Abstract
The rigorous testing of hypotheses on suitable sample cohorts is a major limitation in translational research. This is particularly the case for the validation of protein biomarkers where the lack of accurate, reproducible and sensitive assays for most proteins has precluded the systematic assessment of hundreds of potential marker proteins described in the literature.
Here, we describe a high throughput method for the development and refinement of selected reaction monitoring (SRM) assays for human proteins. The method was applied to generate such assays for more than 1000 cancer-associated proteins, which are functionally related to candidate cancer driver mutations. We used the assays to determine the detectability of the target proteins in two clinically relevant samples, plasma and urine. 182 proteins were detected in depleted plasma, spanning five orders of magnitude in abundance and reaching below a concentration of 10 ng/mL. The narrower concentration range of proteins in urine allowed the detection of 408 proteins. Moreover, we demonstrate that these SRM assays allow the reproducible quantification of 34 biomarker candidates across 84 patient plasma samples. Through public access to the entire assay library, which will also be expandable in the future, researchers will be able to target their cancer-associated proteins of interest in any sample type using the detectability information in plasma and urine as a guide. The generated reference map of SRM assays for cancer-associated proteins is a valuable resource for accelerating and planning biomarker verification studies.
Introduction
The identification of validated disease biomarkers for diverse clinical needs such as prognosis, diagnosis, and patient stratification to follow and guide therapies is a predominant line of inquiry in translational research (1, 2). To establish the value of a protein biomarker it is imperative to reliably identify and reproducibly quantify the protein of interest over multiple samples (3). Due to advances in proteomic and genomic technologies, long lists of biomarker candidates have been generated that contain proteins hypothesized to change in abundance relative to specific diseases or disease states. However, subsequent hypothesis testing in large cohorts of patient specimens needs to be performed in order to verify the clinical utility of such biomarker candidates (4). The preferred specimens for biomarker testing are easily sampled body fluids like plasma and urine (2, 4-6). However, the highly complex proteomes of plasma and urine pose technical challenges for analysis (7, 8). This applies particularly to the verification of biomarker candidates, a process that requires the accurate, sensitive, and reproducible quantification of multiple proteins in complex backgrounds over large cohorts of patients’ specimens (9).
Traditionally, such hypothesis testing has been accomplished using affinity-based assays, such as enzyme-linked immunosorbent assays (ELISA). Major constraints of this approach are the limited availability of validated ELISA for the majority of human proteins, the expensive and time-consuming development of de novo assays and the difficulty of assay multiplexing (10). These limitations preclude the timely verification of the rapidly increasing number of candidate proteins that are derived from high-throughput proteomic and genomic screens (11). Additionally, computational approaches are emerging that use data integration and network inference to generate sets of biomarker candidates (12-17) that further increase the number of candidate proteins that need to be experimentally verified. Therefore, it is anticipated that the rate of hypothesis generation for biomarker research will further increase and consequently, a well-matched analytical platform that allows rapid and high-throughput hypothesis testing is needed.
Targeted mass spectrometry (MS) via SRM has emerged as an alternative to affinity-based measurements of defined protein sets (5, 10, 18-22). The main advantage of SRM is the capacity for faster and cost-efficient assay development (23). SRM has also the feature of being able to quantify multiple proteins in parallel (multiplexing) at a low limit of detection and high accuracy. Additionally, it has been shown that protein quantification by SRM in complex samples using predefined assay coordinates is reproducible across different laboratories and instrument platforms (24). Consequently, SRM-based hypothesis testing is ideally matched with high-throughput hypothesis generation and has the potential to bridge the gap between generating lists of candidates and evaluating their clinical utility (9).
There are two major challenges facing the implementation of SRM in a biomarker verification pipeline. The first is the generation of high quality SRM assays for sets of biomarker candidates, which can include several hundred proteins. Picotti et al. developed a high-throughput method for SRM assay development that is based on the use of crude synthetic peptide libraries as a reference for validating SRM assays (23). Once such an assay has been developed it becomes universally applicable. Therefore, publicly accessible repositories have been generated that contain SRM assays (25). The second challenge is the reliable identification and quantification of several hundred proteins in the sample of interest, usually complex body fluids (10, 20). Recently, two novel data analysis tools, mProphet (26) and SRMstats (27) have been developed to assist in the identification and quantification of peptides and proteins measured by SRM. mProphet is an automated tool that allows the objective and reliable association of groups of SRM signals with their corresponding peptide sequences in complex samples, with and without isotope-labeled internal standards (26). SRMstats is a statistical modeling framework for protein significance analysis based on linear mixed effects models specifically adapted for the SRM data structure (27). Therefore, major bottlenecks impeding the wide use of SRM technology have recently been alleviated.
In this study, we generated a resource of SRM assays for more than 1000 proposed protein biomarker candidates that have been previously associated with cancer. Using a protein functional network, we demonstrate that these proteins are enriched among the interaction partners of genes mutated in cancer. The SRM assays developed via high-throughput peptide synthesis and MS were subsequently applied to determine the detectability of the targeted peptides in clinically relevant samples. Furthermore, we demonstrated the applicability of the assays to reproducibly and accurately quantify biomarker candidates across a large number of patient specimens. This resource of definitive SRM assays for cancer-associated proteins enables and accelerates clinical SRM-based biomarker verification studies.
Results
A comprehensive list of cancer-associated proteins
Our goal was to select a comprehensive list of proteins that have been previously implicated in cancer. Polanski and Anderson compiled an evidence-based list of 1261 proteins, which are differentially expressed in cancer (cancer-associated proteins, CAPs) (28). To compile this list, the authors found reported abundance changes observed at the protein level in human plasma or tissue. They also accepted reported cases with changes at the DNA (ploidy changes) or RNA level in affected tissue. Furthermore, the selection was derived from various sample types and technology platforms. Of the 1261 proteins, we selected 1130, which could be unambiguously associated with UniProt identifiers (http://www.uniprot.org/). We added FDA-approved protein markers if they were not already included in the target list of CAPs (8). In total, the list of CAPs selected for SRM assay development consisted of 1172 proteins (Table S1).
Relation of cancer-associated proteins to candidate cancer driver mutations
It has been demonstrated that cancer origin and progression is driven not by single gene mutation or expression changes, but by coordinated changes in variable subsets of genes (29). Although diverse genomic alterations are observed in different individuals with tumors of the same clinical type, sets of mutated genes can function in the same signaling pathway or sub-network leading to similar or identical phenotypes (12). Proteins that are functionally related to the mutated genes in a sub-network or pathway can potentially be used as a biomarker for the state of pathways or sub-networks perturbed by gene mutations. Therefore, we investigated the functional relationship of CAPs to candidate cancer driver mutations (CDMs), which have been discovered by unbiased whole exome sequencing of multiple human cancers. We compiled a list of 379 CDMs from the whole exome sequencing data of 7 different cancer types (30-35). Their sub-network in the Reactome Functional Protein Interaction Network (RFIN) (36) was examined for the presence of CAPs. Further, we investigated the enrichment of CAPs among the functional interaction partners of CDMs. 43 out of the 1172 CAPs have also been discovered as CDMs by unbiased whole exome sequencing (Table 1). Including the functional interaction partners of CDMs we obtained 608 CAPs that have a direct functional relationship to CDMs (Table 1). We determined that the CAPs are significantly enriched in the sub-network of CDMs compared to random networks of the same size and degree distribution (p-value 4.3e−11) (Table 1) (37). These results demonstrate that the selected target proteins are not only interesting as potential cancer biomarkers but are also relevant for studying perturbed protein networks by genetic mutations that drive cancer development.
Table 1.
Intersection of the candidate cancer driver mutations (CDMs) and their interaction partners in the Reactome Functional Interaction Network with the cancer-associated proteins (CAPs). It is assumed that a CDM can be monitored by measuring the protein encoded by the CDM or in the sub-network of CDMs.
| Cancer associated proteins (CAPs) | Overlap of CDMs with CAPs | # of CAPs in the sub-network of CDMs | p-value (Enrichment of CAPs in the sub-network of CDMs) | # of CDMs that can be monitored by CAPs (out of 379) | |
|---|---|---|---|---|---|
| All | 1172 | 43 | 608 | 4.3e−11 | 175 |
| Detectable (plasma and urine combined) | 473 | 18 | 232 | 5.9e−5 | 143 |
Selection of representative peptides for cancer-associated proteins
The selection of peptides that uniquely represent the target proteins in the proteome (proteotypic peptides, PTPs) is an important step in the development of SRM assays (38, 39). For each protein, we aimed for five tryptic peptides with favorable MS properties and unique occurrence within the human proteome. We primarily used empirical evidence of MS detectability for PTP selection. Therefore, we prioritized peptides for each target protein that have been previously detected in large-scale shotgun MS data collections, namely the Human Peptide Atlas (PA), Human Plasma PA (40, 41) and an extensive dataset produced by in-depth fractionation and MS sequencing of human cell lysates (42). If no or insufficient empirical evidence was available, we computationally predicted PTPs using criteria that have been shown to favor detection by MS (39, 43). In total 5426 peptides were selected to represent the 1172 CAPs (Table S2). 2948 peptides (54%) have been previously observed in shotgun proteomic datasets and 2478 peptides (46%) were predicted (Fig. S1). The majority of the protein targets, numbering 1002, were covered by 5 peptides. 93% of the selected peptides were unique to their respective protein, indicating a high likelihood of developing a specific assay for the majority of proteins.
