Abstract
Inferring antibiotic mechanisms on translation through static structures has been challenging as biological systems are highly dynamic. Dynamic single-molecule methods are also limited to few simultaneously-measurable parameters. We have circumvented these limitations with a multifaceted approach to investigate three structurally-distinct aminoglycosides that bind to the aminoacyl-tRNA site (A site) in the prokaryotic 30S ribosomal subunit: apramycin, paromomycin, and gentamicin. Using several single-molecule fluorescence measurements combined with structural and biochemical techniques, we observed distinct changes to translational dynamics for each aminoglycoside. While all three drugs effectively inhibit translation elongation, their actions are structurally and mechanistically distinct. Apramycin does not displace A1492 and A1493 at the decoding center, as demonstrated by a solution NMR structure, causing only limited miscoding; instead it primarily blocks translocation. Paromomycin and gentamicin, which displace A1492 and A1493, cause significant miscoding, block intersubunit rotation, and inhibit translocation. Our results show the power of combined dynamics, structural, and biochemical approaches to elucidate the complex mechanisms underlying translation and its inhibition.
Introduction
The ribosome is the molecular machine that rapidly and accurately interprets the genetic code on the mRNA to synthesize proteins (Green and Noller, 1997). During elongation, the ribosome repeats the cycle of selecting a tRNA molecule matching the codon in the ribosomal A site, incorporating the amino acid from the selected A-site tRNA into the polypeptide on the P-site tRNA, translocating the A- and P-site tRNAs to the P and E sites, and stepping precisely three bases in the 3′ direction (Korostelev et al., 2008; Wintermeyer et al., 2004; Zaher and Green, 2009). To elongate with an optimal balance of speed and accuracy, the ribosome employs G-protein elongation factors (EF-Tu and EF-G in bacteria) to facilitate key steps during the process (Nilsson and Nissen, 2005). Using the energy from GTP hydrolysis, EF-Tu enhances the rate and specificity of tRNA selection and EF-G catalyzes translocation.
The central role of translation makes the ribosome a rich target for clinically important small-molecule antibiotics. They employ diverse strategies to interfere with translation, and fall into several distinct classes, including macrolides, tetracyclines, and aminoglycosides (Benveniste and Davies, 1973; Bottger, 2006; Davies et al., 1965; Perzynski et al., 1979; Woodcock et al., 1991). Biochemical and structural studies of these antibiotics have shed light on their mechanism, and have in turn provided clues to the molecular workings of the ribosome and its ligands (Yonath, 2005). Here we focus on the clinically important aminoglycosides as a representative group of translational inhibitors.
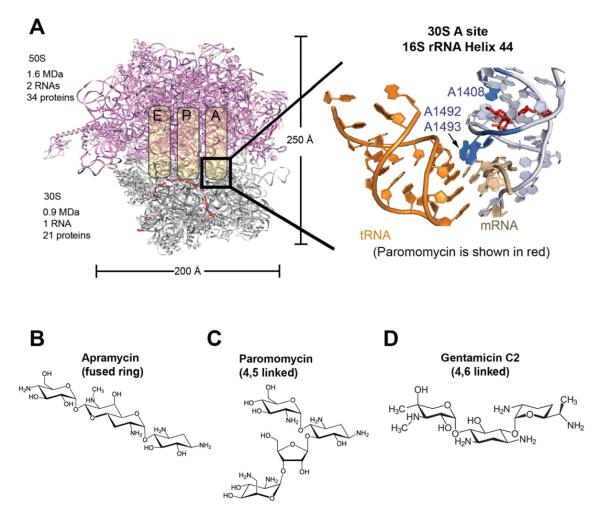
Aminoglycosides contain a central deoxystreptamine ring with amino-sugar modifications (4,5 and 4,6 di-substituted deoxystreptamine), and include neomycin, paromomycin, gentamicin, kanamycin, and the novel fused-ring compound, apramycin. Aminoglycosides disrupt the fidelity of tRNA selection and block translocation (Benveniste and Davies, 1973; Davies and Davis, 1968; Davies et al., 1965). NMR and X-ray crystal structures (Fourmy et al., 1998), as well as biochemical and molecular biological studies (Bottger et al., 2001; Powers and Noller, 1991), have revealed the structural basis for specific aminoglycoside binding to bacterial ribosomes. Aminoglycosides bind in the major groove of the 16S rRNA decoding site, forming specific contacts with conserved nucleotides G1494 and U1495; ring I of the aminoglycosides fits into a prokaryote specific binding pocket formed by universally conserved nucleotides A1492 and A1493, and the prokaryotic-specific nucleotide A1408, imparting their specificity (Bottger et al., 2001; Recht et al., 1999).
These structural studies suggested a mechanism for aminoglycoside action (Figure 1A). In the presence of a cognate codon-anticodon complex, A1492 and A1493 (Moazed and Noller, 1990) make shape-specific contacts in the minor groove of the codon-anticodon A-form helix (Fourmy et al., 1998; Ogle et al., 2001; Yoshizawa et al., 1999). These contacts assist in distinguishing between correct and incorrect codon-anticodon pairings, as shown by both structural and kinetic investigations. In the absence of tRNA or drug, A1492/A1493 are stacked within an asymmetric internal loop at the base of Helix 44 (h44). Upon codon-anticodon interaction, A1492/A1493 are displaced from h44 to make the contacts discussed above. Binding of either 4,5 or 4,6 disubstituted aminoglycosides to the decoding site mimics this conformational effect, displacing A1492 and A1493 towards the minor groove.
Figure 1. Aminoglycosides bind in the 30S A site in helix 44 of the 16S rRNA.
(A) The location that aminoglycosides bind to the bacterial ribosome is shown. The detailed binding site is shown to the right, with the A-site tRNA in orange and paromomycin in red. The three nucleotides in blue are A1408, which is specific to prokaryotes and conveys the specificity of aminoglycosides, and A1492/A1493, which are destacked by aminoglycosides and mimic the conformation of correct codon-anticodon recognition between the mRNA and the A-site tRNA. A-site structure with paromomycin is rendered from PDB 2J00 (Selmer et al., 2006). (B), (C), and (D) show the structures of the three aminoglycosides tested. Biochemical experiments have shown that aminoglycosides induce miscoding and block translocation. Such effects were also seen in our single-molecule assays, as shown in Figure S1.
Aminoglycosides alter the kinetics of tRNA accommodation, supporting these structural predictions (Pape et al., 1998, 2000). Paromomycin significantly reduces the dissociation rate of near-cognate tRNA and increases the rate of GTPase activation on EF-Tu by an order of magnitude. These results suggest that aminoglycosides stabilize near-cognate tRNA binding to the ribosome with a 16S rRNA conformation that mimics cognate codon-anticodon recognition. While the structural basis of aminoglycoside-induced miscoding has become clear, little is known about how they inhibit the subsequent steps of elongation. The relative importance of miscoding and inhibiting translocation to the potency of the drugs has not been determined. Here we use a combined dynamics and structural approach to understand how distinct aminoglycosides disrupt translation.
