are either proinflammatory cytokines or lymphocyte (especially T helper 2 subset)-derived cytokines.
We hypothesized in this study that Math1, together with TNFα and RA, plays an important role in the differentiation of mucous cells in the middle ear. As expected, transfection of mMEEC with Math1 regulated the expression of mucins and TFFs in the presence of TNFα and RA.
MATERIALS AND METHODS
Cell Culture
Mouse middle ear epithelial cells (mMEEC) were made in our laboratory as previously described (22). They were maintained in Ham’s F-12K culture media (American Type Culture Collection, ATCC, Manassas, VA) supplemented with 2 mM L-glutamine and 10 ng/ml epidermal growth factor (EGF), 5 µL/ml insulin-transferrin-sodium selenite (ITS, 100×, Sigma-Aldrich, St. Lewis, MO) solution, 2.7 g/L glucose, 500 ng/ml hydrocortisone, 0.1 mM nonessential amino acids, 50 µg/mL streptomycin, 50 units/mL penicillin and 4% fetal bovine serum, thereafter referred to as full growth medium (FGM, Sigma-Aldrich, St. Lewis, MO). FGM change was made every 3~4 days. During the experiment, EGF in the above media was omitted when various factors were added.
Regulation of Math1 in mMEEC
mMEEC was cultured on 8-well chamber slides or in T25-flasks. Cells were starved in F-12K culture medium for 24 hours, then incubated with 20ng/ml EGF, 20ng/ml TNFα, 10−9 M RA, or TNFα+RA for 12 hours, and harvested for evaluation of Math1 expression.
Transfection of Math 1 in the middle ear mucosa of mice
Full-length Math1 cDNA was cloned into a protein-expressing vector (C2 pEGFP, e.g., empty vector, Clontech, San Diego, CA), as previously described (23). The Math1 cDNA sequence in empty vector in a sense manner was confirmed by sequencing and referred to as sense-Math1 cDNA (hereinafter referred to as Math1). To study the role of Math1 in the differentiation of mucous cells, bilateral bullae of 5 mice were transfected with 10 µL of Math1 and empty vectors (ev), respectively, at 1.4 µg/mL in Opti-MEM containing Lipofectin at 6 µg/mL via the tympanic membrane approach. Transfected animals were sacrified 7 days after Math1 transfection for harvest of the bullae. The bullae were then fixed in 10% formalin and routinely processed for histological sections. Alcian blue-periodic acid Schiffs (AB-PAS) were employed for identification of mucous cells. Mucous cell numbers were quantitatively analyzed by the method as previously described (24).
Establishment of Math 1 stably transfected mMEEC
In our pilot study, cDNA transfection efficiency of mMEEC was low and transient transfection was obviously insufficient for determination of the effect of Math1 on cells. To overcome this problem, Math1 and ev were stably transfected mMEEC. Briefly, cells were cultured in a 12-well plate to 70% confluence and transfected with s-Math1 and ev at 1.4 µg/mL for 16 hours in Opti-MEM (serum-free, Invitrogen, Grand Island, NY) containing 6 µg/mL of Lipofectin, cultured in FGM, incubated with G418 (400 µg/mL, Invitrogen, Grand Island, NY) for two weeks for selection of successfully transfected cells. This procedure was repeated once. Cells persistently resistant to G418 for 3 months were defined as stably transfected cells. To select highly positive cells for GFP, top 5% of GFP+ cells were sorted out by a cell sorter (FACSAria, BD Biosciences, Inc, Becton-Dickinson, Mountain View, CA) as described previously (25) after G418 selection twice. Math1 mRNA and protein expression in these selected cell clones were routinely examined by RT-PCR, immunohistochemistry and/or fluorescence-activated cell sorting (FACS) for verification of Math1 expression.
Induction of mucous cell differentiation in Math1 stably transfected cells
Math1 and ev stably transfected cells were cultured for 24 hours in T-25 flasks or on 8-well chamber slides with a starting cell number at approximately 2.5×104 cells per well in starvation media (2% serum without any growth factors). Cells were incubated with 20ng/mL of TNFα, 10−9 of M RA, or TNFα+RA, respectively, for two weeks in FGM (media and factors were supplied every two days) and then starvation media for 2 days. Cells were then harvested for evaluation of mucous cell differentiation by AB-PAS, immunohistochemistry, and FACS.
Inhibition of mucous cell differentiation with specific inhibitors
To study which signaling pathways are involved in mucous cell differentiation, Math1 and ev stably transfected cells were pre-incubated pathway inhibitors (25 µM PD98059 for the Erk pathway, 20 µM LY294002 for the Akt pathway, and 10µM AG1478 for the EGFR pathway from Calbiochem-Novabiochem, San Diego, CA, 15 µM SB203580 for the p38 MAPK pathway and 20µM SP600125 for the JNK pathway from Sigma-Aldrich, St. Lewis, MO) for two hours and incubated with TNFα, RA, and TNFα+RA for 14 days in the presence of the above inhibitors, followed by the above starvation media for 2 days without TNFα, RA, and TNFα+RA but with inhibitors. Cells were harvested for evaluation of mucous cell differentiation by AB-PAS, immunohistochemistry and FACS.
AB-PAS stain
Cells on chamber slides were fixed with 100% ethanol for 6 minutes in room temperature and incubated with 1% alcian blue (AB) for 30 minutes, 0.5% PAS reagent for 10 minutes, then sulfurous acid for two minutes. Slides were washed in tap water and examined under a light microscope for identification of positive mucous granules. Cells with blue and purple colors were defined as mucous cells. Mucous cell numbers from 4~6 areas on each chamber were counted. Quantitative data are presented as mean±S.E. Total cell numbers were determined by DAPI stain simultaneously and the percentage of AB-PAS positive cells against total cells was calculated. Final mucous cells are presented as % of total cell numbers
Immunohistochemistry
Cells on Lab-Tec chamber slides (Nalge Nunc International, IL) were fixed in 100% ethanol, and incubated with mucous cell marker antibodies: anti-TFF3(1:50, Calbiochem, San Diego, CA) or anti-Math1 (1:100, abcam, Cambridge, MA) at 4°C overnight, washed with PBS, incubated with fluorescein isothiocyanate (FITC)- or tetramethylrhodamine isothiocyanate (TRITC)-conjugated secondary antibodies (Zymed, Santa Cruz, CA) for 60 minutes at room temperature, washed with PBS, and examined under a fluorescence or a confocal microscope. Non-specific IgG was used for immunohistochemistry negative control.