Development of SRM assays for cancer-associated proteins
We developed SRM assays intended for the identification and quantification of CAPs in various sample types and protein backgrounds. Such assays consist of the mass, charge state distribution and chromatographic retention time of the precursor ion and the mass, charge state distribution and relative intensity of the fragment ion signals (25). To establish the assays we used a multi-step process. The first step was the acquisition of full fragment ion spectra for the target peptides using chemically synthesized peptide libraries as described by Picotti et al. (23) (Fig. 1A). Overall, 6787 fragment spectra were confidently assigned to 4821 out of the 5426 synthesized peptides (89%) and used to extract the SRM assay coordinates. In the second step, SRM assay coordinates were refined by determining the precise relative fragment ion signal intensities and the indexed retention time (iRT) (44) in SRM acquisition mode (Fig. 1B), thereby providing important information for scheduling SRM measurements as well as for scoring the SRM data (26). Finally, SRM assays showing a low quality in SRM mode were eliminated by manual inspection. The overall success rate of assay generation at the peptide level was 74% (3996 peptides) and 99% (1157 proteins) at the protein level. For the majority of the CAPs (80%), refined assays are available to target at least three peptides per protein (Fig. 2). Each refined SRM assay was defined by the iRT of the peptide and the relative intensity ratios of its 5 most intense transitions (Table S3). Thus, the generated SRM assays constitute a high quality map for CAPs that can be applied to any biological sample of interest.
Fig. 1.
Workflow outlining SRM assay generation, refinement and application to detect target proteins in plasma and urine. In the first step, a crude synthetic peptide library was used to generate QQQ full fragment ion spectra for the extraction of the preliminary coordinates for SRM assays (A). In the second step, SRM assays were refined by measuring the crude synthetic peptides in SRM mode using the coordinates established in full scan mode (B). This step refined the relative transition intensities specific for the SRM acquisition mode and the iRT in the chromatographic gradient to be used for endogenous peptide detection. The final SRM assay library was then used to detect the CAPs in complex samples (depleted plasma and urine) (C). Decoy transition groups and positive controls were included in the SRM measurements to allow for objective data analysis using the mProphet software tool (26).
Fig. 2.
Number of peptides per protein in the SRM assay library for CAPs.
Detection of cancer-associated proteins in depleted plasma and urine using SRM
To generate a resource for accelerating the verification of CAPs as potential biomarkers, we determined the detectability of the target proteins in widely used clinical samples using the SRM coordinates determined above. We applied the generated SRM assays to human urine and depleted plasma using label-free SRM, i.e. without addition of internal standards (Fig. 1C and Supplementary Result 1). Such measurements in complex samples record numerous interfering transition signals and it is challenging to distinguish between true and false assignments by manual inspection of the data (45, 46). To minimize the number of false positive peptide identifications we evaluated the resulting data using the mProphet software tool, which was modified compared to the original publication to account for the iRT deviation as an additional score in the combined scoring function (26, 44).
Plasma Results
In the depleted plasma sample 302 peptides corresponding to 182 proteins were detected with an FDR of 2% and a sensitivity of 70% (Fig. S2A, B and Table S4). We investigated the concentration range of the detected proteins in depleted plasma using estimated concentrations based on spectral counts derived from Human Plasma PA (41) (Table S1). The detected proteins span five orders of magnitude in plasma reaching a limit of detection (LOD) below 10 ng/mL (Fig. 3A). The distribution of detected CAPs over the concentration range confirms the limited detectability by mass spectrometry for low-abundance proteins in plasma. However, using SRM, we made 83 novel protein observations among the 182 detectable CAPs in plasma in comparison to the 187 CAPs previously observed in the Human Plasma PA by data dependent analysis in crude or depleted plasma (Fig. 3B). In contrast, 88 out of 187 CAPs listed in the Human Plasma PA were not detected using SRM, of which 19 have an estimated concentration above 100 ng/mL. To study the effect of depletion on the detectability of CAPs in plasma, we targeted the SRM assays in a crude plasma digest. Only 73 proteins were detected in the crude digest and, as expected, these proteins were mostly high-abundance plasma proteins (Fig. S3 and Table S5). The abundance of the 73 proteins ranged from 1.6 mg/mL to 35 ng/mL (Fig. S3). These results demonstrate the increased sensitivity of detecting low abundance proteins in plasma using a simple sample preparation such as depletion of the high abundance proteins. However, for detecting a higher number of proteins in the low ng/mL concentration range additional sample preparation steps are needed to decrease sample complexity. The SRM assays developed in this study will be equally applicable to fractionated or enriched plasma samples.
Fig. 3.
Detectability results for depleted plasma and urine. Estimated protein concentrations for the CAPs in plasma were extracted from Human Plasma PA (41). The plotted concentration range shows detected CAPs (blue) and proteins that could not be detected (grey) in depleted plasma (A). Proteins detected by SRM were compared to proteins previously observed by large-scale proteomic experiments derived from Human Plasma PA (including measurements in crude and depleted plasma without additional fractionation) (B). Estimated protein concentrations for the CAPs in urine were extracted from Urine PA (41). The plotted concentration range shows detected CAPs (blue) and proteins that could not be detected (grey) in urine (C). Proteins detected by SRM were compared to proteins previously observed by large-scale proteomic experiments derived from Urine PA combined with protein observations from Adachi et al. (49) (D).
To further investigate the functional characteristics of the detectable CAPs in depleted plasma compared to all targeted CAPs we conducted an enrichment analysis using the functional annotation tool DAVID (http://david.abcc.ncifcrf.gov/) (47, 48). The CAPs detectable in depleted plasma are enriched for extracellular region proteins (117 proteins found; p-value 4.7e−11). They are mainly involved in acute inflammatory response (29; 3.1e−10), complement activation (16; 7.1e−8) and response to wounding (59; 7.5e−7). The majority of the detected CAPs are either annotated in UniProt (www.uniprot.org) as plasma proteins (69; 8.7e−34) or proteins highly expressed in the liver (86; 9.1e−13) and are thus among high abundance proteins in plasma.
Urine Results
In urine we detected 661 peptides corresponding to 408 proteins with an FDR of 3% and a sensitivity of 70% calculated by mProphet on the level of the SRM assay (Fig. S2C, D and Table S6). Different FDR cutoffs were chosen to report detectable proteins for depleted plasma and urine based on a consistent sensitivity for both sample types. We also investigated the detected concentration range in urine by extracting estimated concentrations from the Urine PA (Table S1). The narrower concentration range of proteins in urine allowed us to detect a larger number of CAPs in the low ng/mL range (Fig. 3C). For many of the CAPs detected in urine no estimated concentrations are available (Fig. S3). Nevertheless, we expect that their concentration is in the sub-ng/mL range, which would translate into a dynamic concentration range of detected proteins similar to that of plasma. It is also expected that the distribution of detected proteins thins out towards lower pg/mL concentrations, reflecting the trend observed in plasma. Compared to previously published, large-scale proteomic datasets acquired for urine, derived from the human Urine PA and the dataset of Adachi et al. (49), we detected 169 previously undetected proteins using our SRM assays (Fig. 3D). Similar to depleted plasma the detectable CAPs in urine are enriched for extracellular region proteins (189 proteins found; p-value 5.4e−5). However, in comparison to depleted plasma a larger number of plasma membrane (137 versus 65 proteins) and cytosolic proteins (67 versus 16) could be detected in urine. These proteins could be derived from cells that are shed into urine. Additionally, it has been suggested that plasma membrane proteins are excreted in urine through exosome formation (49). Furthermore, the narrower concentration range of proteins in urine allows the detection of proteins that are usually masked in plasma by the highly abundant classical plasma proteins.
