Abstract
Mycobacteria exploit nonhomologous end-joining (NHEJ) to repair DNA double-strand breaks. The core NHEJ machinery comprises the homodimeric DNA end-binding protein Ku and DNA ligase D (LigD), a modular enzyme composed of a C-terminal ATP-dependent ligase domain (LIG), a central 3’-phosphoesterase domain (PE), and an N-terminal polymerase domain (POL). LigD POL is proficient at adding templated and nontemplated deoxynucleotide and ribonucleotides to DNA ends in vitro and is the catalyst in vivo of unfaithful NHEJ events involving nontemplated single-nucleotide additions to blunt DSB ends. Here, we identify two mycobacterial proteins, PolD1 and PolD2, as stand-alone homologs of the LigD POL domain. Biochemical characterization of PolD1 and PolD2 shows that they resemble LigD POL in their monomeric quaternary structures, their ability to add templated and nontemplated nucleotides to primer-templates and blunt ends, and their preference for rNTPs versus dNTPs. Deletion of polD1, polD2, or both, in an M. smegmatis strain carrying an inactivating mutation in LigD POL failed to reveal a role for PolD1 or PolD2 in templated nucleotide additions during NHEJ of 5’-overhang DSBs or in clastogen resistance. Whereas our results document the existence and characteristics of new stand-alone members of the LigD POL family of RNA/DNA polymerases, they imply that other polymerases can perform fill-in synthesis during mycobacterial NHEJ.
INTRODUCTION
Nonhomologous end joining (NHEJ) is a pathway of DNA double-strand break (DSB) repair, operative in eukarya and many bacteria (1,2), in which the broken ends are sealed without instruction by a homologous DNA. The core components of the eukaryal and bacterial NHEJ systems are the DNA end-binding protein Ku and a dedicated ATP-dependent DNA ligase that joins 3’-OH and 5’-PO4 ends. In eukarya, the Ku protein is a heterodimer of Ku70 and Ku80 subunits and the major NHEJ ligase is DNA ligase IV (Lig4). In bacteria, Ku is a homodimer and the DSB ends are sealed by DNA ligase D (LigD). NHEJ frequently entails the action of DNA polymerases and nucleases that add or subtract nucleotides from the broken ends prior to their sealing by ligase. These end remodeling reactions contribute to the mutagenic quality of NHEJ in vivo. The end-modifying steps of eukaryal NHEJ are performed by a variety of enzymes encoded separately from Lig4 (1). By contrast, the LigD component of the bacterial NHEJ apparatus is a multifunctional enzyme composed of three autonomous catalytic modules – ligase (LIG), polymerase (POL) and 3’-phoshoesterase (PE) – fused into one large polypeptide (2).
The LigD POL domain incorporates dNMPs or rNMPs at DSB ends and gaps prior to strand sealing by the LIG domain (3–10). The LigD PE domain provides a 3’ end-healing function, whereby it converts “dirty” DSBs with 3’-phosphate ends to 3’-OH termini that can serve either as primers for polymerization or as substrates for ligation (6,11–13). The PE domain also trims short 3’-ribonucleotide tracts (produced by POL) to generate the 3’ monoribonucleotide ends that are the preferred substrates for sealing by dedicated bacterial ATP-dependent NHEJ DNA ligases (11–14). Available atomic structures of the component domains of LigD provide insights to their mechanisms, substrate specificities, and evolutionary origins. For example, the crystal structure of the LIG domain (as the covalent ligase–adenylate intermediate) revealed that it is composed of nucleotidyltransferase and OB-fold modules and thus resembles other members of the covalent nucleotidyltransferase superfamily (15). Crystal structures of the LigD POL domain, in complexes with NTPs and/or gapped DNAs, highlighted its membership in the archaeal/eukaryal primase-polymerase family (4,8–10). By contrast, the crystal and NMR structures of the LigD PE module unveiled it as the founding member of a new class of DNA repair enzymes, with a distinctive fold and a unique active site and catalytic mechanism (16–19).
The centrality of Ku and LigD in bacterial NHEJ in vivo has been verified by genetic studies in Mycobacterium smegmatis, facilitated by the development of plasmid-based and chromosomal reporter assays of DSB repair (20–22) that specifically interrogate NHEJ events and allow for characterization of the molecular outcomes of individual events – by phenotypic scoring for restitution of a functional lacZ gene (indicative of error-free NHEJ) and, where NHEJ is unfaithful, by sequencing the repair junctions to identify how the original DSB ends were modified. In this way, it was established that NHEJ of complementary 5’ overhang and blunt DSB ends was inherently unfaithful (∼50% of the outcomes are mutagenic), while NHEJ at complementary 3’ overhangs was error-free (97% fidelity) (21). Analysis of the molecular outcomes of unfaithful NHEJ in vivo in wild-type M. smegmatis highlighted the prevalence of templated insertions at 5’ overhang DSBs and of nontemplated single nucleotide insertions at blunt DSBs (4,20,21). Interrogating NHEJ in M. smegmatis strains bearing deletions or mutations in Ku and LigD illuminated the distinct contributions of LigD’s three catalytic activities (polymerase, ligase and 3’ phosphoesterase) and structural domains (POL, LIG and PE) to the efficiency and molecular outcomes of NHEJ in vivo (21). Most notably, we found that efficient repair of blunt and 5’ overhang DSBs depends stringently on Ku and the LigD POL domain, but not on the LigD polymerase activity, which mainly serves to promote NHEJ infidelity. The essentiality of the POL domain is thought to reflect its structural role in assembly of a DSB-bridging Ku-LigD complex, via direct physical interaction between POL and Ku. The effect of ablating polymerase catalysis (by a double-alanine change in the POL active site) is to eliminate nontemplated single nucleotide insertions at blunt DSBs (21), signifying that LigD POL is the immediate, and likely exclusive, catalyst of the mutagenic single nucleotide additions. By contrast, the templated nucleotide insertions at 5’ overhang DSBs persisted when the LigD polymerase was inactivated (21). Thus, mycobacteria must have another polymerase that incorporates nucleotides at 5’-overhangs during Ku-dependent NHEJ.
In the present study, we identify two mycobacterial proteins as plausible candidates for the role of backup NHEJ polymerase. They are free-standing mycobacterial paralogs of the isolated LigD POL domain, which we named PolD1 and PolD2, respectively. We report here a biochemical and genetic characterization of M. smegmatis PolD1 and PolD2.
MATERIALS AND METHODS
Expression vectors for M. smegmatis PolD1 and PolD2
The MsmPolD1 ORF (Msmeg_6301) was PCR-amplified from Mycobacterium smegmatis genomic DNA with primers designed to introduce an NdeI site at the start codon and a BamHI site 3’ of the stop codon. The MsmPolD2 ORF (Msmeg_0597) was PCR-amplified using primers designed to introduce NdeI sites at the start codon and 3’ of the stop codon. The PCR products were digested with NdeI and BamHI (PolD1) or NdeI alone (PolD2) and inserted into pET16b to generate pET16b-MsmPolD1 and pET16b-MsmPolD2, respectively. Asp-to-Ala mutations in the putative metal-binding sites of PolD2 (D153A) and PolD1 (D140A) were introduced by two-stage overlap extension PCR; the mutated ORFs were cloned into pET16b. The inserts were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning.
Purification of PolD1 and PolD2
The pET-MsmPolD plasmids were transformed into Escherichia coli BL21(DE3). Cultures (1 L) of E. coli BL21(DE3)/pET16b-MsmPolD were grown at 37°C in Luria-Bertani medium containing 0.1 mg/ml ampicillin until the A600 reached 0.6. The cultures were adjusted to 0.5 mM isopropyl-β-D-thiogalactopyranoside and then incubated at 17°C for 15 h. Cells were harvested by centrifugation and the pellets were stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 50 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 1.5 M NaCl, 200 mM Li2SO4, 10% glycerol, 15 mM imidazole). Lysozyme and Triton X-100 were added to final concentrations of 50 µg/ml and 0.1%, respectively. The lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation. The supernatants were applied to 2-ml columns of Ni2+−nitrilotriacetic acid-agarose (Qiagen, Chatsworth, CA, USA) that had been equilibrated with lysis buffer. The columns were washed with 50 ml of lysis buffer and then eluted stepwise with 4-ml aliquots of buffer A (50 mM Tris-HCl, pH 7.5, 0.2 M NaCl, 10% glycerol) containing 50, 100, 200 and 500 mM imidazole. The polypeptide compositions of the fractions were monitored by SDS-PAGE. The MsmPolD1 and MsmPolD2 proteins were recovered predominantly in the 200 mM imidazole eluate fractions, which were then applied to 2-ml DEAE-cellulose columns that had been equilibrated with buffer A. The DEAE-cellulose columns were washed with 10 ml buffer A and then eluted stepwise with 4-ml aliquots of 0.3, 0.4, 0.5, and 1.0 M NaCl in 50 mM Tris-HCl (pH 7.5), 10% glycerol. MsmPolD1 and MsmPolD2 were recovered predominantly in the flow-through fractions, which were then applied to 2-ml columns of phosphocellulose that had been equilibrated with buffer A. The phosphocellulose columns were washed with 10 ml of buffer A and then eluted stepwise with 4-ml aliquots of 0.3, 0.4, 0.5, and 1.0 M NaCl in 50 mm Tris-HCl (pH 7.5), 10% glycerol. MsmPolD1 and MsmPolD2 were recovered predominantly in the flow-through fractions (which contained 1.6 mg and 2.8 mg protein, respectively). The protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard. The MsmPolD1 and MsmPolD2 preparations were stored at −80°C. The MsmPolD1-D140A and MsmPolD2-D153A mutants were purified in a similar fashion.