We probed the mechanism of translational inhibition by an aminoglycoside from each of the three distinct structural classes- apramycin (novel fused-ring), paromomycin (4,5-linked), and gentamicin (4,6-linked) (Figure 1 B-D) using single-molecule, structural, and biochemical approaches. We solved the solution structure of apramycin bound to the bacterial decoding site by NMR spectroscopy, providing a novel structure to the repertoire of drug-RNA complexes. We determined the effects of all three drugs on the dynamics of translation using an array of single-molecule techniques (Aitken et al., 2010): we monitored the global conformation of the ribosome during elongation (Aitken and Puglisi, 2010), the dynamics of tRNA accommodation by fluorescence resonance energy transfer (FRET) (Blanchard et al., 2004a) and used zero-mode waveguides (ZMW) to track multiple tRNAs through several elongation cycles at near-physiological concentrations (Levene et al., 2003; Uemura et al., 2010). We also used bulk kinetics (Johansson et al., 2011) to measure the effects of aminoglycosides on rates of peptide bond formation and subsequent translocation. This combination of single-molecule dynamics, structural, and biochemical techniques reveals the mechanism of aminoglycoside action at an unparalleled level of detail.
Results
Apramycin, paromomycin, and gentamicin target distinct aspects of the elongation cycle
To monitor the global effects of aminoglycosides on elongation, we used intersubunit FRET between site-specifically labeled Escherichia coli 30S and 50S subunits (Aitken and Puglisi, 2010; Dorywalska et al., 2005; Marshall et al., 2008) that enables real-time monitoring of single-ribosomes performing multiple elongation cycles. For each cycle of elongation, the ribosome rotates the 30S body relative to the 50S and then reverses the rotation, controlled respectively by peptide bond formation upon tRNA accommodation and translocation. The non-rotated state is detected by high FRET and the rotated state by a lower FRET value. The resultant FRET cycles reports on the processivity and dynamics of the ribosome at each codon (Figure 2 A & B) (Aitken and Puglisi, 2010). All three aminoglycosides significantly inhibited elongation efficiency (Figure S1 A-C, see supplementary text for details). To probe the mechanistic origins of translation inhibition by each aminoglycoside, we leveraged the ability of this single-molecule FRET approach to detect codon-specific effects on the dynamics of 30S body rotation and counter-rotation during elongation (Figures 2 C-F and S2 A-D) (Aitken and Puglisi, 2010; Marshall et al., 2008).
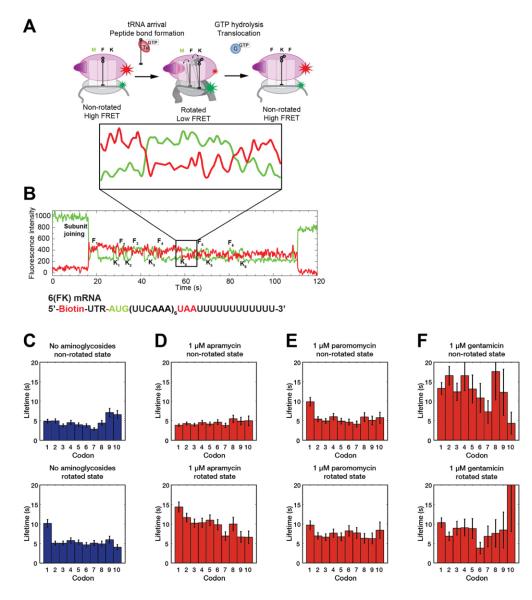
Figure 2. Aminoglycosides inhibits translation elongation at different stages.
(A) The intersubunit FRET signal involves cycles between a high FRET state where the 30S body of the ribosome is not rotated relative to the 50S subunit and a low FRET state, where the 30S body is rotated. From previous studies (Aitken and Puglisi, 2010; Marshall et al., 2008), peptide bond formation rotates the 30S body and causes a transition from high to low FRET. EF-G translocates and counter-rotates the 30S body, causing a low to high FRET transition. (B) An example trace of the intersubunit FRET where the ribosome translated the entire 6(FK) mRNA. The zoomed panel directly below the diagram in panel (A) illustrates the changes in FRET state over one elongation cycle. (C-F) The lifetimes that each ribosome spends in a conformation state over a given codon are fitted to a single exponential decay function and plotted. From left to right, the conditions are no aminoglycosides (C, n = 381), 1 μM apramycin (D, n = 429), 1 μM paromomycin (E, n = 264), and 1 μM gentamicin (F, n = 297). Error bars are s.d. errors. Different aminoglycosides target different aspects of translation elongation. See also Figure S2 for lifetimes from additional experiments.
At 1 μM, apramycin did not measurably affect the lifetime (~4 s) of the high-FRET, or non-rotated state, where the ribosome selects tRNA and performs peptidyl transfer. In contrast, apramycin lengthened the low FRET, or rotated state, where the ribosome waits for EF-G to drive translocation, by 2-fold from 5 s without drugs to ~10 s. Increasing apramycin concentration to 10 μM further increased the rotated ribosome lifetime to 3-fold (~15 s) longer than minus drugs, but did not otherwise introduce any additional effects on elongation. Increasing the EF-G concentration to 160 nM, (from 80 nM, Figure S2 E-G), partially rescued the rotated ribosome lifetime (1.6-fold longer than that minus drugs (~8 s)).
At 1 μM paromomycin, the non-rotated state lifetime of the ribosome at the first codon was increased 2-fold (~10 s), but was unaffected at subsequent codons. Paromomycin also lengthened the rotated state lifetime to a lesser extent, by 1.4-fold (~7 s). Increasing the concentration to 10 μM further lengthened the first non-rotated state lifetime to 3-fold (~15 s) and the rotated state lifetimes to 2-fold of minus drugs (~10 s). Additionally, paromomycin lengthened the rotated state specifically over the first codon to 4-fold (~20 s) from normal. However, this effect is not observed at 1 μM, suggesting a possible alternative binding site (Borovinskaya et al., 2007). Overall, paromomycin displays a more complex set of effects on the elongation cycle, and appears to participate in unique interactions with the ribosome at the first codon of the mRNA.
At 1 μM, gentamicin had the most severe effects on the non-rotated state lifetime, lengthening it by 3-fold (~15 s) across all codons, suggesting a mechanism distinct from paromomycin. However, similar to paromomycin, gentamicin slightly lengthened the rotated lifetime of the ribosome by 1.6-fold (~8 s). Addition of 10 μM gentamicin lead to no further increase in the non-rotated ribosome lifetime; however, similar to paromomycin, a secondary effect of a lengthened rotated ribosome lifetime (to ~14 s, a 2.8-fold increase over no drugs) over the first codon was observed.