Characteristics of detected cancer-associated proteins in body fluids
The final list of 473 CAPs detected in urine and depleted plasma demonstrates the power of SRM in targeting proteins over a large dynamic range in minimally processed complex body fluids. Next, for all detectable peptides we determined the theoretical specificity of the SRM assays. This analysis was based on the uniqueness of the transition mass-to-charge ratios (m/z) that define the assay in a background of all human peptides identified in the Human PA, assuming that this represents the MS-detectable fraction of the human proteome. We used SRMCollider (50), a tool based on the unique ion signature (UIS) approach described by Sherman et al. (51). The analysis revealed high theoretical assay specificity for the detected peptides in urine and plasma in the background of the Human PA. 94% of the peptides were monitored by a unique combination of transitions (Supplementary Result 2 and Fig. S4).
Furthermore, we investigated how many of the detectable CAPs in urine and plasma are functionally related to CDMs. Out of the 43 CDMs that are also reported as CAPs, 19 can be directly monitored in at least one of the body fluids (Table 1). 232 detectable CAPs represent functional interaction partners of the CDMs (Table 1). Assuming that the status of the candidate cancer driver can be deduced either by the direct measurement of the protein encoded in the CDM or by a functionally related protein, the detectable CAPs allow monitoring the status of 143 CDMs in either urine or plasma (Table 1). For all the source cancer types which we used to derive the CDMs we generated individual sub-networks consisting of the CDMs identified in the respective cancer types and the functionally interacting CAPs (Fig. S5-11), thereby demonstrating that they are highly interconnected. Fig. 4 shows the sub-network for pancreatic cancer, highlighting the detectable CAPs in urine and plasma. 30 of the 39 CDMs (77%) that were discovered for pancreatic cancer by unbiased whole exome sequencing are connected to detectable CAPs. While well-studied CDMs such as TP53, KRAS and SMAD4 have many connected CAPs which are also detectable in one of the body fluids, other CDMs are poorly connected in the sub-network or not even part of the sub-network. We propose that the sub-network of these CDMs should be studied as new potential cancer protein biomarkers.
Fig. 4.
Functional interaction network for pancreatic cancer. The diagram depicts functional interactions of the identified candidate cancer driver mutations for pancreatic cancer and the detectable cancer-associated proteins. Nodes represent CAPs (circles), CDMs (squares) and CDMs that are also reported cancer-associated on protein level (triangles). Colors denote the detectability of the proteins in plasma or urine: blue – detectable; pink – not detectable; grey – not targeted in plasma or urine. Functional interactions between the proteins are marked as edges. The figure was generated using Cytoscape (68).
Monitoring biomarkers by SRM in a large patient cohort
Finally, we aimed to demonstrate the capacity of the SRM assays developed in this study to monitor known biomarkers in a larger cohort of patient specimens. In our initial list of CAPs we included proteins for which FDA approved assays exist, for example those in the OVA1 biomarker panel. This panel assesses the ovarian cancer (OC) risk in women diagnosed with an ovarian tumor prior to planned surgery (52, 53). The OVA1 panel analyses five protein biomarkers, cancer antigen 125 (CA125), beta-2-microglobulin (B2MG), apolipoprotein A1 (APOA1), transthyretin (TTHR) and transferrin (TRFE) using antibody-based assays, and combines the results of each test to classify patients into high- or low-risk for ovarian malignancy. In the detectability test we demonstrated that B2MG, APOA1, TTHR and TRFE are accessible by SRM in an unfractionated tryptic digest of the plasma proteome. We chose a cohort of plasma samples derived from OC patients (n=68) and patients with benign ovarian tumors (BOT) (n=16) to confirm that the SRM assays quantify the proteins reproducibly across the patient plasma samples and detect abundance differences between the two patient groups. Plasma samples derived from healthy individuals were not included, since the OVA1 panel specifically detects OC in women already diagnosed with a pelvic mass. To fully explore the capacity of SRM to multiplex protein measurements, we added 30 target proteins to the OVA1 panel. These proteins were selected because they were either proposed as biomarker candidates for OC before or because we predicted them to be functionally related to mutated or epigenetically silenced genes in OC (54) (Table 2). In total we monitored 34 proteins (62 peptides) across plasma derived from 68 OC patients and 16 patients with BOT. mProphet (26) identified the peptides and proteins that were confidently detected across the samples and SRMstats (27) was used for protein significance analysis.
Table 2.
Quantification of selected proteins measured by SRM in plasma of ovarian cancer (OC) patients and patients with benign ovarian tumors (BOT). Statistical analysis was performed using a linear mixed effect model implemented in SRMstats (27). The proteins were selected because they are either part of the OVA1 biomarker panel (OVA1), have been proposed as biomarker candidates for ovarian cancer (literature) or because they functionally interact with epigenetically silenced or mutated genes in ovarian cancer (network).
| Protein | Gene symbol | Fold-change | P-value | Significant | Source (regulation) | Publication |
|---|---|---|---|---|---|---|
| AACT | SERPINA3 | 1.54 | 7.18E-10 | yes | literature (up), network | (73-75) |
| APOA1 | APOA1 | 0.86 | 0.0016 | yes | OVA1 (down) | (53, 55, 56) |
| APOE | APOE | 0.83 | 1.39E-04 | yes | literature (up) | (75) |
| B2MG | B2M | 1.19 | 2.51E-04 | yes | OVA1 (up) | (53, 55, 56) |
| C1R | C1R | 1.16 | 1.19E-05 | yes | network | |
| CFAB | CFB | 1.29 | 8.22E-06 | yes | network | |
| CO5 | C5 | 1.19 | 1.27E-05 | yes | network | |
| CO6 | C6 | 1.11 | 0.0026 | yes | network | |
| CO7 | C7 | 1.22 | 3.55E-04 | yes | network | |
| GELS | GSN | 0.77 | 1.39E-04 | yes | network | |
| HPT | HP | 1.67 | 0 | yes | literature (up), network | (56, 75) |
| IC1 | SERPING1 | 1.18 | 0.0050 | yes | literature (up), network | (73) |
| ITIH4 | ITIH4 | 1.18 | 1.77E-05 | yes | literature (up) | (55, 73, 74) |
| RET4 | RBP4 | 0.76 | 4.11E-07 | yes | network | |
| SHBG | SHBG | 1.47 | 6.96E-11 | yes | network | |
| TETN | CLEC3B | 0.78 | 3.02E-08 | yes | literature (down) | (76) |
| THBG | SERPINA7 | 1.20 | 1.24E-04 | yes | network | |
| TRFE | TF | 0.84 | 2.79E-08 | yes | OVA1 (down) | (53, 55, 56) |
| TTHY | TTR | 0.70 | 2.73E-08 | yes | OVA1 (down) | (53, 55, 56) |
| A2MG | A2M | 0.89 | 0.0605 | no | network | |
| APOA | LPA | 1.33 | 0.0605 | no | literature (up) | (73) |
| APOB | APOB | 1.04 | 0.4771 | no | literature (down) | (77) |
| C1QA | C1QA | 0.98 | 0.6961 | no | network | |
| C1QB | C1QB | 1.08 | 0.0362 | no | literature (up), network | (73) |
| C1QC | C1QC | 0.97 | 0.6555 | no | network | |
| CADH5 | CDH5 | 1.01 | 0.8670 | no | network | |
| CLUS | CLU | 0.95 | 0.0626 | no | literature (up) | (74, 78, 79) |
| CO3 | C3 | 1.09 | 0.0163 | no | network | |
| CO4A | C4A | 1.05 | 0.4255 | no | network | |
| FN1 | FN1 | 0.82 | 0.0181 | no | network | |
| IGF2 | IGF2 | 0.97 | 0.5384 | no | network | |
| PLMN | PLG | 1.00 | 0.9690 | no | network | |
| THRB | F2 | 1.00 | 0.9598 | no | network | |
| VTNC | VTN | 1.03 | 0.4771 | no | literature (up), network | (80) |
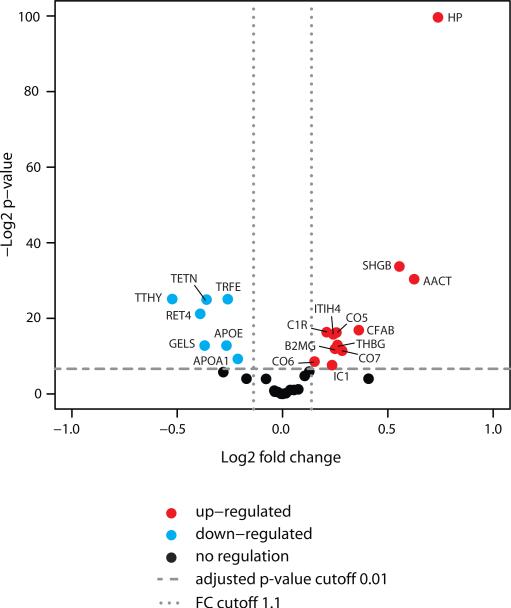
Of the consistently quantified proteins, 19 showed a significant fold-change comparing plasma samples from OC patients and patients with BOT (Fig. 5 and Table 2). The protein significance analysis confirmed a significant abundance difference for the proteins of the FDA-approved OVA1 panel, APOA1, TRFE, B2MG and TTHR for the two patient groups. Furthermore, the direction of abundance change for these proteins was consistent with the results described previously that were generated by immunoassays (55, 56). Of the other 15 proteins with significant abundance change, 3 have been previously suggested as biomarker candidates for OC in literature, 9 proteins were derived from the network analysis of mutated and epigenetically silenced genes in OC and 3 proteins were selected based on both literature and network analysis (Table 2). These results demonstrate that SRM allows the reproducible and accurate quantification of proteins across larger patient cohorts and confirm the capacity of SRM to complement and extend antibody-based assays for expedient verification of biomarker candidates. Furthermore, the significant abundance difference of the proteins predicted by network analysis suggests that this approach could be explored for the discovery of novel biomarkers.