Glycerol Gradient Sedimentation
Aliquots (30 µg) of MsmPolD1 or MsmPolD2 were mixed with catalase (30 µg), BSA (30 µg), and cytochrome c (30 µg) in 200 µl of buffer A. The protein mixtures were applied to 4.8-ml 15–30% glycerol gradients containing 50 mM Tris-HCl (pH 8.0), 0.2 m NaCl, 1 mM EDTA, 2.5 mM DTT, 0.1% Triton X-100. The gradients were centrifuged in a Beckman SW50 rotor at 50,000 rpm for 18 h at 4°C. Fractions (0.2 ml) were collected from the bottoms of the tubes. The polypeptide compositions of the gradient fractions were analyzed by SDS-PAGE. Aliquots of the fractions were assayed for activity as specified in the figure legends.
Polymerase Assays
Polymerase activity was gauged by NTP- or dNTP-dependent extension of an 18-mer 5′ 32P-labeled oligodeoxynucleotide primer. The 18-mer primer strand was end-labeled by reaction with T4 polynucleotide kinase and [γ-32P]ATP, then purified by native gel electrophoresis, and (where indicated) annealed to a 4-fold excess of an unlabeled complementary DNA strand to generate the blunt-ended or 5′-tailed substrates depicted in the figures. The labeled and unlabeled oligonucleotides were mixed in a 0.2 M NaCl solution and then heated at 95°C for 7 min, followed by slow cooling to room temperature. Polymerase reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 7.5 or 8.0), 1.25 mM CoCl2 (or other divalent cations as specified), 1 pmol of 5′ 32P-labeled DNA substrate, NTPs/dNTPs and enzyme as specified were incubated at 37°C for 10 min. The reactions were quenched by adjusting the mixtures to 10 mM EDTA and 48% formamide. The products were resolved by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 M urea in 90 mM Tris-borate, 2.5 mM EDTA. The products were visualized by autoradiography. Where indicated, the fraction of input primer extended was determined by scanning the gel with a Fujifilm BAS-2500 imaging apparatus.
Deletion of polD1 and polD2 in M. smegmatis
The polD2 gene (Msmeg_0597) was deleted in M. smegmatis wild-type and ligD-(D136A–D138A) strains (4,21) by transduction of a polD2::hygR null allele carried in the temperature-sensitive mycobacteriophage phAE87 (23). The null allele was composed of M. smegmatis genomic DNA segments flanking the polD2 locus (the upstream segment consisting of 509 nucleotides 5’ of the start codon plus the first 42 nucleotides of the ORF; the downstream segment comprising the 3’-terminal 63 nucleotides of the ORF plus 527 nucleotides downstream of the stop codon), between which was inserted a hygromycin-resistance gene flanked by loxP sites. Wild-type and ligD-(D136A–D138A) strains were infected with the polD2::hygR phage and hygromycin-resistant transductants were selected for growth on agar medium containing 50 µg/ml hygromycin. Correct allelic replacement was confirmed by diagnostic Southern blotting. A plasmid bearing a Cre-recombinase gene and a kanamycin-resistance marker was introduced into the bacteria to excise the hygR cassette, which was verified by re-acquisition of sensitivity to growth inhibition by hygromycin. Strains that had spontaneously lost the Cre-expressing plasmid were identified by plating individual colonies on agar media containing 20 µg/ml kanamycin and selecting kanamycin-sensitive isolates. The ΔpolD2 allele contains the first 42 and last 63 nucleotides of polD2.
The polD1 gene (Msmeg_6301) was deleted in the wild-type, ligD-(D136A–D138A) and ligD-(D136A–D138A) ΔpolD2 backgrounds using a two-step allelic exchange technique (24). A plasmid that has 5’ and 3’ flanking regions of polD1, two counter-selectable markers (sacB and galK), and hygR was transformed into wild-type, ligD-(D136A–D138A) and ligD-(D136A–D138A) ΔpolD2 strains. The 5’ flank contained 539 nucleotides upstream of polD1 and 61 nucleotides from the open reading frame. The 3’ flank comprised 24 nucleotides from the open reading frame of polD1 and 474 nucleotides downstream of the ORF. Hygromycin-resistant colonies arising via integration of the plasmid at 3’ and 5’ flanks of the polD1 gene were identified by Southern blotting (Fig. 7). The transformants were grown in non-selective LB medium overnight. Dilutions of the culture were counter-selected on 10% sucrose/0.5% 2-DOG and sucrose/2-DOG-resistant, hygromycin-sensitive colonies were examined for the deletion of polD1 by Southern blotting of genomic DNA (Fig. 7).
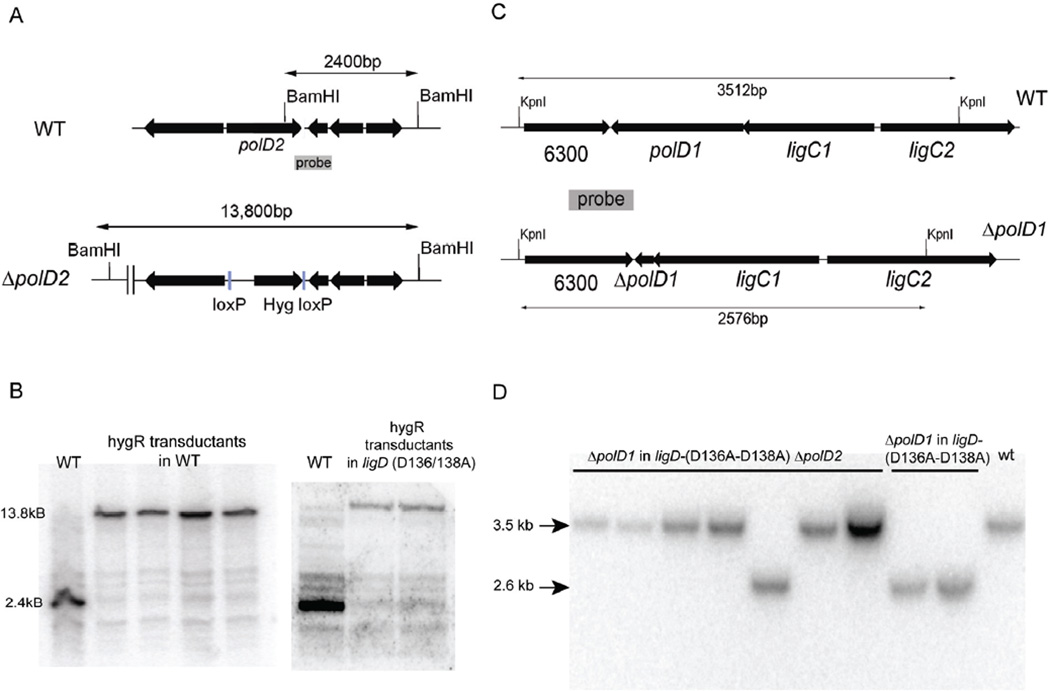
Figure 7. Deletion of polD1 and polD2 from M. smegmatis.
(A) Genomic organization of the polD2 locus. The polD2 loci in wild type (top) and ΔpolD2 hyg (bottom) strains are shown. The location of BamHI restriction sites and probes for Southern hybridization are indicated. The BamHI restriction fragments that will hybridize to the indicated probe are 2400 bp in wild type cells and 13,800 bp in ΔpolD2 cells. (B) Southern blotting. Genomic DNA was digested with BamHI, resolved by agarose gel electrophoresis, transferred to membrane and then hybridized to the 32P-labeled DNA fragment indicated in panel A. An autoradiogram of the blot is shown. The left panel shows wild type DNA alongside that of hygromycin-resistant transductants of wild type cells (to create strain MGM2027). The right panel shows wild type DNA alongside that of two hygromycin-resistant transductants in the ligD-(D136A–D138A) background (to create strain MGM 2028). (C) Genomic organization of the polD1 locus. The polD1 loci in wild-type (top) and ΔpolD1 (bottom) strains are shown. The location of Kpn1 restriction sites and probes for Southern hybridization are indicated. The Kpn1 restriction fragments that will hybridize to the indicated probe are 3512 bp in wild-type cells and 2576 bp in ΔpolD2 cells. (D) Confirmation of the ΔpolD1 strains by Southern blotting. Seven recombinants are shown in the ligD-(D136A–D138A) ΔpolD2 background, six of which have retained the wild-type allele and one of which contains the ΔpolD1 allele (MGM 846). Two recombinants in the ligD-(D136A–D138A) are shown, both of which contain the ΔpolD1 allele (MGM 845).