In addition to impacting different phases of elongation, the three aminoglycosides also induce different levels miscoding as described in the supplementary text (Figure S1 D-F).
Following real-time tRNA transit and sampling in the presence of aminoglycosides using zero mode waveguides (ZMW)
We next probed the dynamics of aminoglycoside action from the perspective of tRNA binding by directly tracking in real-time labeled tRNA binding and transit through the ribosome in zero mode waveguides (ZMW). The sequence, frequency, and length of each tRNA signal report on the fidelity of tRNA selection and on the speed of elongation based on the dwell-time of the tRNA (Figure 3 A & B). We have used this approach previously to track ribosomal compositional dynamics during initiation and elongation (Tsai et al., 2012; Uemura et al., 2010).
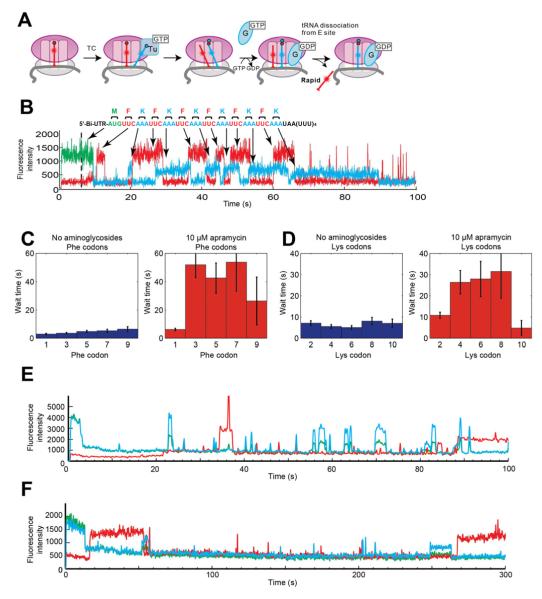
Figure 3. Aminoglycosides alter tRNA binding patterns in ZMW tRNA transit experiments.
(A) Using Phe-(Cy5)tRNAPhe and Lys-(Cy2)tRNALys, tRNA binding behavior and sequence can be observed and tRNA transit through the ribosome can be tracked. The tRNA signal lifetimes, wait times between tRNA events, and the frequency of the events can be tracked.
(B) An example trace of a tRNA transit experiment without aminoglycosides. The pulse sequence up to the last codon on the 6(FK) mRNA is well-behaved and can be analyzed codon-by-codon. Over the stop codon, short tRNA events are observed as tRNA sampling in the ribosomal A site. These types of events are greatly increased in the presence of paromomycin and genatmicin. The waiting times until the next tRNA event over each codon are plotted for the Phe (C) and Lys (D) codons. All error bars are s.d. Paromomycin and gentamicin induce too many short tRNA events with the wrong sequence, which precludes an accurate analysis, as exemplified by the bottom two traces with 10 μM gentamicin. (E) shows many short tRNA binding events whose sequence does not conform to the repeating Phe-Lys sequence encoded in the 6(FK) mRNA used in this experiment. (F) shows a ribosome stalling after the first 1 or 2 codons and does not restart until nearing the end of the 300 s observation time in ZMW experiments. See also Figure S3 for additional analysis of ZMW experiments.
In the absence of aminoglycosides, stable binding and transit of multiple tRNAs on the ribosome were observed as regular pulse cycles of repeating Cy5 followed by Cy2 signals with regular times between pulses of 10~20 s (Figures 3 C-D and S3 A-C), as observed previously (Uemura et al., 2010). In the presence of 10 μM apramycin, the time between pulses lengthened significantly to 30~40 s, a nearly 2-fold increase, where the pulse sequence was still recognizable. Addition of 10 μM paromomycin and gentamicin introduces frequent meta-stable tRNA binding events of the length of 1~5 s and the sequences of those events do not match the codon sequence of the 6(FK) mRNA used for the experiments (Figure 3 E). The abundance of these short-lived events complicates the identification of binding events resulting in accommodation and translocation. See the supplementary text for the detailed analysis on how the aminoglycosides alter tRNA binding behavior (Figure S3 D-F). This result is consistent with our intersubunit FRET experiments that both aminoglycosides severely impact tRNA selection.
Following real-time tRNA dynamics in the presence of aminoglycosides
To delineate the distinct effects of aminoglycosides on tRNA sampling and selection, we followed FRET between P-site tRNA and incoming A-site tRNA in the presence of antibiotics. The evolution of this FRET signal reports on the decoding and accommodation of the A-site tRNA, as well as classical to hybrid state fluctuations of the tRNA after peptide bond formation (Blanchard et al., 2004b) (Figures 4 & S4 A-C).
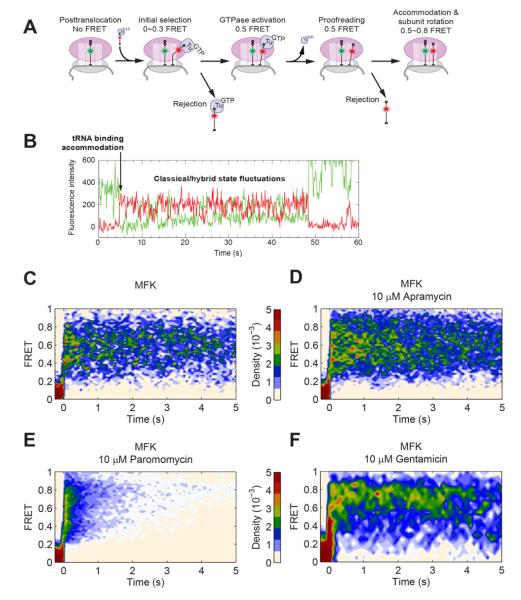
Figure 4. Paromomycin and gentamicin disrupts the selection of the A-site tRNA.
(A) FRET between Cy3 labeled P-site tRNA and incoming Cy5 labeled A-site tRNA can be used to track tRNA selection. The evolution of this FRET signal reports on the dynamic decoding and accommodation of the A-site tRNA, as well as classical to hybrid fluctuations of the tRNA upon peptide bond formation (Blanchard et al., 2004b).
(B) An example trace from a tRNA-tRNA FRET experiment without aminoglycosides on an MFK mRNA. The traces from tRNA-tRNA FRET experiments are then postsynchronized to have the first appearance of FRET to be t = 0. The condition for each panel is listed in the title. Without drugs on the MFK mRNA (C, n = 54), a high FRET (~0.75) signal appears and, within 100 ms the tRNAs begin to fluctuate between the classical (~0.75 FRET) and hybrid (~0.5 FRET) conformations. The FRET signal is present for > 10 s. Apramycin (D, n = 136) does not significantly affect the tRNA selection process. Paromomycin (E, n = 184) severely alters the behavior by introducing short (~1 s) high FRET events and long events (>10 s) with limited classical/hybrid fluctuations. Gentamicin (F, n = 66) also introduces many sub 1-s events and long events with little classical/hybrid fluctuations. See also Figure S4 for additional tRNA-tRNA FRET experiments and Table S1 for lifetimes of tRNA events.