Fig. 5.
Quantification of selected proteins in plasma of ovarian cancer patients and patients with benign ovarian tumors. All proteins with a p-value below 0.01 and a fold-change larger than 1.1 were considered significant.
Discussion
Over the last several years, long lists of proteins have been proposed as potential biomarkers for various cancer types without further evaluation of their clinical utility. The lack of follow-up is due, by large extent, to the lack of a technology for the expedient, reproducible and accurate verification of the proteins as biomarkers. Recent developments in SRM-based targeted proteomics show promise for accelerating the hypothesis testing of multiple biomarker candidates in large cohorts of patient specimens. The aim of this study was the generation of a resource of validated SRM assays for the detection and quantification of cancer associated proteins to assist and accelerate the verification of cancer biomarker candidates in clinical specimens.
We developed definitive SRM assays for 1157 proteins, which have been previously reported to change abundance in various human cancers and which were found to be functionally linked with genetic mutations driving cancer development. Out of the 1157 CAPs for which we generated assays, we detected 182 proteins in depleted plasma and 408 in urine using a label-free SRM strategy. The datasets have been submitted to the PeptideAtlas SRM Experiment Library (PASSEL, http://www.peptideatlas.org/passel/) (57), which allows researchers to extract the SRM assay coordinates, and importantly, to check detectability information for proteins of interest (http://www.peptideatlas.org/PASS/PASS00004 for depleted plasma, PASS00006 for crude plasma, PASS00007 for urine and PASS00041 for the quantification of proteins in plasma of OC patients and patients with BOT). We demonstrated the use of this library to accurately and reproducibly quantify 34 biomarker candidates across a larger cohort of patient plasma samples. The quantified proteins included APOA1, TRFE, B2MG and TTHY which are all part of the FDA-approved OVA1 biomarker panel and for which expected abundance changes were confirmed comparing OC patients and patients with BOT. Furthermore, a subset of the proteins tested that were not included in the OVA1 panel also achieved highly significant separation of OC and BOT plasma samples, thus raising the possibility that the performance of the OVA1 test could be further improved by the inclusion of additional proteins.
[Note to reviewers: Datasets are not publicly viewable until acceptance of the manuscript; reviewers may access all the data for review prior to release by going to http://www.peptideatlas.org/passel/, entering the reviewer password EQ7277p in the entry box in the middle of the web page and opening the “Browse SRM experiments” window. The four datasets are listed as “Human CAP depleted plasma”, “Human CAP crude plasma”, “Human CAP urine” and “Human CAP ovarian cancer plasma”.]
The SRM assays for the CAPs have been generated in a multistep process, in which the SRM coordinates were first extracted from fragment spectra of synthetic peptides and subsequently refined by measuring the synthetic peptides in SRM acquisition mode. While sample-specific interferences may compromise some transitions, we demonstrated the quality of the refined assay library by applying it to two of the most complex proteomes commonly analyzed. Moreover, we calculated a high theoretical specificity for the detected targets in plasma and urine by simulating the instances of interfering transitions assuming a complex proteomic background. This shows that the SRM assays enable researchers to directly target and consistently detect CAPs in any sample type and to thus test their potential as biomarkers. However, the resource does not provide LODs and limits of quantification (LOQs) for the SRM assays. These properties cannot be defined generally because they are dependent on the sample preparation and the instrument platform. They should be determined locally and preferentially using isotope-labeled internal standards before the verification of the proteins in a large cohort of clinical samples.
The initial list of CAPs was assembled from studies of different sample types and various technologies applied at the protein and nucleic acid level (28). Only a subset of the proteins has been observed by MS and an even smaller subset has been detected previously in plasma and urine, usually with low reproducibility in extensively fractionated samples. Therefore, it was anticipated that many of the proteins would not be detectable in plasma or urine by SRM. Nevertheless, in comparison to data extracted from the large-scale shotgun MS datasets in Human Plasma PA, Urine PA and the urine study of Adachi et al. (49) we obtained a high number of novel observations using our targeted proteomics approach, 83 and 169 CAPs for plasma and urine, respectively. However, 88 and 103 CAPs previously detected by shotgun MS in plasma and urine, respectively, were not detected with our method, likely due to the use of alternative sample preparation strategies, different types of MS instruments, or because their abundance may be lower in the samples used in this study. The proteome complexity of plasma and urine is the major limitation for the detectability of target proteins. We demonstrate that the depletion of the 14 highest abundance plasma proteins increases the number of detected CAPs in plasma from 73 to 182, especially increasing the detectability of proteins in the ng/mL concentation range. The results obtained are similar to previously reported studies combining depletion of high abundance plasma proteins and shotgun proteomics (58, 59). While some high-abundance proteins have proven to be clinically usefully, such as those in the OVA1 panel, cancer biomarker studies should still reach the low ng/ml concentration range in plasma routinely (4, 11), since tissue-derived proteins are expected and many current clinically used biomarkers are located in this concentration range (2). The combination of SRM and depletion of the highest abundance proteins in plasma achieves the required sensitivity only for a subset of the CAPs.
It has been previously shown that the sensitivity for detecting low abundance proteins in body fluids can be further improved using other sample preparation regimens that reduce complexity. These include the selective isolation of N-glycosylated peptides (22, 60), fractionation by strong cation exchange chromatography (61), peptide enrichment using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) (10, 62), and the enrichment of low molecular weight and low abundance proteins using nanoparticles (63, 64). The main disadvantage of fractionation is the reduction in throughput, since the number of samples is multiplied by the number of fractions made. The enrichment of low molecular weight proteins has a high potential to detect peptidomic-sized fragments shed or secreted from of tissue proteins, but it also neglects a large part of the proteome. Additional costs for achieving higher sensitivity with enrichment strategies include increasing technical variability with additional handling steps, a dependency on suitable affinity reagents, focusing on a subset of proteins and limited multiplexing capabilities. Therefore, in this study, we used the depletion of highly abundant plasma proteins, a simple sample preparation for detectability testing that does not adversely affect throughput and is commonly used for plasma proteomics. However, the disadvantage of the depletion strategy is the removal of additional proteins that are non-covalently associated with the depleted peptides or proteins (e.g. albumin) and thus potentially disturb the observed protein patterns of the target proteins (65). Furthermore, plasma depletion reaches the required sensitivity for biomarker studies only for a subset of proteins. However, the SRM assays of this resource are not limited to one sample preparation strategy but can be combined with all above-mentioned methods, except N-glycopeptide enrichment, to lower sample complexity and gain sensitivity in detecting low-abundance proteins except for the enrichment of N-glycopeptides. Additionally, the resource can be easily expanded to include detectability information of CAPs for the other sample preparations.
We demonstrated that the CAPs, even though they were compiled from various sources, are highly enriched in the sub-networks of CDMs discovered by unbiased whole exome sequencing. This not only shows the potential clinical importance of the compiled CAPs, but it also suggests a novel systematic approach for discovering new biomarker candidates. Current efforts in unbiased whole exome or full genomic sequencing give more insights into the molecular development of different cancer types. Additionally, a wealth of data is publically available from transcriptomic and proteomic screens. Integrating the available large-scale datasets with rapidly growing protein-protein or functional interaction networks for humans can potentially lead to the identification of pathways and sub-networks that are perturbed in specific cancers. Proteins that are part of the identified pathways or sub-networks can then be tested as potential novel biomarkers. This network-based biomarker candidate prediction is supported by the results that we obtained in the current study. We predicted biomarker candidates combining genes mutated in ovarian cancer and a functional interaction network and using our SRM assay library, could confirmed significant abundance changes for 12 of the proteins comparing plasma samples derived from OC patients and patients with BOT. Such computational network approaches reduce time and resources spent on generating new potential biomarkers lists, so that more efforts can be invested in the verification of their clinical utility.