Assay of NHEJ in M. smegmatis
Plasmid-based assay of NHEJ of 5’-overhang DSBs was performed as described previously (21) in wild-type M. smegmatis and mutant strains ligD-(D136A–D138A), ΔpolD1, ΔpolD2, ligD-(D136A–D138A) ΔpolD1, ligD-(D136A–D138A) ΔpolD2, and ligD-(D136A–D138A) ΔpolD1 ΔpolD2. The NHEJ substrate was generated by digesting circular plasmid pjsm101 with restriction enzymes Asp7181 and PsiI and then purifying the linear plasmid DNA by agarose gel electrophoresis. Three replicate transformations by electroporation were performed for each strain. Dilutions of the electroporated cells were plated on agar media containing kanamycin (20 µg/ml) and X-Gal (50 µg/ml). The numbers of blue and white colonies were counted for linear and circular transformations. NHEJ efficiency was calculated as the ratio of the number of transformants per ng of the linear plasmid to the number of transformants per ng of the uncut circular plasmid. The NHEJ efficiency values for mutant strains were normalized to the efficiency value of wild-type M. smegmatis (defined as 100%). Fidelity was calculated as the number of blue colonies over the total number of colonies. The unfaithful repair events in white colonies were determined by PCR-amplifying the repair junctions in the reporter plasmid. Primers 795 bp upstream and 1252 bp downstream of the original DSB ends were used for colony PCR. The PCR products were sequenced using primers 200 bp downstream, 200 bp upstream, 400 bp upstream, and 500 bp downstream of the original DSB ends.
Assay of sensitivity of M. smegmatis to killing by UV irradiation
The M. smegmatis strains were grown to early log-phase (A600 of 0.2 to 0.4). Serial dilutions of the cultures in phosphate-buffered saline with 0.05% Tween-80 were spotted on LB agar containing 0.5% glycerol and 0.5% dextrose. The plates were irradiated with escalating UV doses using a Stratalinker (Stratagene) fitted with 254 nm bulbs, then shielded from light and incubated at 37°C. Surviving colonies at different 10-fold dilutions were counted after 3 d and fraction survival was calculated by comparison to non-irradiated controls.
Assay of sensitivity of M. smegmatis to killing by ionizing radiation (IR)
M. smegmatis cultures were grown to early log-phase; the cells were harvested by centrifugation and then resuspended in phosphate-buffered saline with 0.05% Tween-80. Aliquots of the resuspended cells were irradiated with a 137Cs source that delivered 10.3 Gy/min, after which serial 10-fold dilutions were plated on agar medium. Surviving colonies were counted after incubation for 3 d at 37°C. Percent survival was calculated after normalization to the colony counts obtained from the unexposed control cells.
RESULTS
Two free-standing LigD-like polymerases in mycobacteria
We identified two paralogous proteins in M. smegmatis as homologs of the LigD POL domain: MsmPolD1, a 350-aa polypeptide encoded by Msmeg_6301, and MsmPolD2, a 426-aa polypeptide encoded by Msmeg_0597. MsmPolD1 and MsmPolD2 are free-standing POL homologs that lack the signature motifs of the LigD LIG and PE domains. The human pathogen M. tuberculosis encodes an equivalent pair of LigD-like POL proteins, which we named MtuPolD1 (the 346-aa product of the Rv3730c gene) and MtuPolD2 (the 397-aa product of the Rv0269c gene). Pairwise comparisons show that: (i) MsmPolD1 and MtuPolD1 share 294 positions of side chain identity/similarity; (ii) MsmPolD2 and MtuPol2 share 319 positions of side chain identity/similarity; and (iii) MsmPolD1 and MsmPolD2 share 192 positions of side chain identity/similarity. Counterparts of PolD1 and PolD2 are present in the proteomes of many other species of the genus Mycobacterium (Table S1) and in many of the genera that comprise the phylum Actinobacteria.
An alignment of the primary structures of Msm and Mtu PolD1 and PolD2 and the POL domain of Pseudomonas aeruginosa LigD (Fig. S1) highlights conservation of the amino acids that, in the case of Pseudomonas or Mycobacterium LigD POL, are responsible for binding the NTP substrate, the two essential manganese ion cofactors, or a gapped DNA template (4,9,10). The conservation of active site and DNA-binding residues raises the questions of (i) whether the PolD1 and PolD2 proteins are bona fide polymerases and, if so, the extent to which their biochemical repertoire resembles that of LigD POL and (ii) whether they play a role in DNA repair in vivo, especially in mutagenic NHEJ as potential agents of the LigD-POL independent templated fill-in events at 5’ overhang DSBs.
To query the biochemistry of MsmPolD1 and MsmPolD2, we produced them in E. coli as His10-tagged derivatives and purified the recombinant proteins from soluble bacterial extracts by sequential nickel-affinity and ion exchange chromatography steps. SDS-PAGE showed that the MsmPolD2 preparation contained a single 45 kDa polypeptide (Fig. 1A). The MsmPolD1 preparation contained a predominant 40 kDa polypeptide (Supplemental Fig. S2A). The results of the biochemical and physical characterization of MsmPolD2 are presented in Fig 2–Fig 6. Analogous studies of MsmPolD1 are described and shown in Fig S2 and Fig S3.
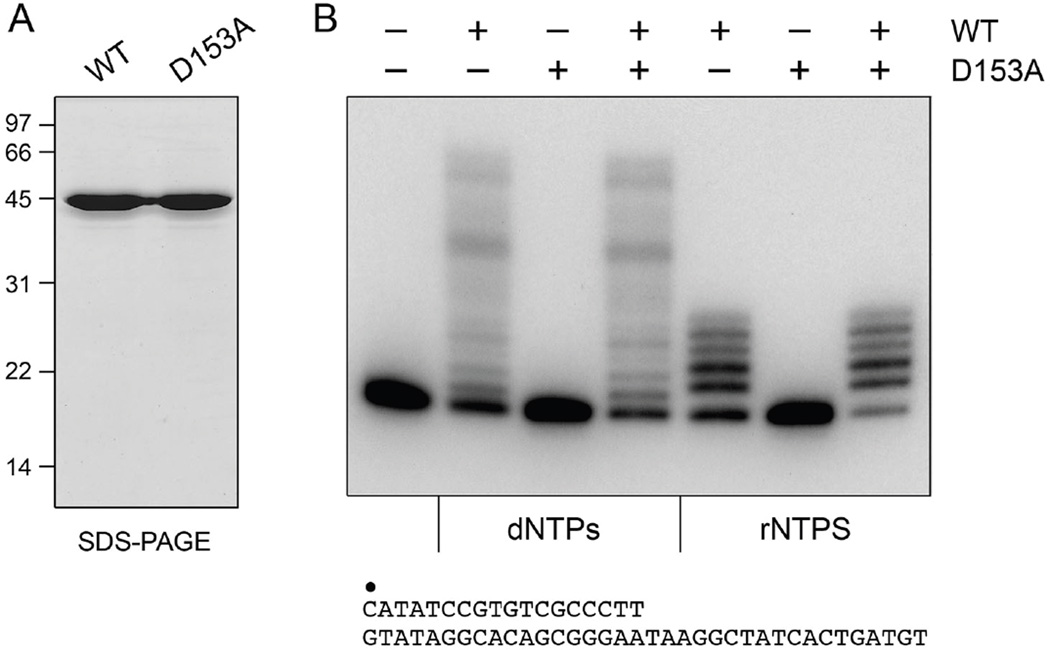
Figure 1. Purification and polymerase activity of MsmPolD2.
(A) Aliquots (5 µg) of the phosphocellulose preparations of wild-type MsmPolD2 and mutant D153A were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Polymerase reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 8.0), 1.25 mM CoCl2, 1 pmol of 32P-labeled primer-template DNA substrate (shown at the bottom, with the 5’ 32P label of the primer strand denoted by •), 100 µM each of dATP, dGTP, dCTP and dTTP (dNTPs) or 100 µM each of ATP, GTP, CTP and UTP (rNTPs), and 200 ng (∼4.4 pmol) of MsmPolD2 (WT or D153A, where indicated by +) were incubated for 10 min at 37°C. The products were resolved by PAGE and visualized by autoradiography.
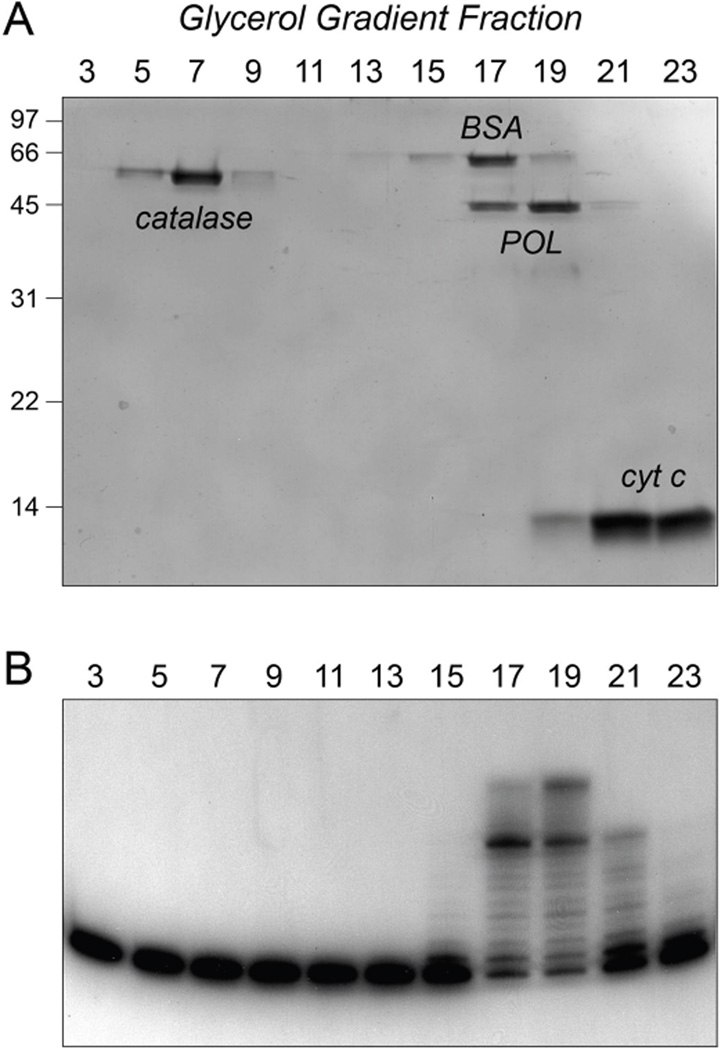
Figure 2. MsmPolD2 sediments as a monomer.