Fluorecence-Activated Cell Sorting (FACS)
Cells were incubated with 0.3% saponin in PBS for 5 minutes, incubated with anti-TFF3 (1:50) or anti-Math1 (1:50) for 30 minutes on ice, washed with 0.3% saponin in PBS twice, incubated with TRITC-conjugated secondary antibody for 30 minutes on ice, washed with 0.3% saponin in PBS twice, and analyzed on a BD FACSCalibur flow cytometer. Cells incubated with non-specific IgG (zymed products), followed by TRITC-conjugated secondary antibodies, served as antibody controls. Data was analyzed with CellQuest Pro (BD Sciences) and FlowJO (7.1 version, Tree Star Inc) for presentation. Experiments were run in duplicate or triplicate. Results are presented as % positive cells over 3,000 cells per sample after subtraction with non-specific IgG background.
RT-PCR
Total RNA was isolated from the above harvested cells using RNA Miniprep Kit (Stratagene). Residual genomic DNA in total RNA samples was digested with DNases according to the manufacturer’s instruction. Specific primers for Math1, TFF3, GAPDH were as follows: Math1 primers: 5’-agatctacatcaacgctctgtc-3’/5’-actggcctcatcagagtcactg-3’ (58.5°C, 452bp, 30cycles); TFF3 primers: 5’-cagattacgttggcctgtctcc-3’/5’-atgcttgctacccttggaccac-3’(60°C, 254bp, 30cycles); and GAPDH primers: 5’-aacgggaagcccatcacc-3’/5’-cagccttggcagcaccag-3’ (61°C, 441bp, 20cycles). The specificity of these primers was by BLAST search using the National Center of Biologic Information web site (fttp://www.ncbi.nlm.nih.gov). RT-PCR was performed as described previously (26, 27). PCR products were analyzed on a 2% agarose gel for evaluation of band sizes and routinely purified for sequence verification. RNA samples omitting reverse-transcription enzyme served as controls.
Microarrays
Affymetrix microarrays were performed using the above total RNA as previously described (28). Briefly, cDNA was prepared from 20 µg total RNA using a T7-dT24 primer. cRNA was synthesized from cDNA and biotinylated using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY) and hybridized with the Rat U34 arrays (Affymetrix, Santa Clara, CA). The microarray data were analyzed as previously described (28).
RESULTS
Math1 transfection induces mucous cell metaplasia in the middle ear mucosa of mice
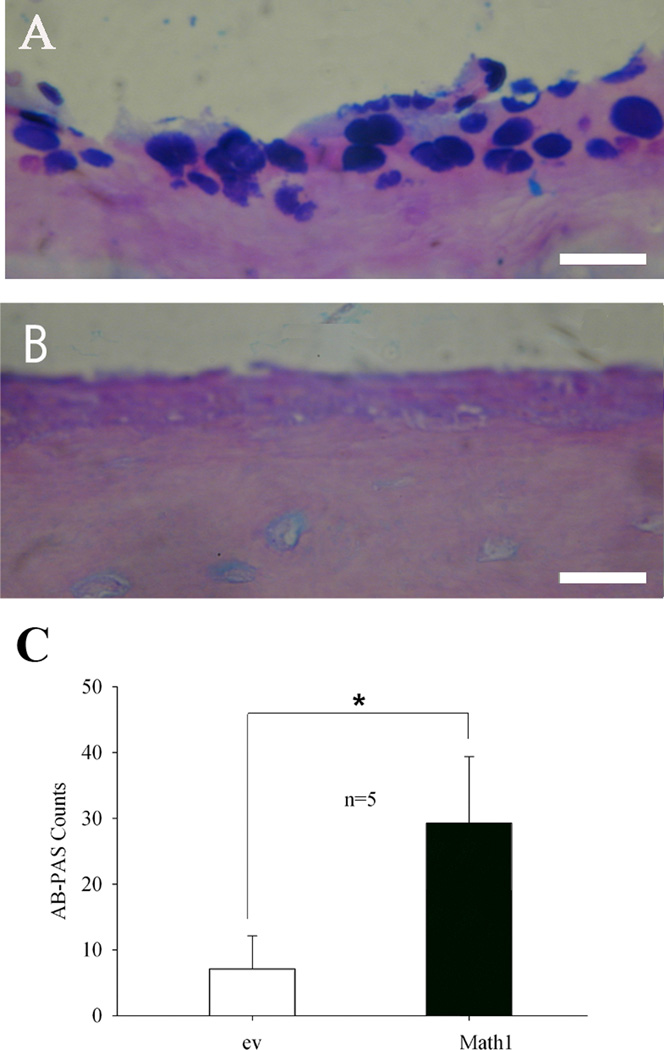
To study the importance of the Math1 gene in differentiation of mucous cells, transfection of Math1 in the middle ear was performed in the 10 bullae of 5 mice. It was found that mucous cell numbers were increased in Math1 transfected middle ear mucosa (Fig 1A) compared with empty vector transfected middle ear mucosa (Fig. 1B). Statistically, AB-PAS positive cells were significantly higher in the bullae transfected with Math1 than those transfected with empty vector (Fig. 1C).
Fig. 1.
Math1 transfection increases the mucous cell numbers in the middle ear mucosa of mice. Math1 transfection had more AB-PAS positive cells in the mouse middle ear mucosa (A) than empty vector (B) transfection. Statistical analysis showed that Math1 significantly increased the AB-PAS cell counts in the middle ear mucosa of 5 mice (C). *p<0.01(n=5); Bar=10 µm.