The experimental procedures used in previous studies to generate lists of biomarker candidates were time and resource consuming and most of the studies concluded after the discovery of cancer-associated proteins without verifying their clinical utility. Recent developments in SRM showed that it is a valuable alternative technology for protein quantification compared to the “gold-standard” ELISA. High throughput, cost efficient and fast SRM assay development as well as multiplexed analyte measurements allow for faster verification of the CAPs in complex samples. In order to translate the research findings into clinical practice, research efforts need to concentrate on hypothesis testing in large clinical cohorts, since it is anticipated that only a few of the proposed markers will have an impact in the current clinical practice. So far it has been shown that most biomedical research has focused on only a few well-studied proteins precisely because the costly analytical tools for their study are already available (66). In contrast, the resource described here has the capacity to facilitate exploratory biomedical research and to drive it towards unstudied parts of the human proteome. For example, on top of the assays for more than 1000 proteins contained in this library, we estimate that the expansion of our SRM resource by an additional 100 proteins could be accomplished in less than one month. Therefore, we envision this SRM resource as a tool well matched with computational candidate prediction.
The resource SRM reference map generated here provides high quality SRM assays enabling direct measurement of the proteins associated with cancer and functionally related to genes frequently mutated in cancer in any sample type. The detectability information indicates where proteins are accessible and thus guides the selection of targets and experimental procedures involved in biomarker verification. Using the outlined workflow, the resource of assays can be rapidly expanded to include more proteins and sample types. A strategy based on this assay library in combination with the software tools, mProphet and SRMstats, for the analysis of large scale SRM datasets, provides the foundation for clinical SRM-based cancer biomarker verification studies. Once candidate cancer biomarker verification is accelerated, validation of clinical utility should follow.
Materials and Methods
Protein Selection
1172 protein targets were selected based on two previously published lists of proteins, one enumerating those associated with cancer (28) and the other containing protein analytes which have FDA approved clinical assays (8). Proteins were identified by their unique UniProt accession number (Table S1). If a protein did not have a UniProt accession number given in the source list, the protein's name was searched in the UniProt database (http://www.uniprot.org/) or the protein's gene symbol was searched in the Gene/Protein Synonyms finder (http://expasy.org/cgi-bin/gpsdb/form), in order to assign a UniProt identifier.
Network analysis
Functional relationships between the CAPs and CDMs were investigated using the RFIN (16, 67). CDMs were obtained from whole-exome sequencing and re-sequencing studies of cancer genomes for 7 human tissue types (30-35). CDMs were projected on the functional interaction network and their interaction partners were explored for the presence of CAPs. 100 random protein networks of the same size and degree-distribution as RFIN were generated using the “switching algorithm” (37) implemented in the Random Network Pulgin for Cytoscape, which enabled assessing the statistical significance of enrichment of CAPs among interaction partners of CDMs. The interactions were obtained using the Reactome Functional Interaction Cytoscape plug-in (67) and the graphs were visualized in Cytoscape (68).
Peptide Selection
For each protein a set of PTPs was selected based on the following criteria: only fully tryptic peptides, with no missed cleavages, unique to a particular protein, and with a length between 6 and 20 amino acids were considered. For proteins listed in large-scale proteomic repositories, like the Human PA (http://www.peptideatlas.org/), the Human Plasma PA (40, 41), and a human cell line MS deep sequencing dataset (42), the five PTPs most frequently observed and fulfilling the selection criteria were chosen. For proteins that were observed in the proteomic repositories by less than five PTPs or that were not previously observed, additional PTPs with good MS properties were selected by bioinformatic prediction. The main criterion for prediction was peptide hydrophobicity estimated using the SSRCalc algorithm (69, 70). Only peptides with a SSRCalc value between 10 and 40 were considered. If less than five uniquely mapping peptides could be selected for a protein, peptides mapping to a maximum of three proteins in the Uniprot database were also considered.
Crude Peptide Library Generation
Selected peptides were synthesized by the SPOT-synthesis technology (71, 72) (JPT Peptide Technologies), recovered from the solid support and used in an unpurified form. Synthesized peptides were lyophilized in 96-well plates with approximately 50 nmol of unpurified peptide material per well. Aliquots of the peptides contained in each well were combined to generate mixes of approximately 100 peptides. Prior to LC-MS/MS analysis the peptide mixes were desalted and concentrated using Vydac C18 silica MicroSpin columns (The Nest Group Inc.). A set of eight synthetic peptides (AAVYHHFISDGVR, HIQNIDIQHLAGK, GGQEHFAHLLILR, TEVSSNHVLIYLDK, TEHPFTVEEFVLPK, NQGNTWLTAFVLK, LVAYYTLIGASGQR, TTNIQGINLLFSSR) with elution times spanning the solvent gradient were spiked into each mixture to enable the correlation of relative retention times between LC-MS/MS runs.
SRM Assay Library Generation
The fragment ion spectral library was assembled using a hybrid triple quadrupole/ion trap mass spectrometer (5500QTRAP, AB Sciex) by triggering the acquisition of a full fragment ion spectrum upon threshold detection of an SRM trace corresponding to the first fragment ion of the y-series with an m/z above the m/z precursor + 20 Thomson (Th), for the doubly and triply charged peptide precursors. The instrument setup and parameters are described in Supplementary Methods. The resulting MS/MS spectra were assigned to peptide sequences using Mascot (Matrix Science, Version 2.3.0). The search results were validated using a cutoff for the Mascot ion score corresponding to a FDR < 1%. All the peptide-spectrum matches taken together constituted the spectral library for target peptides. For SRM assay refinement, the crude peptide mixtures were analyzed on a TSQ Vantage triple-quadrupole mass spectrometer (Thermo Fisher) in scheduled SRM acquisition mode. The QQQ spectral library was used to extract the optimal coordinates for the SRM assays, e.g. the most intense fragments, relative intensities of fragments and peptide elution times. Instrument specific parameters and further method details can be found in Supplementary Methods.
Plasma Handling
Collection, handling and shipping of the plasma sample for the detectability test was carried out by Sera Laboratories International Ltd. Blood was collected from two healthy individuals, one male and one female, using EDTA as an anticoagulant. Plasma was obtained from each sample of blood by centrifuging at 2000 × g for 10 minutes at room temperature. After pooling of the two samples, the resulting plasma was filtered through a 0.2 μm filter, aliquoted and frozen at −80°C for shipping. Upon thawing, Complete Protease Inhibitor Cocktail (Roche) was added.
For the collection of the patient plasma samples all patients signed an informed consent document. Blood was drawn prior to surgery and collected into tubes processed with EDTA to prevent coagulation. Within 30 min blood was centrifuged at the speed of 2000 × g for 10 min to separate the red blood cells, buffy coat and plasma. The plasma was removed, aliquoted in 300 uL amounts and stored at −80°C. The blood sample handling, from drawing to storage, was done within 2 hours.
Plasma Protein Depletion
Plasma was depleted of the 14 most abundant plasma proteins using the multiple affinity removal system (MARS Hu-14 spin cartridge; Agilent Technologies) according to the manufacturer's protocol. Depleted samples were exchanged using Vivaspin 500 concentrators with a 5000 MW cut-off (Sartorius Stedim Biotech) and denatured in 6 M urea, 0.1 M ammonium bicarbonate prior to digestion with trypsin and LC-MS analysis.
Urine Handling
Second morning urine samples were collected from four healthy individuals, two males and two females, in 50 mL conical tubes (Greiner) and spiked with Complete Protease Inhibitor Cocktail (Roche). Urine was centrifuged at 2000 × g for 10 min at room temperature. The supernatants were transferred to a fresh tube and the urinary protein concentration was estimated by pyrogallol assay (Sigma). A single pooled urine sample was prepared from the four healthy individuals with a final concentration of 120 µg/mL. Protein precipitation was achieved by adding TCA (Sigma-Aldrich) to 10 mL of urine to a final concentration of 6%. The sample was mixed and incubated at 4°C for 2h followed by centrifugation at 14000 × g for 15 min. The supernatant was removed and the pellet was washed twice with 100% ice-cold acetone (Sigma-Aldrich) to remove all interfering compounds. The supernatant was removed, the pellet was air-dried, and resuspended in 300 μl of denaturing buffer containing 8 M urea (Sigma-Aldrich) and 0.1 M ammonium bicarbonate (Sigma-Aldrich).
Plasma and Urine Protein Digestion
Following reduction and alkylation with DTT (Sigma-Aldrich) and iodoacetamide (Sigma-Aldrich), the proteins were digested using sequencing grade porcine trypsin (Promega) at a protease:protein ratio of 1:50 for plasma and 1:100 for urine. Digests were desalted and concentrated using Vydac C18 silica MicroSpin columns (The Nest Group Inc.) and Sep-Pak C18 cartridges (Waters), for plasma and urine respectively, prior to LC-MS analysis. The crude plasma digest was prepared in the same way as depleted samples. An aliquot of retention time calibration peptides from RT-kit-WR (Biognosys) was spiked into each sample to allow for the correlation of relative retention times between LC-MS runs. The extracted elution times of the RT peptides was used to calculate an iRT value relative to the RT peptides for each SRM assay according to the vendors’ instructions (44).