Glycerol gradient sedimentation of a mixture of MsmPolD2, catalase, BSA and cytochrome c was performed as described under Experimental Procedures. (A) Aliquots (15 µl) of the odd-numbered gradient fractions were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. (B) Polymerase reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 8.0), 1.25 mM CoCl2, 1 pmol of 32P-labeled primer-template DNA substrate, 100 µM each of dATP, dGTP, dCTP and dTTP, and 2 µl of the odd-numbered gradient fractions were incubated for 10 min at 37°C. The reaction products were resolved by PAGE and visualized by autoradiography.
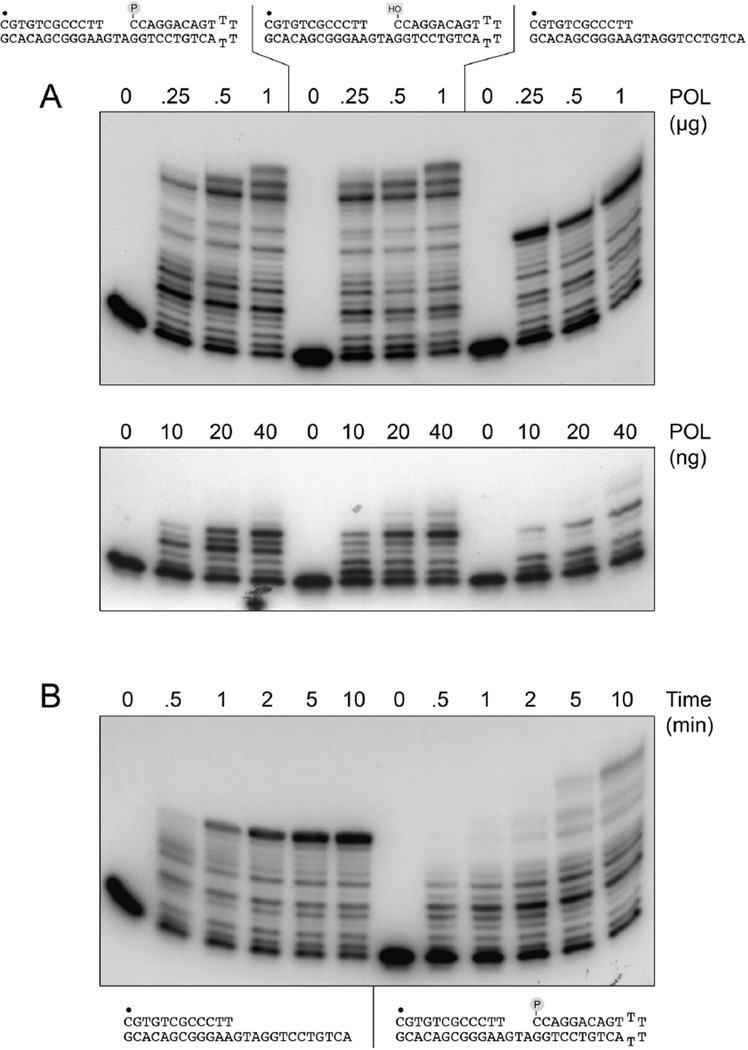
Figure 6. Gap filling by PolD2 extends into the downstream duplex segment.
(A) Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 8.0), 1 pmol of 32P-labeled DNA substrate (gapped duplexes or simple primer-template as illustrated above the lanes), 100 µM each of dATP, dGTP, dCTP and dTTP (dNTPs), 1.25 mM CoCl2, and MsmPolD2 as indicated above the lanes were incubated for 10 min at 37°C. (B) Reaction mixtures (140 µl) containing 50 mM Tris-HCl (pH 8.0), 100 µM each of dATP, dGTP, dCTP and dTTP (dNTPs), 1.25 mM CoCl2,, 7 pmol of 32P-labeled substrate (simple primer-template or gapped duplex as illustrated below the lanes), and 1.2 µg of MsmPolD2 were incubated at 37°C. Aliquots (20 µl) were withdrawn at the times specified and quenched immediately with EDTA/formamide. The products were analyzed by PAGE and visualized by autoradiography.
Polymerase activity of PolD2 and PolD1
MsmPolD2 catalyzed templated DNA or RNA synthesis, as gauged by its ability, in the presence of a divalent cation and the 4 dNTP or rNTPs, to extend a 5’ 32P-labeled 18-mer DNA primer strand annealed to a complementary 36-mer template strand (Fig. 1B). dNMP addition yielded a ladder of radiolabeled products representing the range of fill-in synthesis, up to 18 steps to reach the end of the template strand. By contrast, DNA-primed RNA synthesis was limited to between 1 and 5 steps of rNMP incorporation (Fig. 1B). The MsmPolD1 protein executed the equivalent patterns of “long” and “short” primer-template extensions in the presence of dNTPs and rNTPs, respectively (Fig. S2B). This length restriction on rNMP addition is characteristic of LigD POL (11) and arises because: (i) productive use of the primer 3’-OH depends on contacts with deoxynucleotides at the terminal positions of the primer; and (ii) serial rNMP incorporation events diminish the ability of the ribo-extended primer to undergo the next round of reaction with an rNTP. After 4 ribonucleotides are added, LigD POL activity is suppressed strongly (11). It is likely that the A-form helical conformation of an RNA-DNA hybrid at the primer terminus is responsible for this suppressive effect.
LigD POL employs a two-metal mechanism of nucleotide addition to the 3’-OH primer terminus (4). The metals are coordinated by three aspartate side chains (Asp669, Asp671, Asp759 in PaeLigD) that are conserved in MsmPolD2 (Asp153, Asp155, Asp241) and MsmPolD1 (Asp140, Asp142, Asp231). We produced and purified a mutated version of MsmPolD2 in which the metal-binding Asp153 was replaced by alanine (Fig. 1A) and the equivalent variant of MsmPolD1 in which Asp140 was changed to alanine (Fig. S2A). The D153A change abolished the DNA and RNA polymerase activity of MsmPolD2 (Fig. 1B); the D140A mutation ablated the polymerase activities of MsmPolD1 (Fig. S2B). Mixing experiments showed that the mutant proteins did not inhibit the polymerase activities of the wild-type MsmPolD2 and MsmPolD1 enzymes (Fig. 1B and Fig. S2B). We conclude that the polymerase activities are intrinsic to the recombinant MsmPolD2 and MsmPolD1 proteins.
PolD2 and PolD1 are monomeric enzymes
The quaternary structure of MsmPolD2 was examined by zonal velocity sedimentation in a 15–30% glycerol gradient (Fig. 2). Marker proteins catalase (native size 248 kDa), BSA (66 kDa) and cytochrome c (12 kDa) were included as internal standards in the gradient. After centrifugation, the polypeptide compositions of the odd-numbered gradient fractions were analyzed by SDS-PAGE. PolD2 sedimented as a discrete peak (in fractions 17–19) on the “light” side of the BSA peak (Fig. 2A). The DNA polymerase activity profile was coincident with that of the PolD2 polypeptide (Fig. 2B). Sedimentation analysis of MsmPolD1 is shown in Fig. S3. A discrete peak of PolD1 protein, situated between the BSA and cytochrome c markers (Fig, S3A), coincided with the DNA polymerase activity profile (Fig. S3B). These results are consistent with MsmPolD2 and MsmPolD1 being monomers in solution.