Apramycin did not significantly alter the dynamics of tRNA. In both cases, FRET events proceeded to a high (0.8) FRET state within 100 ms of the appearance of FRET and subsequently fluctuated between high (0.8) and medium (0.5) FRET, consistent with rapid tRNA selection and accommodation, and peptide bond formation followed by classical-hybrid dynamics (Blanchard et al., 2004b). However, apramycin increased the overall tRNA FRET lifetime from 10 s in the absence of drugs to 18 s (Table S1). By blocking of translocation, apramycin could prevent the eventual dissociation of tRNAfMet from the E site, preserving FRET.
In contrast, paromomycin significantly altered the behavior of tRNA on the ribosome and induces many short-lived excursions (<1 s) from zero FRET to a high (> 0.7) FRET level consistent with accommodated tRNA (Figure S4 F). These events dropped the total FRET lifetime to 1.7 s. While these events are too long to be tRNA sampling attempts (<30 ms), they are much shorter than normal tRNA accommodation without drugs. Longer-lived (1~5 s) high-FRET events with little to no classical-hybrid state fluctuations were also observed (Figure S4 G), consistent with single-molecule FRET measurements by Blanchard and coworkers (Feldman et al., 2009). Together with our ZMW measurements, this data suggest that paromomycin stabilizes tRNAs when they sample the A site and shifts the classical-hybrid tRNA conformation equilibrium strongly toward the classical state. The addition of EF-G enriched longer-lived events, as described in the supplementary text (Figure S4C).
Gentamicin induced behavior similar to that of paromomycin in tRNA-tRNA FRET experiments, with short (<1 s) high-FRET events suggesting that the A-site tRNA has proceeded nearly to accommodation. A longer-lived high-FRET population with limited exchanges between the classical and hybrid tRNA conformations was also observed. These high-FRET events are longer-lived than those observed in paromomycin, with a lifetime approaching the photobleaching time of the FRET pair (18 s). The overall fraction of the short-lived population is also lower then paromomycin, as shown in the higher relative density of the long events in the postsynchronized plot and the longer total tRNA lifetime (6.8 s). These differences suggest a mechanism distinct from paromomycin.
Paromomycin induces miscoding
To probe how paromomycin alters tRNA selection fidelity, we repeated our tRNA-tRNA FRET experiments with an mRNA encoding Met-Leu-Lys (MLK), in which the Leu codon is CUU, near-cognate to the Phe UUU codon. Delivery of Phe-(Cy5)tRNAPhe to immobilized ribosomes initiated with fMet-(Cy3)tRNAfMet in the P site tested the ability of the ribosome to distinguish between cognate and near-cognate tRNA in the A site. Without drugs and in the presence of apramycin, few detectable tRNA FRET events were observed. Thus, the ribosome was able to discriminate against the near-cognate tRNA; mRNA-independent binding events did not produce FRET and so were not detected in these experiments (Blanchard et al., 2004a).
In the presence of paromomycin we observed many detectable tRNA FRET events on the near-cognate CUU codon (Figure S4 D & E). Similar to the cognate mRNA experiment, we observed short excursions to high FRET as well as longer-lived (1~5 s) high FRET events that showed very limited classical/hybrid fluctuations. The overall lifetime of tRNA FRET in these experiments (1.4~1.9 s) was unchanged as compared to tRNA FRET lifetimes on the cognate MFK mRNA with paromomycin. In contrast to the MFK cognate experiment, about 10 % of longer-lived events (1~3 s) were locked in a medium FRET state (~0.5), consistent with progression to EF-Tu GTPase activation but not full A-site accommodation (Lee et al., 2007). Pape and coworkers previously observed reduced rates of EF-Tu conformational change and tRNA accommodation after GTPase activation on a near-cognate codon by the addition of paromomycin (Pape et al., 2000). Overall, however, the behavior of tRNA on the cognate MFK and near-cognate MLK mRNAs was remarkably similar in the presence of paromomycin. These results further support the view that paromomycin interferes with the ribosome in distinguishing between cognate and near-cognate tRNA.
The apramycin binding site on the ribosome is distinct from that of other aminoglycosides
To explore the structural origins of apramycin function, we determined the NMR solution structure of apramycin in complex with a 27-nucleotide RNA duplex that mimics the A site of the ribosome and the primary binding site of the aminoglycoside antibiotics (Recht et al., 1996). The NMR structure of paromomycin and gentamicin C with this model A-site oligonucleotide were previously published (Fourmy et al., 1996; Fourmy et al., 1998; Yoshizawa et al., 1998) , and crystal structures with both oligonucleotide and ribosomal particles are available (Carter et al., 2000; Francois et al., 2005). Data on the RNA-apramycin complex were acquired on both unlabeled and uniformly 13C, 15N-labeled RNAs (Lukavsky et al., 2003; Lukavsky and Puglisi, 2005). A total of 623 NOE distance restraints, including 32 drug-RNA NOEs, 111 dihedral restraints and 23 residual dipolar coupling restraints were used in the structure calculations. The conformation of the drug-RNA interface was well defined by the NMR data: the root-mean squared deviation (r.m.s.d.) of the core RNA (U1406-U1410; A1490-U1495) and apramycin for the final 32 lowest energy structures was 1.21 Å (PDB ID ####).
As with all previous aminoglycoside-RNA complexes, including paromomycin and gentamicin C1a, apramycin binding to the A site was guided by interaction of the 2-deoxystreptamine ring with G1494 and U1495 (Figures 5 and S5) through hydrogen bonding interactions with N7 position of G1494 and the O4 position of U1495. However, the rigid structure of apramycin led to a distinct binding mode compared to that in the NMR structures of paromomycin- and gentamicin-RNA (Fourmy et al., 1996; Yoshizawa et al., 1998). Apramycin did not penetrate deeply in to the aminoglycoside binding pocket formed by A1408, A1492, and A1493, instead making stabilizing interactions with the phosphate backbone. As a result, apramycin did not stabilize the extrahelical, or de-stacked, conformation of A1492 and A1493 observed in the presence of paromomycin and other miscoding aminoglycosides. Apramycin binding to the oligonucleotide in solution favors the intrahelical, or stacked, conformation of the A1408, A1492, and A1493 observed in the structure of the free A-site oligonucleotide (Recht et al., 1996). Hygromycin B, another structurally unique, non-archetypal antibiotic, binds the A site but does not cause de-stacking of A1492 and A1493; hygromycin B inhibits translocation without causing significant miscoding (Brodersen et al., 2000; Cabanas et al., 1978).