Target detection in plasma and urine
For target detection in depleted plasma, crude plasma and urine, peptide preparations were analyzed on a TSQ Vantage using the instrument setup and parameters as described in Supplementary Methods. The refined SRM assays from the library, constituted by the relative intensities of the five most intense fragments and the peptide elution time of each target, were used to detect endogenous peptides. Additionally, five assays were selected as positive controls for both plasma and urine, to be monitored in each MS run, and decoy-transition groups were equally distributed over all runs for the subsequent estimation of the FDR (26). Decoy transition groups were generated by subtracting or adding a random integer to Q1 and Q3 m/z values as described by Reiter et al. (26). In each run, around 400 target transitions were monitored resulting in a total number of 60 MS runs per sample to test the detectability of all refined SRM assays. SRM acquisition was performed with Q1 and Q3 operated at a resolution of 0.7 m/z half maximum peak width, with a retention time window of 240 s and a cycle time of 2.0 s. To better cover the isotopic envelope the Q1 values for triply charged precursors were set to average molecular masses. Resulting SRM data was analyzed using mProphet (26). The following subscores of each assay were considered for the calculation of the discriminant score for the detected peak groups: shape score, delta iRT, intensity-correlation-with-assay-score, transition-coelution-score and total-intensity-score. The top-ranked peak group for each target and decoy transition group was used for the FDR estimation as described by Reiter et al. (26).
Monitoring biomarkers in patient plasma
The plasma peptide preparations were analyzed on a 5500QTRAP (AB Sciex) using the instrument setup and parameters as described in Supplementary Methods. For each target peptide a heavy isotope labeled internal standard (JPT Peptide Technology and Thermo Fischer) was spiked in the plasma peptide mixture for accurate quantification. For each peptide three transitions were monitored for the heavy and light version. Resulting SRM data was analyzed using mProphet (26). The following subscores of each assay were considered for the calculation of the discriminant score for the detected peak groups: shape score, intensity-correlation-with-assay-score, transition-coelution-score, total-intensity-score, light_heavy_correlation, var_light_heavy_shape_score and light_heavy_coelution_score. The top-ranked peak group for each target and decoy transition group was used for the FDR estimation as described by Reiter et al. (26). Protein significance analysis was performed using SRMstats (27). In the first step, data preprocessing was performed by transforming all transition intensities into log2-values. Then a constant normalization was conducted based on reference transitions for all proteins, which equalized the median peak intensities of reference transitions from all proteins across all MS runs and adjusted the bias to both reference and endogenous signals. Protein-level quantification and testing for differential abundance in the different patient groups were performed using the linear mixed-effects model implemented in SRMstats. Each protein is tested for abundance differences between ovarian cancer patients and patients with benign ovarian tumors. The p-values were adjusted to control the FDR at a cutoff of 0.05 (27). All proteins with a p-value below 0.01 and a fold-change larger than 1.1 were considered significant.
Supplementary Material
Acknowledgements
We thank L. Reiter and A. Bensimon for helpful discussions, C.-Y. Chang for support with the SRMstats analysis and R. Ossola for measurement help. Funding: The project was supported in part by the Swiss National Science Foundation (Grant# 3100A0-107679) and by the European Research Council (Grant# ERC-2008-AdG 233226). M.S. is the recipient of a Natural Sciences and Engineering Research Council of Canada PGS M award. T.F. and E.W.D are supported by the National Institutes of Health-National Human Genome Research Institute (grant No. HG005805 to R.M.) and the Duchy of Luxembourg Systems Biology initiative (to R.M.).
Footnotes
Author contributions: R.H., M.S., O.R., and R.A., designed experiments; E.N.M. provided the patient plasma samples; R.H., M.S., N.S., U.K. and C.C. performed the experiments; R.H., M.S., N.S., O.R., H.R. and A.S. analyzed data; E.N.M. provided the patient specimen; T.F., E.W.D. and R.L.M. developed the platform for the accessibility of the data; R.H., M.S. and R.A. wrote the paper.
Competing interests: Declaration: O.R. is employee of Biognosys AG. This company funded parts of the work. The other authors declare that they have no competing interests.
Data and materials: All datasets have been submitted to the PeptideAtlas SRM Experiment Library (PASSEL, http://www.peptideatlas.org/passel/).
References
- 1.Ludwig J, Weinstein J. Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nature Reviews Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 2.Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Molecular Oncology. 2009;3:33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson NL, Anderson NG, Pearson TW, Borchers CH, Paulovich AG, Patterson SD, Gillette M, Aebersold R, Carr SA. A human proteome detection and quantitation project: hPDQ. Molecular & Cellular Proteomics. 2009;8:883–886. doi: 10.1074/mcp.R800015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rifai N, Gillette M, Carr S. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechnology. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 5.Surinova S, Schiess R, Hüttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the development of plasma protein biomarkers. Journal of Proteome Research. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 6.Selevsek N, Matondo M, Carbayo MSS, Aebersold R, Domon B. Systematic quantification of peptides/proteins in urine using selected reaction monitoring. Proteomics. 2011;11:1135–1147. doi: 10.1002/pmic.201000599. [DOI] [PubMed] [Google Scholar]
- 7.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Molecular & Cellular Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Anderson N. The Clinical Plasma Proteome: A Survey of Clinical Assays for Proteins in Plasma and Serum. Clinical Chemistry. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 9.Hüttenhain R, Malmström J, Picotti P, Aebersold R. Perspectives of targeted mass spectrometry for protein biomarker verification. Current Opinion in Chemical Biology. 2009;13:518–525. doi: 10.1016/j.cbpa.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiteaker J, Lin C, Kennedy J, Hou L, Trute M, Sokal I, Yan P, Schoenherr R, Zhao L, Voytovich U, Kelly-Spratt K, Krasnoselsky A, Gafken PR, Hogan J, Jones L, Wang P, Amon L, Chodosh L, Nelson P, Mcintosh M, Kemp C, Paulovich A. A targeted proteomics–based pipeline for verification of biomarkers in plasma. Nature Biotechnology. 2011;29:625–634. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The interface between biomarker discovery and clinical validation: The tar pit of the protein biomarker pipeline. Proteomics Clinical Applications. 2008;2:1386–1402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ideker T, Sharan R. Protein networks in disease. Genome Research. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang H, Lee E, Liu Y, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Molecular Systems Biology. 2007;3:10. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nibbe R, Koyutürk M, Chance M, Price N. An Integrative -omics Approach to Identify Functional Sub-Networks in Human Colorectal Cancer. PLoS Computational Biology. 2010;6:e1000639. doi: 10.1371/journal.pcbi.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Demir E, Schultz N, Taylor B, Sander C, Hannenhalli S. Automated Network Analysis Identifies Core Pathways in Glioblastoma. PLoS ONE. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biology. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, Ouellette B, Quackenbush J. CAERUS: Predicting CAncER oUtcomeS Using Relationship between Protein Structural Information, Protein Networks, Gene Expression Data, and Mutation Data. PLoS Computational Biology. 2011;7:e1001114. doi: 10.1371/journal.pcbi.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nature Biotechnology. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 19.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular Systems Biology. 2008;4:14. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Addona T, Shi X, Keshishian H, Mani D, Burgess M, Gillette M, Clauser KR, Shen D, Lewis G, Farrell L, Fifer M, Sabatine M, Gerszten R, Carr SA. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nature Biotechnology. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiteaker J, Zhang H, Zhao L, Wang P, Kelly-Spratt K, Ivey R, Piening B, Feng L, Kasarda E, Gurley K, Eng J, Chodosh L, Kemp C, Mcintosh M, Paulovich A. Integrated Pipeline for Mass Spectrometry-Based Discovery and Confirmation of Biomarkers Demonstrated in a Mouse Model of Breast Cancer. Journal of Proteome Research. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 22.Cima I, Schiess R, Wild P, Kaelin M, Schuffler P, Lange V, Picotti P, Ossola R, Templeton A, Schubert O, Fuchs T, Leippold T, Wyler S, Zehetner J, Jochum W, Buhmann J, Cerny T, Moch H, Gillessen S, Aebersold R, Krek W. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proceedings of the National Academy of Sciences. 2011;108:3342–3347. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nature Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 24.Addona T, Abbatiello S, Schilling B, Skates S, Mani D, Bunk D, Spiegelman C, Zimmerman L, Ham A, Keshishian H, Hall S, Allen S, Blackman R, Borchers C, Buck C, Cardasis H, Cusack M, Dodder N, Gibson B, Held J, Hiltke T, Jackson A, Johansen E, Kinsinger C, Li J, Mesri M, Neubert T, Niles R, Pulsipher T, Ransohoff D, Rodriguez H, Rudnick P, Smith D, Tabb D, Tegeler T, Variyath A, Vega-Montoto L, Wahlander Å, Waldemarson S, Wang M, Whiteaker J, Zhao L, Anderson NL, Fisher S, Liebler D, Paulovich A, Regnier F, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring–based measurements of proteins in plasma. Nature Biotechnology. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picotti P, Lam H, Campbell D, Deutsch E, Mirzaei H, Ranish J, Domon B, Aebersold R. A database of mass spectrometric assays for the yeast proteome. Nature Methods. 2008;5:913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiter L, Rinner O, Picotti P, Hüttenhain R, Beck M, Brusniak MY, Hengartner M, Aebersold R. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nature Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 27.Chang C-Y, Picotti P, Huettenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R, Vitek O. Protein significance analysis in selected reaction monitoring (SRM) measurements. Molecular & Cellular Proteomics. 2012;11:M111.014662. doi: 10.1074/mcp.M111.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomarker Insights. 2007;1:1–48. [PMC free article] [PubMed] [Google Scholar]
- 29.Barabasi A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature Reviews Genetics. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 31.Jones S, Wang T, Shih I, Mao T, Nakayama K, Roden R, Glas R, Slamon D, Diaz L, Vogelstein B, Kinzler K, Velculescu V, Papadopoulos N. Frequent Mutations of Chromatin Remodeling Gene ARID1A in Ovarian Clear Cell Carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons D, Jones S, Zhang X, Lin J, Leary R, Angenendt P, Mankoo P, Carter H, Siu I, Gallia G, Olivi A, Mclendon R, Rasheed B, Keir S, Nikolskaya T, Nikolsky Y, Busam D, Tekleab H, Diaz L, Hartigan J, Smith D, Strausberg R, Marie S, Shinjo S, Yan H, Riggins G, Bigner D, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu V, Kinzler K. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons D, Li M, Zhang X, Jones S, Leary R, Lin J, Boca S, Carter H, Samayoa J, Bettegowda C, Gallia G, Jallo G, Binder Z, Nikolsky Y, Hartigan J, Smith D, Gerhard D, Fults D, Vandenberg S, Berger M, Marie S, Shinjo S, Clara C, Phillips P, Minturn J, Biegel J, Judkins A, Resnick A, Storm P, Curran T, He Y, Rasheed B, Friedman H, Keir S, Mclendon R, Northcott P, Taylor M, Burger P, Riggins G, Karchin R, Parmigiani G, Bigner D, Yan H, Papadopoulos N, Vogelstein B, Kinzler K, Velculescu V. The Genetic Landscape of the Childhood Cancer Medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones S, Zhang X, Parsons D, Lin J, Leary R, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong S, Fu B, Lin M, Calhoun E, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith D, Hidalgo M, Leach S, Klein A, Jaffee E, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman J, Kern S, Hruban R, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu V, Kinzler K. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao Y, Shi C, Edil B, De Wilde R, Klimstra D, Maitra A, Schulick R, Tang L, Wolfgang C, Choti M, Velculescu V, Diaz L, Vogelstein B, Kinzler K, Hruban R, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR Pathway Genes Are Frequently Altered in Pancreatic Neuroendocrine Tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu R, Hendrixlucas N, Kuick R, Zhai Y, Schwartz D, Akyol A, Hanash S, Misek D, Katabuchi H, Williams B. Mouse Model of Human Ovarian Endometrioid Adenocarcinoma Based on Somatic Defects in the Wnt/β-Catenin and PI3K/Pten Signaling Pathways. Cancer Cell. 2007;11:321–333. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Milo R, Kashtan R, Itzkowitz S, Newman MEJ, Alon U. On the uniform generation of random graphs with prescribed degree sequences. arXiv:cond-mat/0312028v2 [condmat.stat-mech] 2004 [Google Scholar]
- 38.Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nature Reviews Molecular Cell Biology. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 39.Mallick P, Schirle M, Chen S, Flory M, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Computational prediction of proteotypic peptides for quantitative proteomics. Nature Biotechnology. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 40.Deutsch E, Eng J, Zhang H, King N, Nesvizhskii A, Lin B, Lee H, Yi E, Ossola R, Aebersold R. Human Plasma PeptideAtlas. Proteomics. 2005;5:3497–3500. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- 41.Farrah T, Deutsch E, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, Watts JD, Lin B, Zhang H, Moritz RL, Aebersold RH. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Molecular Systems Biology. 2011;7 doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fusaro V, Mani D, Mesirov J, Carr S. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nature Biotechnology. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, MacCoss MJ, Rinner O. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012 doi: 10.1002/pmic.201100463. pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherman J, Mckay M, Ashman K, Molloy M. How specific is my SRM?: The issue of precursor and product ion redundancy. Proteomics. 2009;9:1120–1123. doi: 10.1002/pmic.200800577. [DOI] [PubMed] [Google Scholar]
- 46.Duncan M, Yergey A, Patterson S. Quantifying proteins by mass spectrometry: The selectivity of SRM is only part of the problem. Proteomics. 2009;9:1124–1127. doi: 10.1002/pmic.200800739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang D, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Huang D, Sherman B, Lempicki R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adachi J, Kumar C, Zhang Y, Olsen J, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biology. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Röst H, Malmström L, Aebersold R. Detecting and avoiding redundancy in selected reaction monitoring (SRM) – A computational approach to design selective SRM assays. submitted [Google Scholar]
- 51.Sherman J, Mckay M, Ashman K, Molloy M. Unique Ion Signature Mass Spectrometry, a Deterministic Method to Assign Peptide Identity. Molecular & Cellular Proteomics. 2009;8:2051–2062. doi: 10.1074/mcp.M800512-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fung E. A Recipe for Proteomics Diagnostic Test Development: The OVA1 Test, from Biomarker Discovery to FDA Clearance. Clinical Chemistry. 2010;56:327–329. doi: 10.1373/clinchem.2009.140855. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Chan D. The Road from Discovery to Clinical Diagnostics: Lessons Learned from the First FDA-Cleared In Vitro Diagnostic Multivariate Index Assay of Proteomic Biomarkers. Cancer Epidemiology Biomarkers & Prevention. 