Metal requirements for templated and non-templated polymerase reactions of PolD2
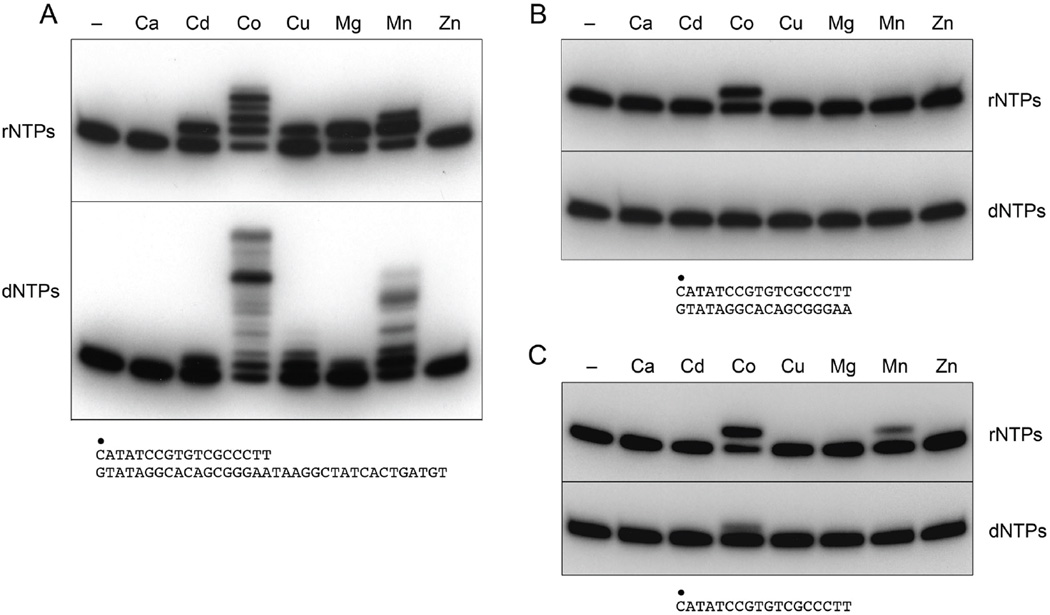
MsmPolD2 was strictly dependent on a divalent cation cofactor for its ability to add dNMPs or rNMPs to the 18-mer/36-mer DNA primer-template (Fig. 3A). The metal requirement was satisfied best by cobalt, which supported the highest level of primer utilization (i.e., the fraction of input primer extended) and the longest tracts of nucleotide synthesis (Fig. 3A). Manganese was the next best metal in terms of primer utilization or tract length (Fig. 3A). Under the same reaction conditions, magnesium, cadmium and copper supported a single step of dNMP or rNMP addition (Fig. 3A). Calcium and zinc were ineffective as cofactors for the templated polymerase activity (Fig. 3A).
Figure 3. Metal cofactor requirements for templated and non-templated nucleotide addition reactions of MsmPolD2.
Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 8.0), either 1 pmol of 32P-labeled primer-template DNA substrate (panel A), 1 pmol of 32P-labeled blunt duplex DNA substrate (panel B), or 1 pmol of 32P-labeled single-strand primer (panel C), 100 µM each of dATP, dGTP, dCTP and dTTP (dNTPs) or 100 µM each of ATP, GTP, CTP and UTP (rNTPs), 200 ng MsmPolD2, and either no added metal (lanes –) or 1.25 mM of the indicated divalent cations were incubated for 10 min at 37°C. The reaction products were resolved by PAGE and visualized by autoradiography.
LigD POL is adept at non-templated additions to blunt DSBs in vitro, a property that is pertinent to its in vivo function in mutagenic NHEJ (3,4). Here we gauged the ability of MsmPolD2 to add deoxynucleotides or ribonucleotides to the radiolabeled primer strand of a blunt-ended 18-bp DNA duplex. We found that PolD2 was able to incorporate a single non-templated ribonucleotide in the presence of cobalt (Fig. 3B). Only a trace amount of rNMP addition was detected in manganese and no dNMP addition was seen with any of the metals tested (Fig. 3B). Preference for rNTPs versus dNTPs as substrates for nontemplated blunt-end addition is characteristic of LigD POL and is typically restricted to a single step of rNMP incorporation (3).
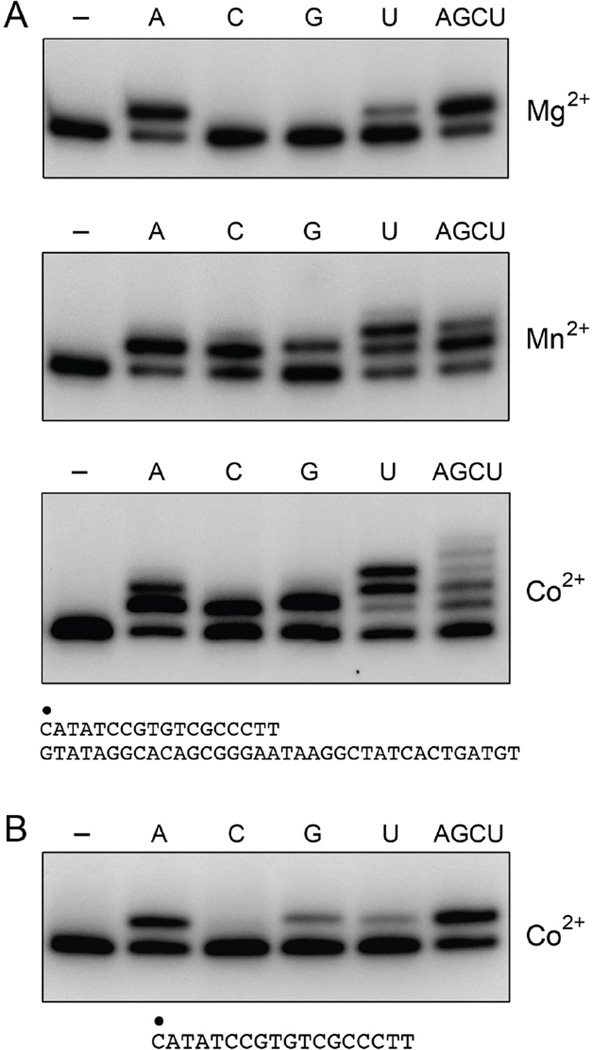
As noted previously for LigD POL (3,8), we found that MsmPolD2 was able to add a single non-templated ribonucleotide to a single-stranded DNA primer (Fig. 3C). The metal requirement for this rNMP addition was satisfied by cobalt and, to a lesser extent, by manganese. rNTPs were preferred versus dNTPs as substrates for cobalt-dependent end-addition to a single-stranded DNA primer (Fig. 3C). Testing individual rNTPs instead of an rNTP mixture showed that ATP was the preferred substrate for cobalt-dependent extension of the primer DNA strand (Fig. 4B).
Figure 4. MsmPolD2 ribonucleotide preferences for templated and nontemplated additions.
(A) Templated additions. Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 8.0), 1 pmol of 32P-labeled primer-template DNA substrate (shown at bottom), either no added nucleotide (lanes –), 100 µM of a single rNTP as specified, or a mixture of 100 µM each of ATP, GTP, CTP and UTP (AGCU), 200 ng MsmPolD2, and either 1.25 mM MgCl2, MnCl2, or CoCl2 as specified were incubated for 10 min at 37°C. (B) Nontemplated additions. Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 8.0), 1,25 mM CoCl2, 1 pmol of 32P-labeled single-strand primer (shown at bottom), either no added nucleotide (lanes –), 100 µM of a single rNTP as specified, or a mixture of 100 µM each of ATP, GTP, CTP and UTP (AGCU), and 200 ng MsmPolD2 were incubated for 10 min at 37°C. The reaction products were resolved by PAGE and visualized by autoradiography.
Fidelity of templated nucleotide addition by PolD2
LigD POL is a relatively unfaithful nucleic acid polymerase that is capable of misincorporating nucleotides during templated DNA or RNA synthesis (5). Here we tested the fidelity of ribonucleotide addition by MsmPolD2 on the 18-mer/36-mer primer-template by comparing the reaction outcomes in the presence of only the correctly templated substrate ATP versus only one of the “incorrect” substrates (CTP, GTP and UTP) and versus a mixture of all four rNTPs (Fig. 4A). Fidelity was gauged in the presence of different metal cofactors: magnesium, manganese, or cobalt. Control experiments verified that no primer extension occurred in the absence of exogenous NTPs (Fig. 4A, lanes –).
MsmPolD2 displayed the highest degree of fidelity and the shortest tract length of NMP addition in the presence of magnesium, whereby: (i) only a single step of AMP addition occurred when ATP was the sole substrate provided, and (ii) inclusion of all four NTPs did not allow for more than one cycle of NMP addition (Fig. 4A, top panel). No primer extension occurred in magnesium in the presence of either CTP or GTP. A low level of extension by one nucleotide was observed in the presence of UTP (Fig. 4A, top panel), which likely reflects either forward slippage or backward slippage of the primer terminus by one nucleotide so that the incoming UTP substrate can pair with an A nucleobase in the template strand. Facile slippage and realignment of the primer on the template is characteristic of LigD POL (5).
The fidelity of ribonucleotide addition was relaxed in the presence of manganese such that the primer was extended by one nucleotide step in the presence of ATP, CTP or GTP and by one or two nucleotide steps in the presence of UTP or all four NTPs (Fig. 4A, middle panel). CTP misincorporation likely arose by forward slippage of the 3’-TTOH of the primer strand to pair with the AA dinucleotide of the 3’-AAG sequence of the template strand. The sequential misincorporation of two UTPs likely reflects forward or reverse slippage of the primer terminus to place it adjacent to AA dinucleotides on the template strand.
MsmPolD2 fidelity was relaxed further and the tract length was increased in the presence of cobalt (Fig. 4A, bottom panel). For example, a fraction of the primers were extended by two nucleotides in the presence of ATP alone, the consequence of one correct AMP addition and a subsequent misincorporation. Again, we observed single step of misincorporation of CTP or GTP. In the presence of UTP, the major projects were extended by two or three nucleotides.