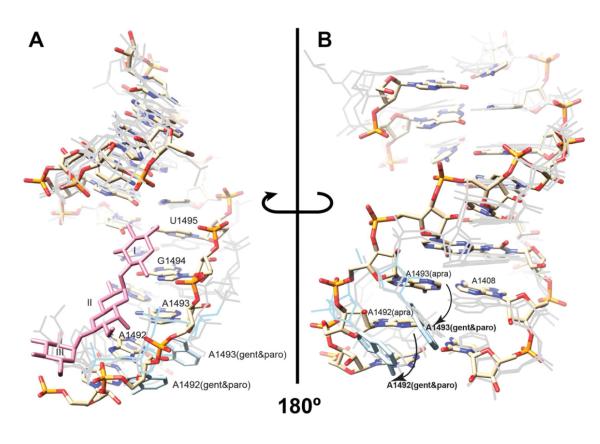
Figure 5. Apramycin bound to the A site does not destack A1492/A1493.
(A) The NMR solution structure with apramycin shown as lines and the 16S rRNA model shown as sticks (PDB ID ####). The three rings of apramycin (plum-colored) are labeled with roman numerals. The conformation of A1492 and A1493 with paromomycin and gentamicin bound is shown in light gray for comparison.
(B) The same NMR solution structure as (A) but viewed from the opposite site of helix 44. The apramycin-RNA complex structure is colored explicitly by atom. A1492 and A1493 (highlighted in light blue) with paromomycin and gentamicin are shifted with respect to their positions in the apramycin complex (indicated by the arrows). The overall structures of the paromomycin and gentamicin RNA structures are also shown in light gray for comparison, but apramycin is not shown for clarity. See also Figure S5 and Table S2.
Effects of apramycin, paromomycin, and gentamicin on peptide bond formation rates
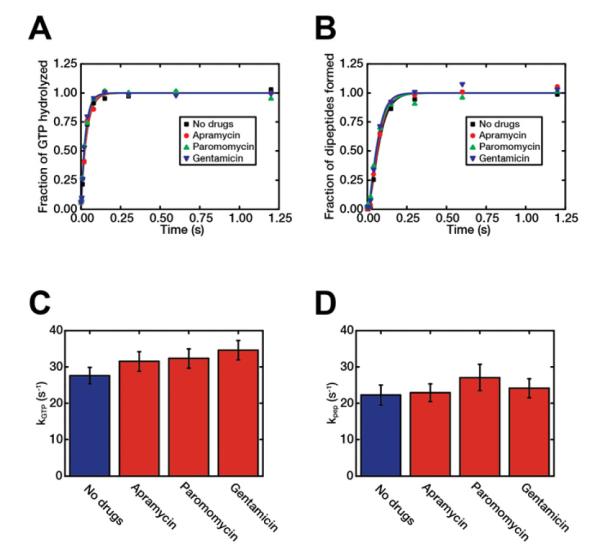
The intersubunit FRET experiments show that paromomycin and gentamicin inhibit 30S body rotation during elongation. This could result from delays in peptide bond formation due to disruption in tRNA selection or steps subsequent to peptide bond formation. Accordingly, we assayed the effect of the three aminoglycosides on peptide bond formation kinetics in bulk experiments, as previously described (Johansson et al., 2011). The rates of GTP hydrolysis (kGTP) and dipeptide formation (kpep) for all three drugs remained unchanged compared to no drugs. As a control, we also measured the rate of formation of tripeptide (fMet-Phe-Phe) in each case (Figure S6). The rate of disappearance of f[3H]Met, reflecting the rate of dipeptide formation, was also the same in each case. The rate of formation of tripeptides was, however, significantly decreased by the presence of each one of the drugs, in line with all three drugs slowing down the ribosome in elongation. These results suggest that the fraction of drug-bound ribosomes was close to 100 % in the experiments.
Discussion
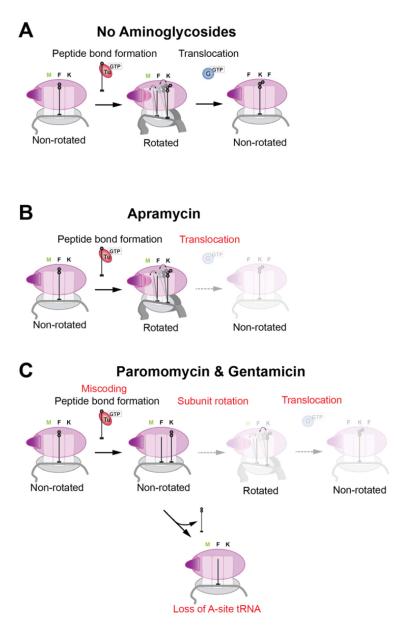
Aminoglycoside are thought to affect translation by decreasing the fidelity of tRNA selection and blocking translocation. We employed a battery of complementary single-molecule methods, supported by structural and biochemical techniques, to form a coherent picture of the mechanism of action for three structurally distinct antibiotics: paromomycin, gentamicin C and apramycin (Figure 7).
Figure 7. Aminoglycosides disrupt various steps of elongation.
(A) In a normal elongation cycle, ribosome rotation occurs quickly after peptide bond formation between the A- and P-site tRNAs, allowing the tRNAs to fluctuate between the classical and hybrid states. EF-G then translocates and counter-rotates the ribosome, moving the tRNAs into the P and E sites to reset the ribosome for another round of elongation.
(B) Apramycin does not hinder the rotation of the ribosome upon peptide bond formation; however, it blocks the translocation step where EF-G is involved. This mode of action differs from other members of the aminoglycoside family.
(C) Paromomycin and gentamicin display a more complex set of effects on elongation, slowing down both the rotation of the ribosome and translocation, as well as inducing severe miscoding. With the ribosome stalled, the A-site tRNA could eventually dissociate with the short peptide chain due to a finite off-rate for tRNAs on the ribosome, leading to a stalled ribosome.
Apramycin slows translation via one primary inhibition mechanism
Apramycin has the clearest but the most atypical mode of action. The NMR solution structure of the apramycin-ribosomal RNA complex suggested a distinct structural mechanism for inhibition of translocation. The apramycin structure shows that the drug binds in the major groove of the 16S rRNA decoding site similar to other aminoglycosides. However, the drug does not induce the de-stacked conformation of A1492/A1493 as observed during correct codon-anticodon pairing. This differs with X-ray crystal structures showing A1492/A1493 de-stacked for all aminoglycosides (see supplementary text). However, the recent crystal structure of apramycin bound to the 30S subunit-tRNA-mRNA complex suggests that the drug would block unstacking of A1492 (Matt et al., 2012), consistent with this interpretation. The bias for a stacked conformation of A1492/1493 in the presence of apramycin is supported by lower level of miscoding observed here and by others (Matt et al., 2012). This bias could additionally reduce the impact of apramycin on elongation dynamics over cognate codons prior to 30S body rotation, as evidenced by the normal non-rotated lifetime in intersubunit FRET, behavior in tRNA-tRNA FRET that is indistinguishable from no drugs, low tRNA sampling frequency in the ZMW (see supplementary text), and lack of effect on cognate kpep.