Math1 transfection alone in vitro dose not induce mucous cell differentiation
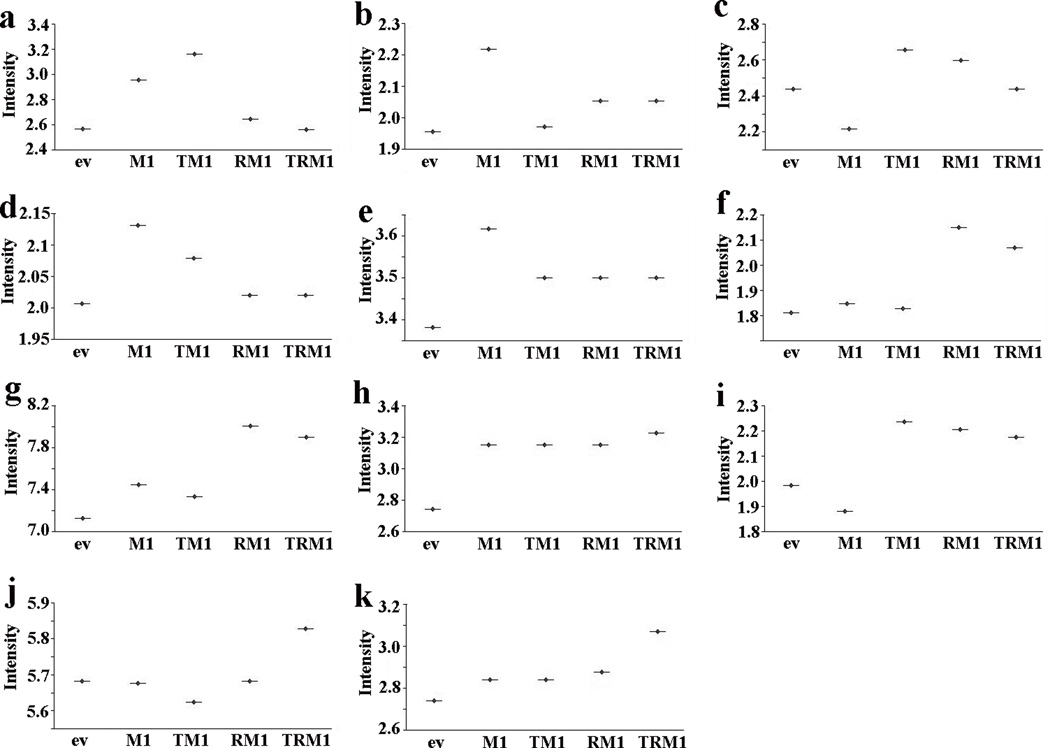
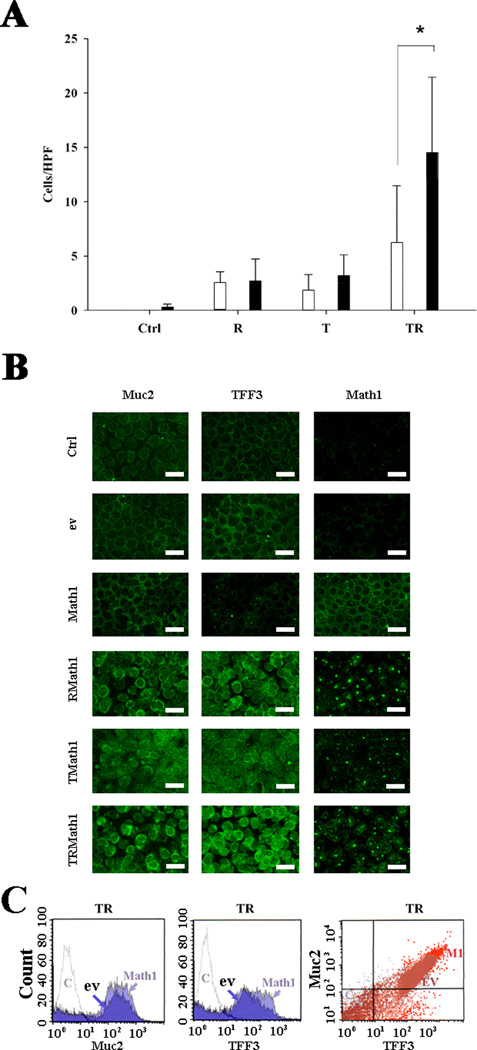
To investigate the role of Math1, cultured mMEEC were stably transfected with Math1 and empty vector, respectively. After incubation with G418 for more than 3 months, survived cells were submitted for selecting GFP highly positive cells. Math1 was weakly expressed in empty vector-transfected cells. Math1 transfection successfully increased the expression of Math1 by RT-PCR, FACS, and immunohistochemistry (Supplemental Figure 1, online), suggestive of efficient transfection of Math1 in cultured mMEEC. To study whether Math1 is involved in the developmet of mucous cells, Math1 and empty vector stably transfected cells were incubated with various factors for 14 days and then without these factors for 2 days before harvesting for evaluations. Microarrays data confirmed that Math1 alone incresed the mRNA of mucins (including Muc1, Muc2, Muc4 and Muc5ac but not Muc3 and Muc10), and mucin shaperones (TFF1 and TFF2 but not TFF3) compared with empty vector (Fig. 2). However, Math1 transfection alone did not significantly increase the AB-PAS positive cell numbers (Fig. 3A), nor the expression of Muc2 protein in the cells as judged by immunohistochemistry (Fig. 3B). Math1increased the % of Muc2 and TFF3 positive cells when used together with TNFα and RA in comparison with empty vector as judged by FACS (Fig. 3C).
Fig. 2.
Math1 regulates the expression of mucous cell markers at the mRNA level in cultured mMEEC. Microarray data confirmed that the upregulation of mucins: Muc1(a), Muc2 (b), Muc4 (d), Muc5ac (e), but not Muc3 (c) and Muc10 (f) and mucin chaperones:TFF1(g) and TFF2 (h) but not TFF3 (i) at the mRNA level. TNFα+RA+Math1 (TRM1) obviously increased the expression of TFF1 (g), TFF2 (h), TFF3 (i), EGFR (j), and AKT (k) at the mRNA level. ev, empty vector; TM1, TNFα+Math1; RM1, RA+Math1; TRM1, TNFα+RA+Math1.
Fig. 3.
TNFα+RA+Math1 synergistically increased mucous granules (AB-PAS positive cells) in cultured cells. Cells were treated with various factors for 14 days and then without these factors for 2 days. RA and TNFα alone induced few AB-PAS positive cells and Math1 transfection alone did not significantly increase the AB-PAS positive cells compared with empty vector (A, AB-PAS staining). However TNFα+RA+Math1 significantly increase the % of AB-PAS positive cells compared with TNFα and RA (A, *p<0.05, n=6). empty bar, ev; solid bar, Math1; T, TNFα; R, RA; TR, TNFα+RA. Immunohistochemistry showed that TNFα+RA+Math1 cocktail obviously increased the expression of both Muc2 and TFF3 (B). bar=10 µm. FACS verified that Math1 transfection increased the % positive cells for Muc2 and TFF3 compared with empty vector in the presence of RA and TNFα, and cells positive for Muc2 were also positive for TFF3 (C). HPF, high power field; Ctrl, control (untransfected cells); ev, empty vector; TMath1, TNFα+Math1; RMath1, RA+Math1; TRMath1, TNFα+RA+Math1.