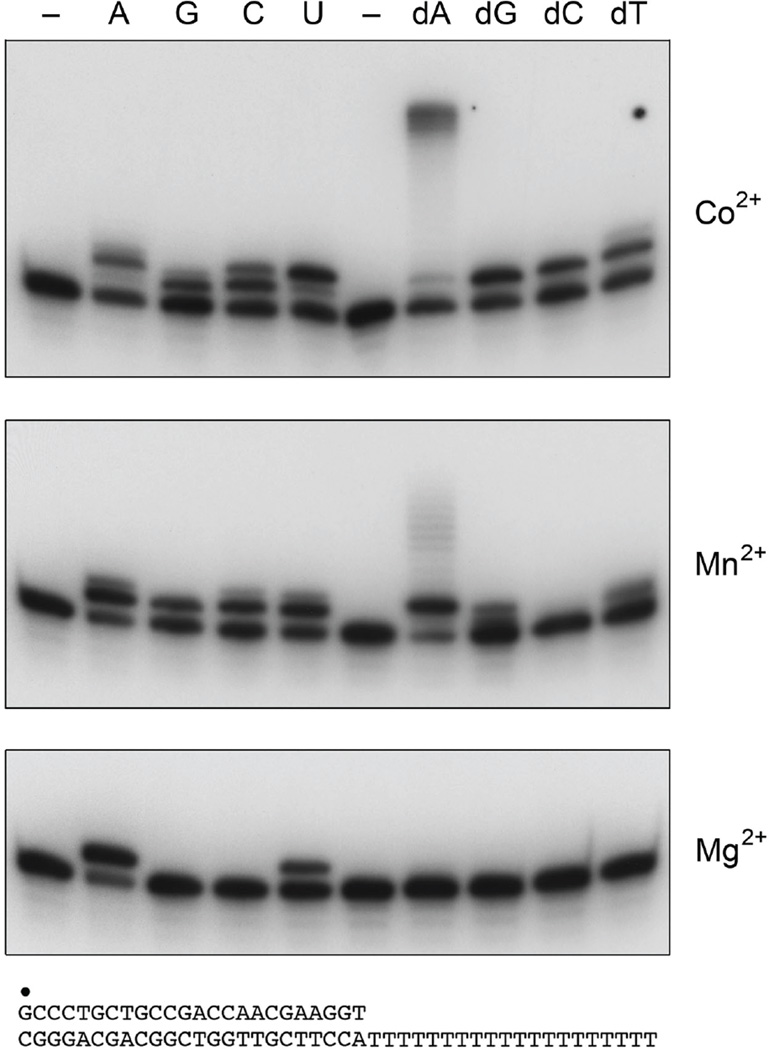
In an effort to diminish forward slippage as a factor in the assessment of MsmPolD2 fidelity, we exploited a different primer-template consisting of a 5’ 32P-labeled 24-mer primer annealed to a complementary 44-mer template DNA strand. The resulting 24-bp duplex contained a 20-nucleotide homogeneous deoxythymidylate tail as the template for primer extension (Fig. 5). We compared the products of primer extension in reactions containing a single rNTP or dNTP substrate and either cobalt, manganese or magnesium as the metal cofactor (Fig. 5). Control reactions verified that no primer extension occurred in the absence of added nucleotide. Magnesium enforced the highest degree of fidelity for incorporation of the correctly templated ATP substrate and the most stringent specificity for templated ribonucleotide incorporation (Fig. 5, bottom panel). The lower extent of UTP incorporation likely reflected backward slippage of the primer end and UMP addition across from the template A nucleobase. Manganese and cobalt were progressively better at enabling the templated DNA polymerase activity of MsmPolD2. The predominant outcome of “correct” manganese-dependent templated synthesis was the addition of single dAMP, with only scant additions directed by the remaining nucleobases of the T20 template strand (Fig. 5, middle panel). By contrast, cobalt-dependent dATP addition proceeded to the end of the available template to yield a discrete fill-in product of the expected size and relatively few intermediates (Fig. 5, top panel). Cobalt promoted misincorporation of single dG, dC and dT nucleotides, thereby attesting to the infidelity of DNA synthesis under these conditions. Manganese and cobalt also relaxed the fidelity of templated RNA synthesis, by allowing misincorporation in the presence of GTP and CTP (Fig. 5).
Figure 5. NTP/dNTP choice and infidelity on a primed homopolymer template.
Reaction mixtures (20 µl) containing 50 mM Tris-HCl (pH 8.0), 1 pmol of 32P-labeled primer-template DNA substrate (shown at bottom), either no added nucleotide (lanes –), 100 µM of a single rNTP or dNTP as specified, 200 ng MsmPolD2, and either 1.25 mM MgCl2, MnCl2, or CoCl2 as specified (at right) were incubated for 10 min at 37°C. The reaction products were resolved by PAGE and visualized by autoradiography.
Gap filling activity of PolD2
LigD POL is thought to bridge broken DNA ends via anchoring contacts with a 5’-phosphate-terminated duplex DNA segment downstream of the captured primer strand terminus (9,10), a scenario analogous to gap repair. Indeed, the gap repair activity of LigD POL in vitro is more efficient than simple templated primer extension and relies on a 5’-phosphate group on the distal gap strand end to confer apparent processivity in filling short gaps (7,8). Mutations of the side chains implicated in DNA 5’-phosphate binding eliminated the preferential filling of 5’-phosphate gaps by LigD POL (7). As several of the DNA-binding residues are conserved in the mycobacterial PolD1 and PolD2 proteins, we undertook to compare the activity of MsmPolD1 in gap repair synthesis versus extension of a simple primer-template. The 3-nucleotide gapped DNA substrates were the same ones used previously to characterize gap-filling by Pseudomonas LigD POL (7). They were prepared by annealing a 5’ 32P-labeled 12-mer primer strand to the 3’ tail of a 39-mer 3’-tailed hairpin duplex oligonucleotide. Two otherwise identical hairpin duplexes were employed, that had either a 5’-OH or a 5’-phosphate terminus at the distal boundary of the gap (Fig. 6A). The gapped substrates were tested for primer extension activity with MsmPolD2, in parallel with a standard primer-template formed by annealing the same radiolabeled primer to a complementary single-stranded DNA oligonucleotide identical in sequence to the template strand of the gapped DNAs (Fig. 6A). The primer extension and gap filling reactions were performed in the presence of 100 µM dNTPs with increasing amounts of input MsmPolD2. At low (10–40 ng) levels of input PolD2 (enzyme:DNA ratios <1), the simple primer-template was extended predominantly by 1, 2, or 5 steps and the fraction of template extended was low (Fig. 6A, bottom panel). Whereas the yield of elongated product was enhanced by on the 5’-OH gapped DNA, the initial product distribution still favored +1 and +2 extensions at low enzyme and the accumulation of a +5 extension product at 40 ng PolD2 (Fig. 6A, bottom panel). The product distribution was appreciably different with the 5’-phosphate gapped DNA, whereby the +3 extension predominated over shorter intermediates at 10–20 ng of input PolD2, signifying that the 5’-phosphate favored processive filling of the 3-nucleotide gap. However, the enzyme readily progressed two-nucleotides into the downstream duplex to yield a predominant +5 extension product at 40 ng of input PolD2.
Extension into the distal duplex of the gapped DNAs was more pronounced at higher (0.25 to 1 µg) levels of input PolD2 (enzyme:DNA ratios of ∼5 to 20), as was complete fill-in of a significant fraction of the simple primer-template, though the ladder of intermediates persisted in all cases (Fig. 6A, top panel). The sizes of the ladder of extension products made on the gapped DNAs indicated that PolD2 was ultimately able to displace and copy most, if not all, of hairpin segment of the template strand. The kinetics of fill-in of the simple primer-template and 5’-phosphate gapped DNAs under conditions of PolD2 excess are shown in Fig. 6B. Complete fill-in of the primer template was apparent at 1 min and the fill-in product accumulated steadily at 2 and 5 min to attain an apparent endpoint that was notable for the persistence of the ladder of smaller products even though the majority of the input primers had been extended. This pattern suggested that PolD2 acts distributively during templated DNA synthesis on a single-stranded 5’ overhang. (It is conceivable that the persistence of some of the smaller products reflects incorporation of mismatched 3’-terminal dNMPs that impede further extension.) The time course of gap-filling synthesis was distinctive, insofar as there was a clear delay (compared with the simple primer-template) in progression through the duplex portion of the hairpin downstream of the gap. The polymerase paused prominently at the +5 position from 0.5 to 10 min, and also was delayed in progressing past +7 (Fig. 6B). At the high Pol2:DNA ratio used in this experiment, we did not observe at 30 s (the earliest time sampled) a prominent +3 extension product corresponding to gap fill-in with no extension into the flanking duplex. These properties of MsmPolD2 are consonant with a role in short fill-ins of 5’-overhang DSBs or the repair of short gaps in duplex DNAs.
Deletion of polD1 and polD2 from the M. smegmatis genome
The preceding biochemical characterization of MsmPolD2 (and MsmPolD1) highlighted their similarities to the LigD POL domain, which is proficient at nontemplated and templated addition and prefers to add ribonucleotides. Accordingly, we regarded PolD2 and PolD1 as potential agents of NHEJ in vivo, especially as candidate catalysts of the templated nucleotide additions that occur during 5’-overhang NHEJ in the M. smegmatis ligD-(D136A-D136A) strain that lacks the LigD polymerase activity (4,21). This notion was attractive in light of the fact that the polD1 gene of M. smegmatis is the distal ORF of a putative two-gene operon, of which the proximal ORF is ligC (Fig. 7C). LigC1 and LigC2 are paralogous ATP-dependent DNA ligases (20) encoded by adjacent M. smegmatis genes that are oppositely transcribed in head-to-head orientation. LigCs plays a back-up strand sealing role in mycobacterial NHEJ when the LigD ligase active site is mutated (20,21). (M. tuberculosis has a single LigC enzyme that is located adjacent to its polD1 gene in an oppositely transcribed head-to-head orientation.) The M. smegmatis polD2 gene is not part of an operon and is not located next to any gene known to be involved in DNA repair.