Apramycin mainly blocks translocation; it significantly lengthens the rotated state lifetime, where the 70S complex is waiting for EF-G in order to translocate. While a previous single-molecule work (Feldman et al., 2009) has correlated inhibition in translocation to perturbations in tRNA conformational dynamics, apramycin breaks this trend as it strongly inhibits translocation without inducing any noticeable increase in the occupancy of the classical tRNA conformational state in our tRNA-tRNA FRET experiment. Apramycin may instead lock the A site in an unfavorable conformation for EF-G, effectively increasing the energy barrier that EF-G must overcome to translocate. Similar effects have been observed for the translocation inhibitor spectinomycin, which binds the 30S subunit (Aitken and Puglisi, 2010). Apramycin inhibits the rate of elongation for about 10 codons into the message, yet it gradually wears off such that the rotated-state lifetime has mostly recovered by the end of our 6(FK) mRNA. This suggests that the early phase of translation is more vulnerable to antibiotic action.
Paromomycin and gentamicin exhibit more complex modes of action
Paromomycin disrupts multiple aspects of elongation, consistent with the conformational changes observed by structural methods. By favoring the de-stacked A1492/A1493 conformation (Fourmy et al., 1998), it causes severe miscoding in our stop codon readthrough assay. From tRNA-tRNA FRET experiments with a near-cognate codon (MLK) mRNA, the many observable events suggests that paromomycin slows the dissociation of an incorrect tRNA from the A site, consistent with prior kinetic measurements (Pape et al., 2000). Strikingly, paromomycin still retained half of its miscoding effect in our miscoding assay where only the non-cognate tRNAPhe (to the stop codon) was present. As the total tRNA concentration in the experiment is halved, this result suggests tentatively that de-stacking of A1492/A1493 can induce miscoding with any tRNA available. The incorrect pulse sequence in ZMW experiments with paromomycin further demonstrates that the ribosome has lost its ability to distinguish correct codon-anticodon interactions.
The effects of paromomycin on elongation appear during both global conformational states of the ribosome and sometimes in a codon-specific manner. The frequent short tRNA FRET events indicate that the ribosome goes through many futile cycles where it accepts a tRNA into the A site, bypassing initial selection, but eventually rejects it. The high tRNA FRET level suggests that the A-site tRNA has proceeded beyond GTP hydrolysis by EF-Tu. Normal bulk peptide bond formation rate suggests that the tRNA proceeds as far as transferring the peptide chain from the P-site tRNA to the A-site tRNA. Combined with the increased non-rotated state lifetime in intersubunit FRET experiments, this suggests that paromomycin significantly increases the energy barrier to 30S body rotation, decoupling it from peptide bond formation. If a ribosome stalls early in elongation with a short peptide chain, it could risk losing the A-site tRNA by dissociation. Such ribosomes are left with a deacylated P-site tRNA and are unable to translocate normally even when a correct tRNA binds in the A site, as indicated by the frequent short-lived binding events observed in the ZMW for stalled traces. Eventually, the ribosome might translocate spontaneously with an acylated A-site tRNA bound and reset itself with an amino acid on the P-site tRNA; however, accumulation of prematurely released peptidyl-tRNA could be toxic. Consistent with that hypothesis, some traces that halted at the first codon in both intersubunit FRET and ZMW experiments subsequently resumed accommodating tRNAs (Figure 3 F). These severely malfunctioning ribosomes can also become candidates termination by release factors (RFs) in vivo (Dunkle and Cate, 2010), aborting translation after significant energy expenditure.
Paromomycin additionally prolongs the rotated state in general; however, this effect is weaker than apramycin. Paromomycin limits the _ENREF_22classical/hybrid tRNA FRET fluctuations, favoring the classical state, which is not conducive for translocation, consistent with observations of Feldman et al. _ENREF_27(Feldman et al., 2009). Alternatively, the stabilized, destacked conformation of A1492 and A1493 may conflict with EF-G function to reset the ribosome (Taylor et al., 2007). For both mechanisms, paromomycin effectively increases the energy barrier for the ribosome to translocate. Additionally, paromomycin can significantly lengthen the rotated lifetime of the first codon, but this is only observed at 10 μM paromomycin concentration. This suggests the presence a secondary binding site; however, the mechanism for this codon-specific effect is not clear.
Gentamicin C, which also promotes the de-stacked A1492/A1493 conformation upon binding, induces miscoding as paromomycin. Gentamicin causes a similar increase in rotated ribosome lifetimes, as well as locking the tRNAs into the classical conformation in tRNA-tRNA FRET experiments (Feldman et al., 2009). Additionally, gentamicin induces short-lived tRNA binding events like paromomycin in ZMW experiments. Unlike paromomycin, gentamicin increases the energy barrier to ribosome rotation across all codons, resulting in more severe disruptions than paromomycin. Gentamicin is 4,6 linked, and its ring III element contacts G1405 in the decoding site (Yoshizawa et al., 1998), whereas the additional ring contacts in 4,5 di-substituted aminoglycosides occur to the helical stem closed by C1409-G1491 (Fourmy et al., 1996). These different contacts and their effects on local and global ribosomal dynamics may explain the different mechanistic observations. Alternatively, secondary binding sites may also contribute. Intriguingly, gentamicin has the strongest inhibitory effect, and is the most important of the 3 drugs in clinical applications.
Dissecting the complex effects of aminoglycosides from multiple perspectives
By employing complementary single-molecule techniques, buttressed by structure and biochemical assays, our results provide a comprehensive understanding of aminoglycoside mechanisms. Their mode of action is far from simple and involves significant modifications to the delicate energy landscape of elongation. Aminoglycosides have distinct profiles of elongation inhibition and their structural diversity indeed translates into a diversity of mechanisms. While aminoglycosides are thought to be effective by inducing miscoding and hindering translocation, we have observed significant differences in the level of miscoding and effects on steps not directly linked to translocation, which for paromomycin and gentamicin were stronger than their effects on translocation. Despite the complexity of their mechanism, a common theme emerges where they significantly increase energy barriers for the ribosome subunits to rotate and to translocate. As the 1.5 to 4-fold inhibitions over each codon to various steps of the elongation cycle accumulate across multiple codons, these drugs can effective stall the ribosome and cause severe disruptions to elongation. In fact, even with our 12-codon model mRNA, we already see a dramatic decrease in the number of codons translated during our observation timeframe. These observations agree with the original discussions of Kurland (Kurland, 1987) and others that aminoglycosides are bacteriocidal because of their inhibition of elongation efficiency, not miscoding. How the local conformational changes caused by aminoglycoside binding to the 30S decoding site affect the global behavior of the ribosome and how miscoding aminoglycosides modify the balance of tRNA selection fidelity versus efficiency remain open questions (Johansson et al., 2012).