RA+TNFα+Math1 synergistically induces formation of mucous granules in cultured mMEEC
Under inflammatory conditions, various cytokines and factors exist in the middle ear. To study whether they synergize in triggering the differentiation of mucous cells, mMEEC cell cultures were incubated with and without inflammatory cytokines and factors for 14days, then without any factors for 2 days to allow them to differentiate. It was found that TNFα+RA were able to synergistically increase the number of AB-PAS positive cells whereas RA and TNFα alone were unable to do so (Supplemental Figure 2, online). To study whether Math1 is involved in the differentiation of mucous cells under inflammatory conditions, Math1 and empty vector stably transfected cells were incubated with TNFα+RA for 14 days and then without TNFα+RA for 2 days. It was found that TNF α+RA+Math1 had significantly higher AB-PAS positive cell numbers compared with TNFα+RA+empty vector (Fig. 3A). Immunohistochemistry verified that TNFα+RA+Math1 obviously increased the expression of both Muc2 and TFF3 in cells compared with controls (empty vector, Math1, RA+Math1, and TNFα+Math1, Fig. 3B). It has been noted that translocation of Math1 protein to the nucleus was observed in the presence of RA and/or TNFα(namely, RMath1 or TMath1). While Math1 transfection alone did not have sufficient Math1 protein in the nucleus (Math1), suggesting that TNFα and/or RA activate Math1 in the nuclei. This explains why TNFα+RA+Math1 cocktail is essential for induction of mucous cell differentiation. FACS showed that Math1 transfectants had an increase of the % of both Muc2 and TFF3 positive cells compared with empty vector transfectants in the presence of TNFα+RA(Fig. 3C), namely, TNFα+RA+Math1 increased the % of both Muc2 and TFF3 positive cells. It is noted that Muc2 positive cells were also TFF3 positive cells (Fig. 3C).
RA activates Math1 in mMEEC
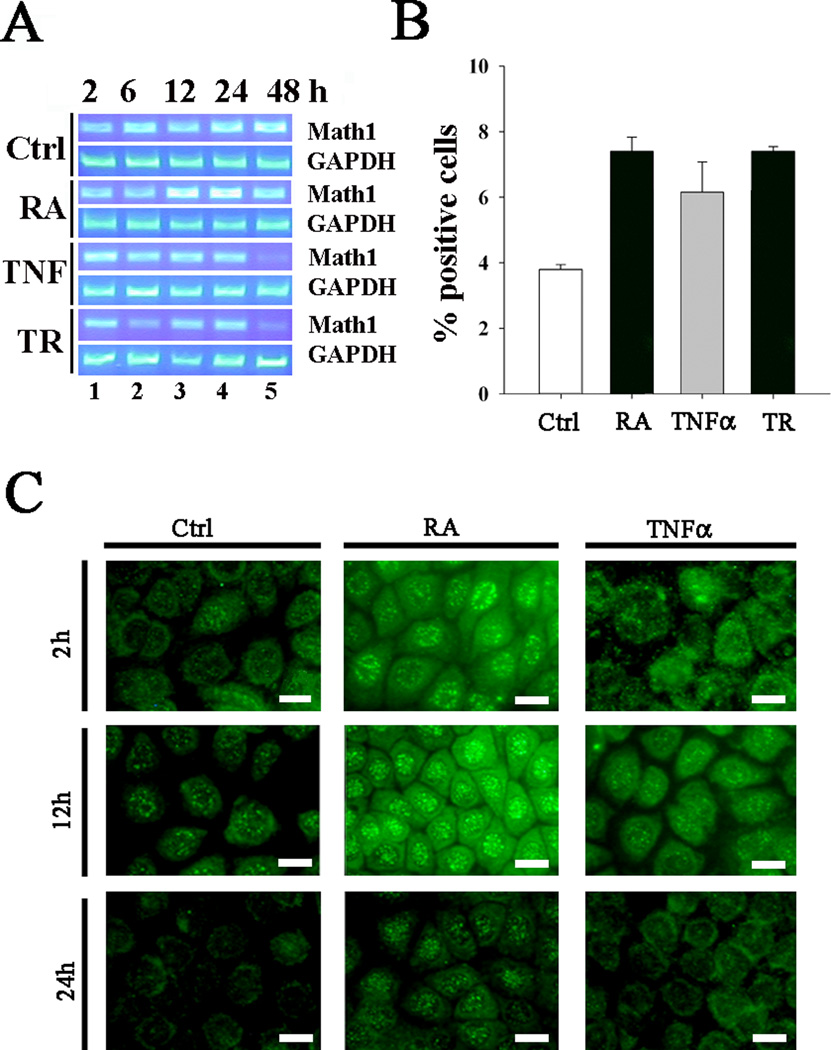
To study whether RA activates Math1, cells were incubated with various factors (with and without RA, TNFα, and TNFα+RA for 2~48 hours) and harvested for RT-PCR, immunohistochemistry, and FACS. It was demonstrated that RA treatment for 12~24 hours increased the expression of Math1 mRNA transcripts by RT-PCR (Fig. 4A). PCR negative controls were all negative. FACS demonstrated that RA, alone or in combination with TNFα, increased the % Math1-positive cells (Fig. 4B). Immunohistochemistry verified that RA increased the expression of the Math1 protein in a time-dependent manner and translocated into the nuclei (Fig. 4C).
Fig. 4.
RA and RA+TNFα strengthen the expression of Math1 in cultured mMEEC. Incubation of cells with RA for 12~24 hours increased the expression of the Math1 mRNA transcripts by RT-PCR (A). FACS verified that RA, TNFα, and RA+TNFα for 12 hours increased Math1-positive cell numbers (B, n=2). Immunohistochemistry further confirmed that an increase of the Math1-positive cells and translocation into the nuclei after treatment with RA and TNFα in a time-dependent manner (within 2~12 hours, C). It is noted that both RA and TNFα had no effects on the Math1 translocation to the nuclei at 24 hour. EGF=20ng/mL, TNFα=20ng/mL, RA=1µM. Ctrl, control; bar=5 µm.
Multiple signaling pathways are involved in mucous cell differentiation by TNFα+RA+Math1
It remains unclear how RA, TNFα, Math1 alone and in combination affect differentiation of mucous cells. To probe the likely signaling pathways for mucous cell differentiation, we evaluated the effects of various chemical inhibitors on mucous granule formation. All the inhibitors except for JAK1 significantly inhibited TNFα+RA+Math1 induced mucous granulation in cultured mMEEC (Fig. 5A). Immunohistochemistry verified that in comparison with DMSO, PD98059, SB203580, LY294002, AG1478, and SP600125 reduced the expression of mucous cell marker TFF3 in Math1 stably trasnfected and TNFα+RA -treated cells (Fig. 5B). Taken together, the data showed that multiple pathways are involved in the differentiation of mucous cells induced by TNFα+RA+Math1.
Fig. 5.