To query the role of PolD1 and PolD2 in vivo, we constructed M. smegmatis strains in which the polD1 and polD2 genes were deleted, singly and pairwise. The ΔpolD1 and ΔpolD2 mutations were introduced into wild-type M. smegmatis and also into a ligD-(D136A–D136A) strain. The ΔpolD2 allele was constructed by inserting the 5’ and 3’ flanking regions of polD2 next to the loxP-hygR-loxP cassette to create a polD2::loxP-hygR-loxP knockout cassette. A temperature-sensitive mycobacteriophage containing the knockout cassette was constructed and used to transduce M. smegmatis to hygromycin resistance. Successful chromosomal targeting of the polD2::loxP-hygR-loxP allele was confirmed by Southern blotting (Fig. 7) and the hygR marker was excised by transfection of a plasmid encoding Cre recombinase. A truncated ΔpolD1 was used to replace the polD1 gene by two-step allelic exchange using the galK/sacB counterselection method (24). Incorporation of the ΔpolD1 allele into wild type, ligD-(D136A–D138A) and ligD-(D136A–D138A) ΔpolD2 strains was confirmed by Southern blotting (Fig. 7). All of the desired knockouts were obtained and they grew as well as the wild-type parental strain in liquid culture and on agar medium, signifying that PolD1 and PolD2 are inessential, singly or together, for growth of M. smegmatis under laboratory conditions, even when the LigD polymerase is inactive. The strain genotypes are shown in Table S2.
Effect of ablating PolD1 and PolD2 on NHEJ
To test the participation of PolD1 and PolD2 in unfaithful NHEJ, we performed NHEJ assays using a linear plasmid substrate with an internal kanamycin-resistance marker and complementary 5’ overhang DSB ends generated by restriction endonuclease cleavage within a lacZ reporter gene. Transfection of the linear plasmids into M. smegmatis will result in the growth of kanamycin-resistant transformants only if the linear DNA is converted into a circular via NHEJ. Restitution of a functional lacZ gene by faithful joining of the original DSB ends will give rise to blue colonies on kanamycin/X-gal plates. White kanR colonies arise via unfaithful NHEJ. The efficiency and fidelity of NHEJ in wild-type M. smegmatis and in mutant strains ΔpolD1, ΔpolD2, ligD-(D136A–D138A) ΔpolD1, ligD-(D136A–D138A) ΔpolD2, and ligD-(D136A–D138A) ΔpolD1 ΔpolD2 were determined and are compiled in Table 1. As reported previously (21), inactivation of LigD POL via the D136A–D138A mutation caused an increase in NHEJ fidelity to 74% compared to 33% for wild-type cells. The fidelity of 5’-overhang NHEJ in the ΔpolD1 and ΔpolD2 single-deletion strains (36% for ΔpolD2 and 22% for ΔpolD1) was similar to that of wild-type M. smegmatis. The instructive findings were that combining the ligD-(D136A–D138A) allele with ΔpolD1, ΔpolD2 or both ΔpolD1 and ΔpolD2 elicited no further increases in fidelity of 5’-overhang NHEJ (70–72%) compared to the ligD-(D136A–D138A) single mutant (Table 1).
Table 1.
Probing the Role of PolD1 and PolD2 in NHEJ of 5’-Overhang DSBs In Vivo
| Strain | NHEJ Efficiency (%) |
NHEJ Fidelity (%) |
Junctions Sequenced |
Deletions | Templated Fill-ins |
|---|---|---|---|---|---|
| Wild-type | 100 | 33 | 22* | 8* | 21* |
|
ligD-(D136A- D138A) |
33 | 74 | 10 | 10 | 7 |
| ΔpolD2 | 153 | 36 | |||
| ΔpolD1 | 117 | 22 | |||
|
ligD-(D136A- D138A) ΔpolD1 |
21 | 72 | |||
|
ligD-(D136A- D138A) ΔpolD2 |
49 | 70 | |||
|
ligD-(D136A- D138A) ΔpolD1 ΔpolD2 |
12 | 71 | 15 | 13 | 11 |
The efficiencies of linear transformation with the Asp7181-cut reporter plasmid are expressed relative to wild-type (defined as 100%). Fidelity is calculated as [blue colonies/(blue+white colonies)] × 100. Each value for efficiency and fidelity is the mean of three independent plasmid transfections and is representative of 2–3 independent experiments. Using primers upstream and downstream of the repair junctions, the repair junctions for non-faithful repair events (white colonies) were amplified by colony PCR. The junctions were sequenced and the repair events were characterized. The number of events with terminal deletions and templated additions are indicated.
The junction sequence data from wild type M. smegmatis is reproduced from ref. 21.
To gauge whether the spectrum of mutagenic NHEJ outcomes is affected by the absence of PolD1 and PolD2, we determined the junction sequences of the repaired 5’ overhang reporter plasmids derived from independent lacZ– kanR transformants of the ligD-(D136A–D138A) single mutant, and the ligD-(D136A–D138A) ΔpolD1 ΔpolD2 triple mutant. In our prior work (21), the most prevalent outcome of 5’ overhang NHEJ in wild-type mycobacteria involved templated addition of nucleotides at the single-strand 5’ tails, which was observed in 21/22 of the junctions (Data reproduced in Table 1). The individual NHEJ outcomes from lacZ– kanR transformants of the ligD-(D136A–D138A) and ligD-(D136A–D138A) ΔpolD1 ΔpolD2 strains are shown in Fig. 8 and categorized in Table 1. Unfaithful NHEJ in both strains entailed a mixture of templated fill-ins and deletions. Of the 10 independent repair events examined in ligD-(D136A–D138A) cells, 7 involved templated fill-in of 4 nucleotides on one side of the break (yielding a blunt end) and a deletion of varying length on the other side of the break (Fig. 8). In the remaining three unfaithful events, both DSB ends suffered deletions prior to ligation (Fig. 8). Of the 15 unfaithful repair events analyzed in the ligD-(D136A–D138A) ΔpolD1 ΔpolD2 strain, 9 involved templated nucleotide additions at one of the 5’-overhang ends, while 2 events entailed templated fill-in at both of the original ends (Fig. 8). Ten of the fill-in outcomes were 4-nucleotide additions; three others were 3-nucleotide additions (Fig. 8). These results indicate that templated fill-in of 5’ overhang DSBs in the absence of the LigD polymerase activity is unaffected by loss of either PolD1 or PolD2.
Figure 8. Molecular outcomes of NHEJ in the presence or absence of freestanding PolD enzymes.
The original DSB sequence is pictured above panel A with the right side of the Asp718I site colored red and the left side colored blue. Templated nucleotide additions are colored green and the number of double stranded nucleotides deleted from one or both sides of the break are denoted with a Δ symbol.
Effect of ablating PolD1 and PolD2 on sensitivity to UV and ionizing radiation
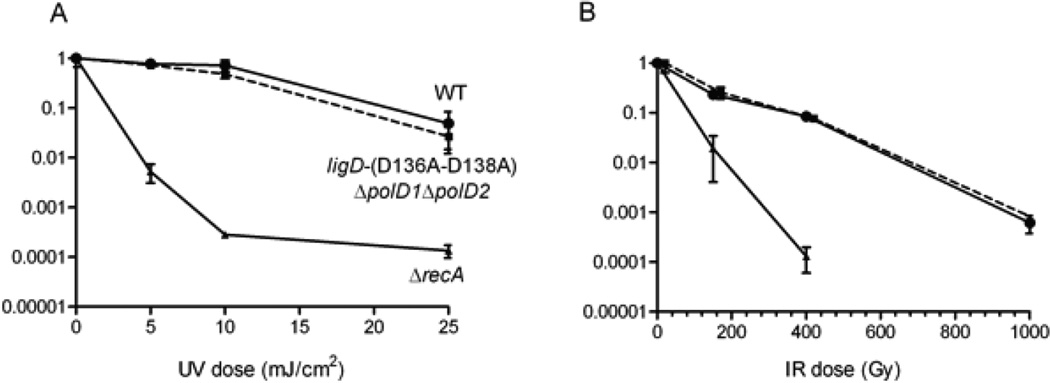
In light of the findings that PolD1 and PolD2 are not required for templated nucleotide additions during mycobacterial NHEJ, we asked whether these enzymes might participate in other repair pathways – by comparing the sensitivity of the ligD-(D136A–D138A) ΔpolD1ΔpolD2 and wild-type M. smegmatis strains to killing by ultraviolet (UV) and ionizing radiation (IR). A clastogen-sensitive M. smegmatis ΔrecA mutant was tested in parallel as a control. We found that there was no difference in the UV or IR sensitivity of the wild-type and ligD-(D136A–D138A) ΔpolD1ΔpolD2 strains (Fig. 9).
Figure 9. UV and IR sensitivity of a ligD-(D136A–D138A) ΔpolD1 ΔpolD2 strain.