Experimental procedures
Single-molecule reagents and buffers
Fluorescently labeled 30S (Cy3 or Cy3B) and 50S (Cy5) subunits, wild-type 30S and 50S subunits, translation factors, S1, mRNA, fluorescently labeled tRNA (fMet-(Cy3)tRNAfMet, Phe-(Cy5)tRNAPhe, and Lys-(Cy2)tRNALys), and unlabeled tRNA (fMet-tRNAfMet, tRNAPhe, and Lys-tRNALys) were prepared as previously described (Blanchard et al., 2004b; Dorywalska et al., 2005; Marshall et al., 2008). All experiments were conducted in a Tris-based polymix buffer consisting of 50 mM Tris-acetate (pH 7.5), 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, 0.5 mM EDTA, 5 mM putrescine-HCl and 1 mM spermidine. All experiments also contained 4 mM GTP. The aminoglycosides used in this study were purchased from Sigma as dry powders and dissolved in water to make stock solutions.
The 5′ biotinylated mRNAs for the intersubunit FRET and ZMW experiments contained the 5′ UTR and Shine-Dalgarno sequence of T4 gene 32 followed by either 3 Phe (UUU) codons (3F) or 6 alternating Phe (UUC) and Lys (AAA) codons (6(FK)). Both 6(FK) and 3F have a stop codon after the end of the message followed by an additional 4 Phe (UUU) codons(Aitken and Puglisi, 2010). The mRNAs for tRNA-tRNA FRET experiments either contain the full T4 gene 32 sequence up to the 7th translated codon (MFK) or have the second translated codon changed from Phe (UUU) to a Leu (CUU) (MLK). All mRNAs were chemically synthesized by Dharmacon.
NMR solution structure of apramycin in complex with A-site oligonucleotide mimic
Near-complete assignment of the proton resonances from the apramycin-RNA complex was accomplished using standard multidimensional, homonuclear and heteronculear experiments with unlabeled RNA and uniformly 13C, 15N – labeled RNA samples (Lukavsky et al., 2003; Lukavsky and Puglisi, 2005). A total of 612 Nuclear Overhauser Effect (NOE) distance restrains were determined from 2D, and 3D NOESY experiments, of which 32 were intermolecular RNA-apramycin distance restraints. Distance restraints were categorized based on peak intensity into four groups with the following inter-proton distances: strong (1.8-3.5 Å), medium (2.0-4.5 Å), weak (2.2-6.0 Å), and very weak (3.0-7.0 Å). Dihedral restrains, β, ε, χ, were determined from 3D HCP, 3D HMQC TOCSY, and 2D NOESY experiments, respectively. Residual dipolar coupling restraints from aromatic N-H and C-H bond vectors were determined by, comparing J-coupling constants collected from 2D 13C/15N TROSY spectra in the presence and absence of pf1 bacteriophage. RDCs were determined according to the following formula:
where 1DXH is the dipolar coupling and 1JXH is the one bond scalar coupling for X = 13C or 15N. The magnitude of C-H RDCs was normalized to those collected for N-H bonds. A break down of the restraints used to determine the structure of the apramycin-RNA complex is shown in Supplementary Table 2. Structures were calculated according to a previously published multi-step simulated annealing protocol using only distance and dihedral restraints during the first stage implemented in XPLOR (Wimberly et al., 1993). RDCs were incorporated into the structure determination during a second stage as described previously in CNS (Lynch et al., 2003).
Intersubunit FRET experiments
We mixed 0.25 μM Cy3B-30S, pre-incubated with stoichiometric S1, 1 μM IF2, 1 μM fMet-tRNAfMet, 1 μM 5′ biotinylated mRNA, and 4 mM GTP to form 30S PICs in the Tris-based polymix buffer described above to form 30S pre-initiation complexes (PICs) (Dorywalska et al., 2005). The PIC mixture was incubated at 37 °C for 5 min.
Before immobilizing the 30S PICs on a slide surface, we diluted the mixture with the Tris-based polymix buffer with 1 μM IF2 and 4 mM GTP. The PICs were then immobilized on the surface of a biotin-PEG derivatized quartz slide via biotin-nuetravidin interactions. We then washed excessive unbound material with the Tris-based polymix buffer containing 1 μM IF2, 4 mM GTP, 1 mM Trolox (to stabilize Cy5 photophysics), and an oxygen scavenging system (2.5 mM 3,4-dihydroxybenzoic acid and 250 nM protocatechuate deoxygenase (Aitken et al., 2008)). We delivered 50 nM Cy5-50S, 1 μM IF2, 80 or 160 nM EF-G, 80 nM ternary complexes, and aminoglycosides (where applicable at the indicated concentration) in the Tris-based polymix buffer using a controlled syringe pump to initiate translation. Phe-tRNAPhe and Lys-tRNALys ternary complexes with EF-Tu(GTP) were formed immediately before delivery as previously described (Marshall et al., 2008).
ZMW experiments
The preparation for ZMW experiments was the same as intersubunit experiments with the following exceptions. The first incubation mixture contains 0.25 μM unlabeled wildtype 30S, 0.25 μM wildtype 50S, 1 μM fMet-(Cy3)tRNAfMet, 1 μM 5′ biotinylated mRNA, and 4 mM GTP to form 70S initiation complexes (ICs). The 70S ICs are then immobilized using the same procedure as in intersubunit FRET experiments. The delivery mixture contained 1 μM IF2, 200 nM EF-G, 200 nM Phe-(Cy5)tRNAPhe ternary complexes, 200 nM Lys-(Cy2)tRNALys ternary complexes, and 10 μM paromomycin, apramycin, or gentamicin, comparable to intersubunit FRET conditions.
tRNA-tRNA FRET experiments
The preparation for tRNA-tRNA FRET experiments was the same as ZMW experiments with the following exception. The delivery mixture contains 1 μM IF2, 500 nM EF-G (where applicable), 25 nM Phe-(Cy5)tRNAPhe ternary complexes, and 10 μM aminoglycosides (where applicable).