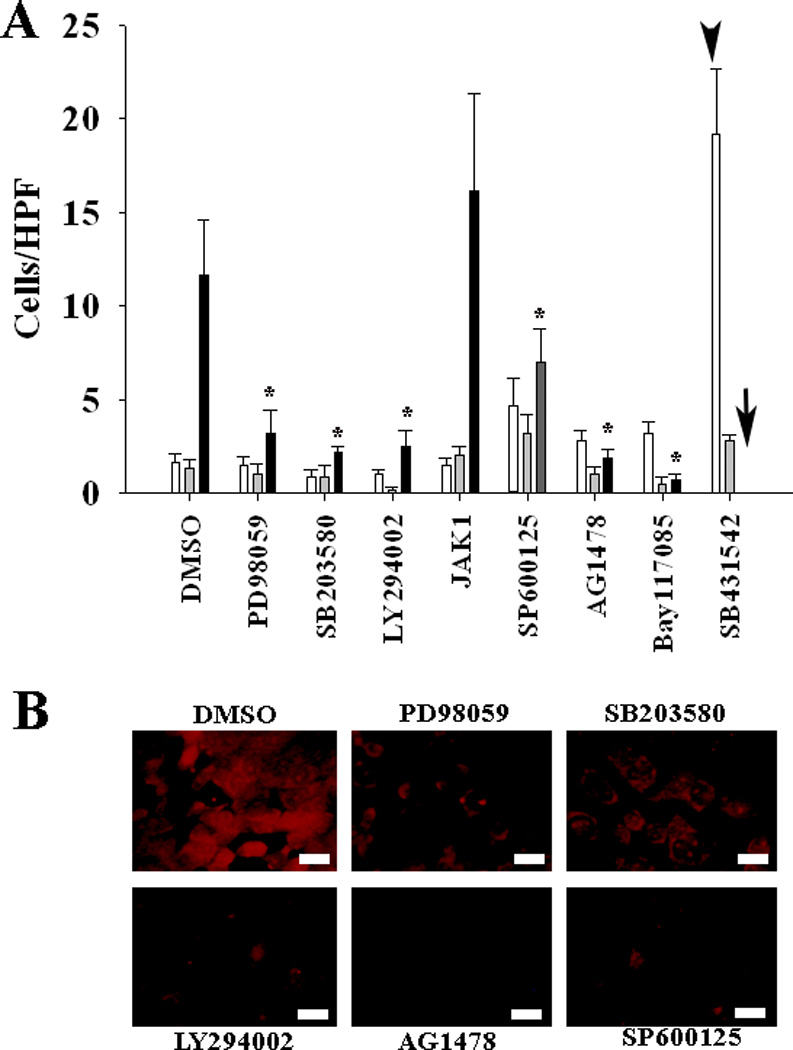
Multiple signaling pathways are involved in mucous cell differentiation by TNFα+RA+Math1. All the inhibitors used in this study, except JAK1, significantly inhibited the mucous granule formation induced by TNFα+RA+Math1(A, white bar, TMath1; grey bar, RMath1; black bar, TRMath1). JAK1 did not affect the mucous cell differentiation triggered by TNFα+RA+Math1 whereas SB431542 did not affect the mucous cell differentiation triggered by TNFα+Math1 (arrow head) but toxic to TNFα+RA+Math1 treated cells (black arrow). Immunohitochemistry demonstrated that PD98059, SB203580, LY294002, AG1478, and SP600125 inhibited the expression of TFF3 in TNFα+RA+Math1 treated cells (B). bar=5 µm. *p<0.05 (n=6)
DISCUSSION
In this study we demonstrated for the first time that mMEEC have the potential to differentiate into mucous-like cells under certain circumstances. The inductive factors are triad master regultors for mucous cell metaplasia: Math1, TNFα, and RA. There is an expression baseline of Math1 in mMEEC. RA or retinoids are also available in the middle ear mucosa. Under normal conditions, these mMEEC become epithelial cells, instead of mucous cells. But under inflammatory conditions, TNFα and other cytokines are induced and released. This results in the synergy of the above three important factors: Math1, TNFα, and RA on the middle ear epithelial cells. Math1 alone has some effects on the expression of mucous cell markers such as mucins and TFFs at the mRNA level (Fig. 2) but limited at the protein level (Fig. 3). Math1, TNFα, and RA synergistically regulate mucins and mucin chaperones (TFFs) at the both mRNA and protein levels. This indicates that mucous cell metaplasia requires converged signaling pathways from multiple factors (Fig. 6). Mucous granules are formed when Math1, RA, and TNFα reach to a certain level, so called threshold for MCM. These mucous granules are AB-PAS positive and cells containing abundant mucous granules and they are mucous cells by nature. Rather, these cells in vitro are not mature mucous cells but are en route to become mucous cells.
Fig. 6.