(A) Fractional survival of M. smegmatis wild-type, ligD-(D136A–D138A) ΔpolD1 ΔpolD2 and ΔrecA strains is plotted on a log scale as a function of the dosage of exposure to UV (mJ/cm2) (panel A) or IR (Gy) (panel B).
DISCUSSION
Previous studies had shown that LigD POL is the direct and only catalyst of nontemplated single-nucleotide insertions at blunt DSB ends during plasmid NHEJ in vivo in M. smegmatis, but that templated fill-in of 5’-overhangs during NHEJ could be independent of LigD POL (21). Here we identified and characterized two stand-alone LigD POL-like enzymes encoded by M. smegmatis. MsmPolD1 and MsmPolD2 are structural homologs of LigD POL with DNA and RNA polymerase activities similar to those documented previously for native LigD and the isolated LigD POL domain. As with LigD POLs, the PolD1 and PolD2 polymerases display their optimal activity with a divalent cation cofactor other than magnesium. For example, Pseudomonas LigD POL favors manganese and cobalt and is less active with zinc (3), whereas Agrobacterium LigD POL prefers cobalt and manganese and, as seen here for MsmPolD2, synthesizes longer tracts of DNA on a primer-template in the presence of cobalt than with manganese (6). We find that MsmPolD2 is capable of both templated and nontemplated nucleotide addition and that it displays a strong preference for rNTPs versus dNTPs as substrates for nontemplated addition to a blunt DSB. MsmPolD1 has similar properties (data not shown). Yet, notwithstanding their biochemical similarity to LigD POL, and their attractiveness, in principle, as candidate back-up agents of templated nucleotide insertion during mutagenic NHEJ when LigD polymerase activity is disabled, we saw no failure of fill-in of 5’-overhang DSBs during NHEJ in vivo when PolD1 and PolD2 were deleted in the context of a polymerase-defective LigD protein. Thus, it is either the case that: (i) PolD1 and PolD2 do not participate in mycobacterial NHEJ or (ii) their role in mutagenic NHEJ is obscured by the existence of other mycobacterial DNA polymerases, of which there are several (26–29), that can add nucleotides to DSB primer-templates. In M. smegmatis, these include a homolog of Pol I (MSMEG_3839), two homologs of Pol III (MSMEG_3178 and MSMEG_1633), and three homologs of Pol IV (MSMEG_2294, MSMEG_6443, and MSMEG_3172).
Bacterial LigD is composed of three different enzymatic domains with known (LIG and POL) or imputed (PE) functions in DSB repair. Although the tri-functional LigD protein and its collaborator Ku are the signatures of the bacterial NHEJ apparatus found in several hundred bacterial taxa, the order of the domains within the LigD polypeptide varies. For example, mycobacterial LigD is arranged as POL–PE–LIG, whereas Pseudomonas and Agrobacterium LigD domains are ordered as PE–LIG–POL. This plasticity suggests that LigDs may have evolved by fusion of previously free-standing DNA repair genes. This idea is supported by studies here and elsewhere that document the coexistence in certain bacterial proteomes of a multifunctional LigD enzyme and free-standing homologs of one or more of the components domains. This was first evident in the identification and characterization of LigC enzymes as ATP-dependent DNA ligases, with properties similar to the LIG domain of LigD, in M. tuberculosis, M. smegmatis, and Agrobacterium tumefaciens (6,20,25). Stand-alone homologs of the PE domain of LigD are found in a variety of bacterial and archaeal taxa (16), including mycobacterial species that have LigD (e.g., M. marinum, M. ulcerans, M. kansasii). Our demonstration that two stand-alone POL enzymes co-exist with LigD and LigC in mycobacteria completes the circle. It is likely that the paralogs of POL, LIG and PE arose via duplications of ancestral catalytic modules. We assume that they play some role in bacterial genome integrity, perhaps redundantly to other repair enzymes (as is the case for LigC) or under circumstances that are not interrogated by available repair assays or laboratory growth conditions.
Supplementary Material
Acknowledgements
This research was supported by grant AI64693 from the U.S. National Institutes of Health. SS is an American Cancer Society Research Professor.
Footnotes
SUPPLEMENTARY DATA
Supporting Information available includes Supplementary Figures 1–3 and Supplementary Tables 1 and 2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nature Rev. Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 3.Zhu H, Shuman S. A primer-dependent polymerase function of Pseudomonas aeruginosa ATP-dependent DNA ligase (LigD) J. Biol. Chem. 2005;280:418–427. doi: 10.1074/jbc.M410110200. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Nandakumar J, Aniukwu J, Wang LK, Glickman MS, Lima CD, Shuman S. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc. Natl. Acad. Sci. USA. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yakovleva L, Shuman S. Nucleotide misincorporation, 3’-mismatch extension, and responses to abasic sites and DNA adducts by the polymerase component of bacterial DNA ligase D. J. Biol. Chem. 2006;281:25026–25040. doi: 10.1074/jbc.M603302200. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H, Shuman S. Characterization of Agrobacterium tumefaciens DNA ligases C and D. Nucleic Acids Res. 2007;35:3631–3645. doi: 10.1093/nar/gkm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu H, Shuman S. Gap filling activities of Pseudomonas LigD polymerase and functional interactions of LigD with the DNA end-binding Ku protein. J. Biol. Chem. 2010;285:4815–4825. doi: 10.1074/jbc.M109.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 10.Brissett NC, Martin MJ, Pitcher RS, Bianchi J, Juarez R, Green AJ, Fox GC, Blanco L, Doherty AJ. Structure of a preternary complex involving a prokaryotic NHEJ DNA polymerase. Mol. Cell. 2011;41:221–231. doi: 10.1016/j.molcel.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 11.Zhu H, Shuman S. Novel 3’-ribonuclease and 3’-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J. Biol. Chem. 2005;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Wang LK, Shuman S. Essential constituents of the 3’-phosphoesterase domain of bacterial DNA ligase D, a nonhomologous end-joining enzyme. J. Biol. Chem. 2005;280:33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Shuman S. Substrate specificity and structure-function analysis of the 3’-phosphoesterase component of the bacterial NHEJ protein, DNA Ligase D. J. Biol. Chem. 2006;281:13873–13881. doi: 10.1074/jbc.M600055200. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3’-OH monoribonucleotide. J. Biol. Chem. 2008;283:8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akey D, Martins A, Aniukwu J, Glickman MS, Shuman S, Berger JM. Crystal structure and nonhomologous end joining function of the ligase domain of Mycobacterium DNA ligase D. J. Biol. Chem. 2006;281:13412–13423. doi: 10.1074/jbc.M513550200. [DOI] [PubMed] [Google Scholar]
- 16.Nair PA, Smith P, Shuman S. Structure of bacterial LigD 3’-phosphoesterase unveils a DNA repair superfamily. Proc. Natl. Acad. Sci. USA. 2010;107:12822–12827. doi: 10.1073/pnas.1005830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith P, Nair PA, Das U, Zhu H, Shuman S. Structures and activities of archaeal members of the LigD 3’-phosphoesterase DNA repair enzyme superfamily. Nucleic Acids Res. 2011;39:3310–3320. doi: 10.1093/nar/gkq1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das U, Smith P, Shuman S. Structural insights to the metal specificity of an archaeal member of the LigD 3’-phosphoesterase DNA repair enzyme family. Nucleic Acids Res. 2012;40:828–836. doi: 10.1093/nar/gkr767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natarajan A, Dutta K, Temel DB, Nair PA, Shuman S, Ghose R. Solution structure and DNA-binding properties of the phosphoesterase domain of DNA ligase D. Nucleic Acids Res. 2012;40:2076–2088. doi: 10.1093/nar/gkr950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of non-homologous end joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nature Struct. Mol. Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 21.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta R, Barkan D, Redelman-Sidi G, Shuman S, Glickman MS. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 2011;79:316–330. doi: 10.1111/j.1365-2958.2010.07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkan D, Hedhli D, Yan HG, Huygen K, Glickman MS. Mycobacterium tuberculosis lacking all mycolic acid cyclopropanation is viable but highly attenuated and hyperinflammatory in mice. Infect. Immun. 2012;80:1958–68. doi: 10.1128/IAI.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkan D, Stallings CL, Glickman MS. An improved counterselectable marker system for mycobacterial recombination using galK and 2-deoxy-galactose. Gene. 2011;470:31–36. doi: 10.1016/j.gene.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 26.Mizrahi V, Huberts P. Deoxy- and dideoxynnucleotide discrimination and identification of critical 5’ nuclease domain residues of the DNA polymerase I from Mycobacterium tuberculosis. Nucleic Acids Res. 1996;24:4845–4852. doi: 10.1093/nar/24.24.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boshoff HIM, Reed MB, Bary CE, Mizrahi V. DNAE2 polymerase contribute to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 28.Kana BD, Abrahams GL, Sung N, Warner DF, Gordhan BG, Machowski EE, Tsenova L, Sacchettini JC, Stoker NG, Kaplan G, Mizrahi V. Role of the DinB homologsRv1537 and Rv3056 in Mycobacterium tuberculosis. J. Bacteriol. 2010;192:2220–2227. doi: 10.1128/JB.01135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma A, Nair DT. MsDpo4 – a DinB homolog from Mycobacterium smegmatis – is an error-prone DNA polymerase that can promote G:T and T:G mismatches. J. Nucleic Acids. 2012;2012:285481. doi: 10.1155/2012/285481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.