Bulk kinetic experiments
Bulk kinetic experiments were performed at 20°C and pH 7.5 in polymix buffer (95 mM KCl, 5 mM NH4Cl, 5 mM Mg(OAc)2, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate and 1 mM DTE) complemented with an energy supply and regeneration system consisting of 2 mM (ATP+GTP), 10 mM phosphoenolpyruvate, pyruvate kinase and myokinase (Johansson et al., 2011). In the experiments for simultaneous monitoring of dipeptide formation and GTP hydrolysis, pre-initiated 70S ribosomal complexes, carrying f[3H]Met-tRNAfMet in P site and displaying the Phe codon UUC in A site, were rapidly mixed with preformed Phe-tRNAPhe:EF-Tu:[3H]GTP ternary complex in a quench-flow instrument (RQF-3, KinTek Corp.) to a final concentration of 200 nM. The reactions were quenched with formic acid at different incubation times and the fractions of products, [3H]GDP/([3H]GDP + [3H]GTP) and f[3H]Met-Phe/( f[3H]Met-Phe + f[3H]Met), were estimated by HPLC as described (Johansson et al., 2011). The rates of GTP hydrolysis and dipeptide formation were measured in the presence (10 μM) and absence of each drug. The rate of GTP hydrolysis, kGTP, including the association of ternary complex to the ribosome, and the rate of tRNA accommodation/peptidyl transfer, kpep, obtained from the inverse of the difference between the dipeptide formation time and GTP hydrolysis time (Johansson et al., 2011), were estimated in drug absence and in the presence of each one of the drugs. Tripeptide formation was performed by rapid mixing and quenching of mixtures similar to those used for dipeptide bond formation, but with EF-G and EF-Ts added to the ternary complex mix and use of unlabeled, rather than labeled GTP.
Supplementary Material
Highlights.
Combining single-molecule dynamics, structural, and biochemical approaches to probe the mechanism of aminoglycosides.
Apramycin inhibits translation elongation by blocking translocation
Paromomycin and gentamicin disrupt ribosomal rotation upon tRNA selection and translocation
Mechanisms are correlated to how each aminoglycoside structurally remodel the 30S A site
Aminoglycosides disrupt translation through inhibiting the dynamics of elongation
Figure 6. No effect of aminoglycosides on peptide bond formation rates.
The extent of hydrolysis of EF-Tu bound GTP (A) and dipeptide formation (B) were monitored over time upon mixing of Phe-tRNAPhe:EF-Tu:GTP ternary complex with pre-initiated 70S ribosomes in the presence or absence of each type of drug, as indicated in the figure. The rate of GTP hydrolysis, kGTP, (C) and the rate of tRNA accommodation/peptidyl transfer, kpep, (D) obtained from the inverse of the difference between the dipeptide formation time and GTP hydrolysis time, were estimated for each case. Error bars in panels C and D represent s.d. as calculated from the fitting procedure averaged over two separate experiments (Johansson et al., 2011). See also Figure S6.
Acknowledgements
Supported by NIH grant GM51266 (JDP), Japan Science and Technology Agency (SU), the Swedish Research Council (ME), and the Knut and Alice Wallenberg Foundation (ME). We thank Alexey Petrov (Stanford), Jin Chen (Stanford), and Seán O’Leary (Stanford) for valuable discussions.
Footnotes
Competing financial interests J.K. is an employee and stock option holder, and J.D.P. a consultant, of Pacific Biosciences, a company commercializing sequencing technologies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Petrov A, Puglisi JD. Single ribosome dynamics and the mechanism of translation. Annu Rev Biophys. 2010;39:491–513. doi: 10.1146/annurev.biophys.093008.131427. [DOI] [PubMed] [Google Scholar]
- Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste R, Davies J. Structure-activity relationships among the aminoglycoside antibiotics: role of hydroxyl and amino groups. Antimicrobial agents and chemotherapy. 1973;4:402–409. doi: 10.1128/aac.4.4.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004a;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004b;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Bottger EC. The ribosome as a drug target. Trends in biotechnology. 2006;24:145–147. doi: 10.1016/j.tibtech.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Bottger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO reports. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Jr., Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Cabanas MJ, Vazquez D, Modolell J. Dual interference of hygromycin B with ribosomal translocation and with aminoacyl-tRNA recognition. Eur J Biochem. 1978;87:21–27. doi: 10.1111/j.1432-1033.1978.tb12347.x. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Davies J, Davis BD. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J Biol Chem. 1968;243:3312–3316. [PubMed] [Google Scholar]
- Davies J, Gorini L, Davis BD. Misreading of RNA codewords induced by aminoglycoside antibiotics. Molecular pharmacology. 1965;1:93–106. [PubMed] [Google Scholar]
- Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annu Rev Biophys. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2009;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Fourmy D, Yoshizawa S, Puglisi JD. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J Mol Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
- Francois B, Russell RJ, Murray JB, Aboul-ela F, Masquida B, Vicens Q, Westhof E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33:5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, Noller HF. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- Johansson M, Ieong KW, Trobro S, Strazewski P, Aqvist J, Pavlov MY, Ehrenberg M. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci U S A. 2011;108:79–84. doi: 10.1073/pnas.1012612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci U S A. 2012;109:131–136. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG. The error catastrophe: a molecular Fata Morgana. BioEssays : news and reviews in molecular, cellular and developmental biology. 1987;6:33–35. doi: 10.1002/bies.950060109. [DOI] [PubMed] [Google Scholar]
- Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci U S A. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Puglisi JD. Structure determination of large biological RNAs. Methods Enzymol. 2005;394:399–416. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- Lynch SR, Gonzalez RL, Puglisi JD. Comparison of X-ray crystal structure of the 30S subunit-antibiotic complex with NMR structure of decoding site oligonucleotide-paromomycin complex. Structure. 2003;11:43–53. doi: 10.1016/s0969-2126(02)00934-6. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt T, Ng CL, Lang K, Sha SH, Akbergenov R, Shcherbakov D, Meyer M, Duscha S, Xie J, Dubbaka SR, et al. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc Natl Acad Sci U S A. 2012;109:10984–10989. doi: 10.1073/pnas.1204073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol. 1990;211:135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Nissen P. Elongation factors on the ribosome. Curr Opin Struct Biol. 2005;15:349–354. doi: 10.1016/j.sbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr., Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nature structural biology. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- Perzynski S, Cannon M, Cundliffe E, Chahwala SB, Davies J. Effects of apramycin, a novel aminoglycoside antibiotic on bacterial protein synthesis. Eur J Biochem. 1979;99:623–628. doi: 10.1111/j.1432-1033.1979.tb13295.x. [DOI] [PubMed] [Google Scholar]
- Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht MI, Douthwaite S, Puglisi JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht MI, Fourmy D, Blanchard SC, Dahlquist KD, Puglisi JD. RNA sequence determinants for aminoglycoside binding to an A-site rRNA model oligonucleotide. J Mol Biol. 1996;262:421–436. doi: 10.1006/jmbi.1996.0526. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy F.V.t., Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012;487:390–393. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B, Varani G, Tinoco I., Jr. The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993;32:1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochemical Society transactions. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Woodcock J, Moazed D, Cannon M, Davies J, Noller HF. Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J. 1991;10:3099–3103. doi: 10.1002/j.1460-2075.1991.tb07863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Fourmy D, Puglisi JD. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the codon-anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.