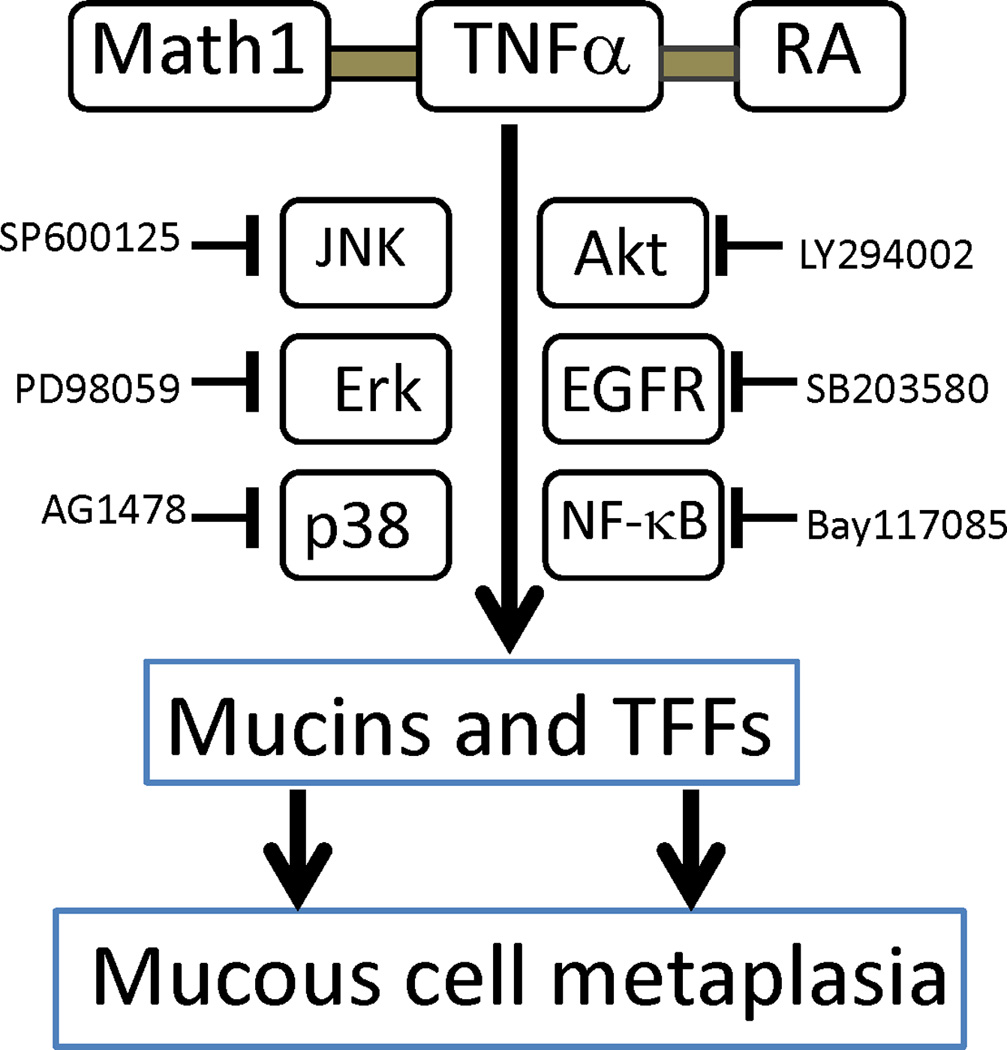
A flow chart summary for the signaling pathways involved in mucous cell metaplasia by TNFα+RA+Math1 in mMEEC. As judged by AB-PAS and immunohistochemistry data (see Fig. 5), the Erk, p38MAPK, Akt, EGFR, JNK and NF-κB signaling pathways are involved in TNFα+RA+Math1 induced mucous-like cells (AB-PAS positive). In other words, Math1,TNFα and RA may act through the above signaling pathways synergistically to increase the proliferation and differentiation of mucous cells in cultured mMEEC and blockage of the above pathways may lead to therapeutic interventions of mucous cell metaplasia in the middle ear milieu.
How TNFα, RA and Math1 make this happen? First, each of these factors plays a role in the regulation of mucins: hallmark proteins for mucous cells. These upregulated mucins are multiple: soluble mucins such as Muc2, Muc5ac, Muc5b and memberane-bound mucins such as Muc1 and Muc4. Math1 has an potential effects on the expression of Muc1, Muc2, Muc4 and Muc5ac, but not Muc3, Muc10, based upon the microarray data (Fig 2) and, in fact, Math1 transfection increases the expression of Muc2 protein in the presence of RA+TNFα (Fig 3C). Consistent with this, Leow and coworkers reported that Math1 is related to MUC2 mucin upregulation (29). Sekine and coworkers reported that overexpression of Math1 in the gastric cancer cells enhanced MUC5AC mRNA and knockdown of Math1 by RNA interference decreased MUC5AC gene (30). In our previous studies,TNFα stimulated the expression of mucins (Muc2) in the rat middle ear mucosa and promotes the differentiation of mucous cells (12). In line with this, TNFα in vitro regulates the expression of Muc2 (Fig 3B), and slightly stimulates the differentiation of mucous cells in vitro. In terms of RA, it has been reported that the expression of MUC2, MUC5AC and MUC5B are depondent on the presence of RA because these mucin mRNA transcripts were not detected in the RA-deficient cultures (31). Our current study demonstrated that RA treatment induces the expression of Muc2 as judged by immunohistochemistory(Fig 3B). Second, in the presence of Math1, TNFα and/or RA regulate the expression of mucin (Muc2) and mucin shaperones TFF3 (Fig. 3B). TFFs are essential for packing mucins into mucus granules (9) which stain positive for AB-PAS. Neither mucins nor TFFs alone make any mucous granules, so the upregulation of both mucins and TFFs at the same time is a prerequisite for the formation of mucous granules and differentiation of cells into mucous-like cells. Math1 transfection had increased the expression of Math1 in the cytosol. However, these upregulated protiens were not translocated to the nuclei without presence of TNFα and/or RA (Fig 3B). This may explain why Math1 transfection alone has a limited or no effect on the expression of TFF3 (Fig. 2). Math1 is thought to synergize the expression of TFF3 because Math1 transfection strengthened the effects of TNFα+RA on TFF3 expression (Fig 3C). The pathway to TFF3 is still poorly understood. It has been reported that the PI3-K/Akt pathway links to TFF3 expression in TH29 cells under confluence (32). Consitent with this, an Akt specific inhibitor, LY294002, inhibited the expression of TFF3 by TNFα+RA(Fig. 5B). TFFs are known to stimulate their own release as well as other family members and this auto- or cross-induction of TFFs requires indirect activation of the EGFR (33). This may be one of reasons why EGFR tyrosin kinase inhibitor, AG1478, inhibited the expression of TFF3 by TNFα+RA(Fig 5B). Our microarray data shown the remakable upregulation of AKT(AF124142) and EGFR was observed in TNFα+RA+Math1 treated cells (Fig. 2). These signaling pathways induced by the combination of factors may explain the synergistic effects of TNFα+ RA+Math1 on TFF3 expression or mucous cell differentiation.
RA has been shown to regulate epithelial cell differentiation related to mucin gene regulation in rabbit tracheal-epithelial cells in vitro (34) and restore to a mucous cell phenotype from squamous cells when cells are derived of retinoids (35). In this study, we demonstrate that RA mainly regulates the expression and activation of Math1, which further potentiate the expression of both mucins and mucin chaperones. The upregulation of mucins and mucin chaperones to a certain level may trigger the differentiation of epithelial cells to mucous cells. Although either TNFα or RA alone is capable of upregulating the expression of mucins or TFF3 in vivo, it is not sufficient for differentiation of mucous cells in a cell culture system. Math1, TNFα, and RA synergistically promote mucous cell differentiation in vitro, suggesting that they are therapeutic targets for mucous cell metaplasia in OM or asthma in the airway.
It is poorly understood how many signaling pathways are involved in the differentiation of mucous cells. The data collected from the current study indicate that the complex effects of Math1+TNFα+RA on mucous cell differentiation are blocked by several specific inhibitors such as SB203580, LY294002, AG1478, and SP60012. It suggests that TNFα+RA+Math1 synergized mucous cell differentiation is a result of the interactions between different pathways. Cross-talks between cytokines and factors are essential for mucous cell differentiation. Further studies are warranted to clarify the specific signaling pathways for individual cytokines in order to design specific therapeutic interventions relevant to inflammatory cytokines.
Supplementary Material
Supplemental Figure 1. Math1 transfection in mMEEC increases the expression of Math1 by RT-PCR, FACS, and immunohistochemistry. A, Math1 mRNA transcripts were markedly increased in mMEEC transfected with Math1 cDNA (lane 1) but barely detected in mMEEC transfected with empty vector (ev, lane 2) by RT-PCR. Lane 3 represents the negative control for RT-PCR by omitting reverse transcriptase. B, Math1 protein was obviously increased in mMEEC transfected with Math1 in comparison with ev by FACS. C, Math1 protein increased in mMEEC transfected with Math1 but not in mMEEC transfected with ev by immunohistochemistry.
Supplemental Figure 2. Various factors are involved in the differentiation of mucous cells. Cells were treated with various factors for 2 weeks (media and factors were supplied on a daily basis) and then without any factors for 2 days. The number of AB-PAS positive cells including faintly stained was counted under a microscope per high power field (×200). It was found that RA+TNF or RA+TNF+IGF treated mMEEC cultures had a significant increase of AB-PAS positive cells compared with untreated cultures (Ctrl), single factor treated (RA, TNFα, EGF, PDGF, IGF1), and dual factor treated (EGF+TNF, RA+EGF, PDGF+TNF, RA+PDGF, IGF1+TNFα (*p<0.05, n=4). Factor concentrations: EGF 20ng/ml, TNFα 20ng/ml, RA 2µM, PDGF 20ng/ml, IGF-1 50ng/ml. TNF, TNFα.
Acknowledgments
Supported in part by the National Institute of Health (NIH) grant #R01 DC008165 and Supplement #00010055 from the National Institute of Deafness and Other Communication Disorders (NIDCD), and the National Organization for Hearing Research (NOHR), and the 5M Lions International Hearing Foundation.
Footnotes
There are no any conflicts of interest involved in this study.
REFERENCES
- 1.Kawano H, Paparella MM, Ho SB, et al. Identification of MUC5B mucin gene in human middle ear with chronic otitis media. Laryngoscope. 2000;110:668–673. doi: 10.1097/00005537-200004000-00024. [DOI] [PubMed] [Google Scholar]
- 2.Lin J, Tsuboi Y, Rimell F, et al. Expression of mucins in mucoid otitis media. JARO. 2003;4:384–393. doi: 10.1007/s10162-002-3023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tos M, Bak-Pedersen K. Histopathology and goblet-cell density in the Eustachian tube and middle ear in children. J Laryngol Otol. 1976;90:475–485. doi: 10.1017/s0022215100082335. [DOI] [PubMed] [Google Scholar]
- 4.Tos M, Bak-Pedersen K. Goblet cell population in the pathological middle ear and eustachian tube of children and adults. Annals of Otology, Rhinology & Laryngology. 1977;86:209–218. doi: 10.1177/000348947708600212. [DOI] [PubMed] [Google Scholar]
- 5.Rinaldo AFA. The pathology and clinical features of "glue ear": a review. Eur Arch Otorhinolaryngol. 2000;257:300–303. doi: 10.1007/s004050000238. [DOI] [PubMed] [Google Scholar]
- 6.Tos M, Caye-Thomasen P. Mucous glands in the middle ear - what is known and what is not. ORL J Otorhinolaryngol. 2002;64:86–94. doi: 10.1159/000057786. [DOI] [PubMed] [Google Scholar]
- 7.Alper CM, Bluestone CD, Buchman C, et al. Recent advances in otitis media. 3. Middle ear physiology and pathophysiology. Ann Otol Rhinol Laryngol. 2002;(Suppl 188):26–35. [PubMed] [Google Scholar]
- 8.Lin J, Tsprun V, Kawano H, et al. Characterization of mucins in human middle ear and eustachian tube. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1157–L1167. doi: 10.1152/ajplung.2001.280.6.L1157. [DOI] [PubMed] [Google Scholar]
- 9.Wiede A, Jagla W, Welte T, Kohnlein T, Busk H, Hoffmann W. Localization of TFF3, a new mucus-associated peptide of the human respiratory tract. Am J Respir Crit Care Med. 1999;159:1330–1335. doi: 10.1164/ajrccm.159.4.9804149. [DOI] [PubMed] [Google Scholar]
- 10.Kawano H, Haruta A, Tsuboi Y, et al. Induction of mucous cell metaplasia by tumor necrosis factor alpha in rat middle ear: the pathologic basis for mucin hyperproduction in mucoid otitis media. Ann Otol Rhinol Laryngol. 2002;111:415–422. doi: 10.1177/000348940211100506. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Haruta A, Kawano H, et al. Induction of mucin gene expression in middle ear of rats by tumor necrosis factor-a: potential cause for mucoid otitis media. J Infect Dis. 2000;182:882–887. doi: 10.1086/315767. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Kim Y, Juhn SK. Increase mucous glycoprotein secretion by tumor necrosis factor alpha via a protein kinase C-dependent mechanism in cultured chinchilla middle ear epithelial cells. Ann Otol Rhinol Laryngol. 1998;107:213–219. doi: 10.1177/000348949810700305. [DOI] [PubMed] [Google Scholar]
- 14.Levine SJ, Larivee P, Logun C, Angus CW, Ognibene FP, Shelhamer JH. Tumor necrosis factor-a induces mucin hypersecretion and MUC-2 gene expression by human airway epithelial cells. Am J Respir Cell Mol Biol. 1995;12:196–204. doi: 10.1165/ajrcmb.12.2.7865217. [DOI] [PubMed] [Google Scholar]
- 15.Dabbagh K, Takeyama K, Lee HM, Ueki IF, Lausier JA, Nadel JA. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J Immunol. 1999;162:6233–6237. [PubMed] [Google Scholar]
- 16.Lee CG, Homer RJ, Cohn L, et al. Transgenic overexpression of interleukin (IL)-10 in the lung causes mucus metaplasia, tissue inflammation, and airway remodeling via IL-13-dependent and -independent pathways. J Biol Chem. 2002;277:35466–35474. doi: 10.1074/jbc.M206395200. [DOI] [PubMed] [Google Scholar]
- 17.Smirnova MG, Birchall JP, Pearson JP. In vitro study of IL-8 and goblet cells: possible role of IL-8 in the aetiology of otitis media with effusion. Acta Otolaryngol. 2002;122:146–152. doi: 10.1080/00016480252814144. [DOI] [PubMed] [Google Scholar]
- 18.Longphre M, Li D, Gallup M, et al. Allergen-induced IL-9 directly stimulates mucin transcription in respiratory epithelial cells. J Clin Invest. 1999;104:1375–1382. doi: 10.1172/JCI6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laoukili J, Perret E, Willems T, et al. IL-13 alters mucociliary differentiation and ciliary beating of human respiratory epithelial cells. J Clin Invest. 2001;108:1817–1824. doi: 10.1172/JCI13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo M, Tamaoki J, Takeyama K, Nakata J, Nagai A. Interleukin-13 induces goblet cell differentiation in primary cell culture from Guinea pig tracheal epithelium. Am J Respir Cell Mol Biol. 2002;27:536–541. doi: 10.1165/rcmb.4682. [DOI] [PubMed] [Google Scholar]
- 21.Whittaker L, Niu N, Temann UA, et al. Interleukin-13 mediates a fundamental pathway for airway epithelial mucus induced by CD4 T cells and interleukin-9. Am J Respir Cell Mol Biol. 2002;27:593–602. doi: 10.1165/rcmb.4838. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya K, Kim Y, Ondrey FG, Lin J. Characterization of a temperature-sensitive mouse middle ear epithelial cell line. Acta Otolaryngol. 2005;125:823–829. doi: 10.1080/00016480510031533. [DOI] [PubMed] [Google Scholar]
- 23.Hu X, Huang J, Feng L, Fukudome S, Harmajima Y, Lin J. Sonic Hedgehog (SHH) Promotes the Differentiation of Mouse Cochlear Neural Progenitors via the Math1–Brn3.1 Signaling pathway in vitro. J Neurosci Res. 2010;88:927–935. doi: 10.1002/jnr.22286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuboi Y, Kim Y, Giebink GS, et al. Induction of mucous cell metaplasia in middle ear of rats by a three-step method: an improved model for otitis media with mucoid effusion. Acta Otolaryngol. 2002;122:153–160. doi: 10.1080/00016480252814153. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Pujol S, Marker PH, Key NS. Platelet microparticles are heterogeneous and highly dependent on the activation mechanism: studies using a new digital flow cytometer. Cytometry A. 2007;71:38–45. doi: 10.1002/cyto.a.20354. [DOI] [PubMed] [Google Scholar]
- 26.Ozeki M, Duan L, Obritch W, Lin J. Establishment and characterization of progenitor hair cell lines in rats. Hear Res. 2003;179:43–52. doi: 10.1016/s0378-5955(03)00077-7. [DOI] [PubMed] [Google Scholar]
- 27.Ozeki M, Hamajima Y, Feng L, et al. Id1 induces the proliferation of cochlear sensorineural epithelial cells via the NF-kB/cyclin D1 pathway in vitro. J Neurosci Res. 2006;85:515–524. doi: 10.1002/jnr.21133. [DOI] [PubMed] [Google Scholar]
- 28.Lin J, Tsuboi Y, Pan W, Giebink SG, Adams GL, Kim Y. Analysis by cDNA microarrays of altered gene expression in middle ears of rats following penumococcal infection. Int J Pedatr Otorhinolaryngol. 2002;65:203–211. doi: 10.1016/s0165-5876(02)00130-1. [DOI] [PubMed] [Google Scholar]
- 29.Leow CC, Romero MS, Ross S, Polakis P, Gao WQ. Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res. 2004;64:6050–6057. doi: 10.1158/0008-5472.CAN-04-0290. [DOI] [PubMed] [Google Scholar]
- 30.Sekine A, Akiyama Y, Yanagihara K, Yuasa Y. Hath1 up-regulates gastric mucin gene expression in gastric cells. Biochem Biophys Res Commun. 2006;344:1166–1171. doi: 10.1016/j.bbrc.2006.03.238. [DOI] [PubMed] [Google Scholar]
- 31.Gray T, Koo JS, Nettesheim P. Regulation of mucous differentiation and mucin gene expression in the tracheobronchial epithelium. Toxicology. 2001;160:35–46. doi: 10.1016/s0300-483x(00)00455-8. [DOI] [PubMed] [Google Scholar]
- 32.Durual S, Blanchard C, Estienne M, et al. Expression of human TFF3 in relation to growth of HT-29 cell subpopulations: involvement of PI3-K but not STAT6. Differentiation. 2005;73:36–44. doi: 10.1111/j.1432-0436.2005.07301006.x. [DOI] [PubMed] [Google Scholar]
- 33.Taupin D, Wu DC, Jeon WK, Devaney K, Wang TC, Podolsky DK. The trefoil gene family are coordinately expressed immediate-early genes: EGF receptor- and MAP kinase-dependent interregulation. J Clin Invest. 1999;103:R31–R38. doi: 10.1172/JCI3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna B, Ashbaugh P, Bhattacharyya SN. Retinoic acid-regulated cellular differentiation and mucin gene expression in isolated rabbit tracheal-epithelial cells in culture. Inflammation. 1995;19:489–502. doi: 10.1007/BF01534582. [DOI] [PubMed] [Google Scholar]
- 35.Koo JS, Yoon JH, Gray T, Norford D, Jetten AM, Nettesheim P. Restoration of the mucous phenotype by retinoic acid in retinoid-deficient human bronchial cell cultures: changes in mucin gene expression. Am J Respir Cell Mol Biol. 1999;20:43–52. doi: 10.1165/ajrcmb.20.1.3310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Math1 transfection in mMEEC increases the expression of Math1 by RT-PCR, FACS, and immunohistochemistry. A, Math1 mRNA transcripts were markedly increased in mMEEC transfected with Math1 cDNA (lane 1) but barely detected in mMEEC transfected with empty vector (ev, lane 2) by RT-PCR. Lane 3 represents the negative control for RT-PCR by omitting reverse transcriptase. B, Math1 protein was obviously increased in mMEEC transfected with Math1 in comparison with ev by FACS. C, Math1 protein increased in mMEEC transfected with Math1 but not in mMEEC transfected with ev by immunohistochemistry.
Supplemental Figure 2. Various factors are involved in the differentiation of mucous cells. Cells were treated with various factors for 2 weeks (media and factors were supplied on a daily basis) and then without any factors for 2 days. The number of AB-PAS positive cells including faintly stained was counted under a microscope per high power field (×200). It was found that RA+TNF or RA+TNF+IGF treated mMEEC cultures had a significant increase of AB-PAS positive cells compared with untreated cultures (Ctrl), single factor treated (RA, TNFα, EGF, PDGF, IGF1), and dual factor treated (EGF+TNF, RA+EGF, PDGF+TNF, RA+PDGF, IGF1+TNFα (*p<0.05, n=4). Factor concentrations: EGF 20ng/ml, TNFα 20ng/ml, RA 2µM, PDGF 20ng/ml, IGF-1 50ng/ml. TNF, TNFα.