2010;19:2995–2999. doi: 10.1158/1055-9965.EPI-10-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell D, Berchuck A, Birrer M, Chien J, Cramer D, Dao F, Dhir R, Disaia P, Gabra H, Glenn P, Godwin A, Gross J, Hartmann L, Huang M, Huntsman D, Iacocca M, Imielinski M, Kalloger S, Karlan B, Levine D, Mills G, Morrison C, Mutch D, Olvera N, Orsulic S, Park K, Petrelli N, Rabeno B, Rader J, Sikic B, Smith-Mccune K, Sood A, Bowtell D, Penny R, Testa J, Chang K, Dinh H, Drummond J, Fowler G, Gunaratne P, Hawes A, Kovar C, Lewis L, Morgan M, Newsham I, Santibanez J, Reid J, Trevino L, Wu Y, Wang M, Muzny D, Wheeler D, Gibbs R, Getz G, Lawrence M, Cibulskis K, Sivachenko A, Sougnez C, Voet D, Wilkinson J, Bloom T, Ardlie K, Fennell T, Baldwin J, Gabriel S, Lander E, Ding L, Fulton R, Koboldt D, Mclellan M, Wylie T, Walker J, O'laughlin M, Dooling D, Fulton L, Abbott R, Dees N, Zhang Q, Kandoth C, Wendl M, Schierding W, Shen D, Harris C, Schmidt H, Kalicki J, Delehaunty K, Fronick C, Demeter R, Cook L, Wallis J, Lin L, Magrini V, Hodges J, Eldred J, Smith S, Pohl C, Vandin F, Raphael B, Weinstock G, Mardis E, Wilson R, Meyerson M, Winckler W, Getz G, Verhaak R, Carter S, Mermel C, Saksena G, Nguyen H, Onofrio R, Lawrence M, Hubbard D, Gupta S, Crenshaw A, Ramos A, Ardlie K, Chin L, Protopopov A, Zhang J, Kim T, Perna I, Xiao Y, Zhang H, Ren G, Sathiamoorthy N, Park R, Lee E, Park P, Kucherlapati R, Absher D, Waite L, Sherlock G, Brooks J, Li J, Xu J, Myers R, Laird P, Cope L, Herman J, Shen H, Weisenberger D, Noushmehr H, Pan F, Triche T, Jr, Berman B, Van Den Berg D, Buckley J, Baylin S, Spellman P, Purdom E, Neuvial P, Bengtsson H, Jakkula L, Durinck S, Han J, Dorton S, Marr H, Choi Y, Wang V, Wang N, Ngai J, Conboy J, Parvin B, Feiler H, Speed T, Gray J, Levine D, Socci N, Liang Y, Taylor B, Schultz N, Borsu L, Lash A, Brennan C, Viale A, Sander C, Ladanyi M, Hoadley K, Meng S, Du Y, Shi Y, Li L, Turman Y, Zang D, Helms E, Balu S, Zhou X, Wu J, Topal M, Hayes D, Perou C, Getz G, Voet D, Saksena G, Zhang J, Zhang H, Wu C, Shukla S, Cibulskis K, Lawrence M, Sivachenko A, Jing R, Park R, Liu Y, Park P, Noble M, Chin L, Carter H, Kim D, Karchin R, Spellman P, Purdom E, Neuvial P, Bengtsson H, Durinck S, Han J, Korkola J, Heiser L, Cho R, Hu Z, Parvin B, Speed T, Gray J, Schultz N, Cerami E, Taylor B, Olshen A, Reva B, Antipin Y, Shen R, Mankoo P, Sheridan R, Ciriello G, Chang W, Bernanke J, Borsu L, Levine D, Ladanyi M, Sander C, Haussler D, Benz C, Stuart J, Benz S, Sanborn J, Vaske C, Zhu J, Szeto C, Scott G, Yau C, Hoadley K, Du Y, Balu S, Hayes D, Perou C, Wilkerson M, Zhang N, Akbani R, Baggerly K, Yung W, Mills G, Weinstein J, Penny R, Shelton T, Grimm D, Hatfield M, Morris S, Yena P, Rhodes P, Sherman M, Paulauskis J, Millis S, Kahn A, Greene J, Sfeir R, Jensen M, Chen J, Whitmore J, Alonso S, Jordan J, Chu A, Zhang J, Barker A, Compton C, Eley G, Ferguson M, Fielding P, Gerhard D, Myles R, Schaefer C, Mills Shaw K, Vaught J, Vockley J, Good P, Guyer M, Ozenberger B, Peterson J, Thomson E. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, Bast RC, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, de Bruijn HW, van der Zee AG, Jacobs IJ, Fung ET, Chan DW. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Research. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 56.Rai AJ, Zhang Z, Rosenzweig J, Shih I, Pham T, Fung ET, Sokoll LJ, Chan DW. Proteomic approaches to tumor marker discovery. Archives of pathology & laboratory medicine. 2002;126:1518–1526. doi: 10.5858/2002-126-1518-PATTMD. [DOI] [PubMed] [Google Scholar]
- 57.Farrah T, Deutsch EW, Kreisberg R, Sun Z, Campbell DS, Mendoza L, Kusebauch U, Brusniak M-Y, Hüttenhain R, Schiess R, Selevsek N, Aebersold R, Moritz RL. PASSEL: The PeptideAtlas SRM Experiment Library. Proteomics. 2012 doi: 10.1002/pmic.201100515. pmic.201100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng L-C, Lorentzen TD, Schoenherr RM, Keane JF, Holzman T, Fitzgibbon M, Lin, Zhang H, Cooke K, Liu T, Camp DG, Anderson L, Watts J, Smith RD, McIntosh MW, Paulovich AG. Head-to-Head Comparison of Serum Fractionation Techniques. Journal of Proteome Research. 2006;6:828–836. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- 59.Tu C, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJC, Liebler DC. Depletion of Abundant Plasma Proteins and Limitations of Plasma Proteomics. Journal of Proteome Research. 2010;9:4982–4991. doi: 10.1021/pr100646w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High Sensitivity Detection of Plasma Proteins by Multiple Reaction Monitoring of NGlycosites. Molecular & Cellular Proteomics. 2007;6:1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Keshishian H, Addona T, Burgess M, Kuhn E, Carr S. Quantitative, Multiplexed Assays for Low Abundance Proteins in Plasma by Targeted Mass Spectrometry and Stable Isotope Dilution. Molecular & Cellular Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuhn E, Addona T, Keshishian H, Burgess M, Mani D, Lee R, Sabatine M, Gerszten R, Carr S. Developing Multiplexed Assays for Troponin I and Interleukin-33 in Plasma by Peptide Immunoaffinity Enrichment and Targeted Mass Spectrometry. Clinical Chemistry. 2009;55:1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luchini A, Geho D, Bishop B, Tran D, Xia C, Dufour R, Jones C, Espina V, Patanarut A, Zhou W, Ross M, Tessitore A, Petricoin EF, Liotta LA. Smart hydrogel particles: biomarker harvesting: one-step affinity purification, size exclusion, and protection against degradation. Nano Letters. 2008;8:350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fredolini C, Meani F, Alex Reeder K, Rucker S, Patanarut A, Botterell P, Bishop B, Longo C, Espina V, Petricoin EF, Liotta LA, Luchini A. Concentration and preservation of very low abundance biomarkers in urine, such as human growth hormone (hGH), by Cibacron Blue F3G-A loaded hydrogel particles. Nano Research. 2008;1:502–518. doi: 10.1007/s12274-008-8054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bellei E, Bergamini S, Monari E, Fantoni L, Cuoghi A, Ozben T, Tomasi A. High-abundance proteins depletion for serum proteomic analysis: concomitant removal of non-targeted proteins. Amino Acids. 2010;40:145–156. doi: 10.1007/s00726-010-0628-x. [DOI] [PubMed] [Google Scholar]
- 66.Edwards AM, Isserlin R, Bader GD, Frye SV, Willson TM, Yu FH. Too many roads not taken. Nature. 2011;470:163–165. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- 67.Haw R, Hermjakob H, D'Eustachio P, Stein L. Reactome Pathway Analysis to Enrich Biological Discovery in Proteomics Datasets. Proteomics. 2011;11:3598–3613. doi: 10.1002/pmic.201100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shannon P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krokhin OV. Sequence-specific retention calculator. Algorithm for peptide retention prediction in ion-pair RP-HPLC: application to 300- and 100-A pore size C18 sorbents. Analytical Chemistry. 2006;78:7785–7795. doi: 10.1021/ac060777w. [DOI] [PubMed] [Google Scholar]
- 70.Krokhin OV, Ying S, Cortens JP, Ghosh D, Spicer V, Ens W, Standing KG, Beavis RC, Wilkins JA. Use of peptide retention time prediction for protein identification by off-line reversed-phase HPLC-MALDI MS/MS. Analytical Chemistry. 2006;78:6265–6269. doi: 10.1021/ac060251b. [DOI] [PubMed] [Google Scholar]
- 71.Wenschuh H, Volkmer-Engert R, Schmidt M, Schulz M, Schneider-Mergener J, Reineke U. Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides. Biopolymers. 2000;55:188–206. doi: 10.1002/1097-0282(2000)55:3<188::AID-BIP20>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 72.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports--principles and applications. Journal of Immunological Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 73.Amon LM, Law W, Fitzgibbon MP, Gross JA, O'Briant K, Peterson A, Drescher C, Martin DB, Mcintosh M. Integrative proteomic analysis of serum and peritoneal fluids helps identify proteins that are up-regulated in serum of women with ovarian cancer. PLoS ONE. 2010;5:e11137. doi: 10.1371/journal.pone.0011137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuk C, Kulasingam V, Gunawardana C, Smith C, Batruch I, Diamandis E. Mining the Ovarian Cancer Ascites Proteome for Potential Ovarian Cancer Biomarkers. Molecular & Cellular Proteomics. 2009;8:661–669. doi: 10.1074/mcp.M800313-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boylan KL, Andersen JD, Anderson LB, Higgins L, Skubitz AP. Quantitative proteomic analysis by iTRAQ(R) for the identification of candidate biomarkers in ovarian cancer serum. Proteome Sci. 2010;8:31. doi: 10.1186/1477-5956-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Begum F, Høgdall E, Kjaer S, Blaakaer J, Christensen I, Christensen L, Høgdall C. Preoperative serum tetranectin, CA125 and menopausal status used as single markers in screening and in a risk assessment index (RAI) in discriminating between benign and malignant ovarian tumors. Gynecologic Oncology. 2009;113:221–227. doi: 10.1016/j.ygyno.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 77.Gadomska H, Grzechocińska B, Janecki J, Nowicka G, Powolny M, Marianowski L. Serum lipids concentration in women with benign and malignant ovarian tumours. European Journal of Obstetrics & Gynecology and Reproductive Biology. 2005;120:87–90. doi: 10.1016/j.ejogrb.2004.02.045. [DOI] [PubMed] [Google Scholar]
- 78.Faça V, Ventura A, Fitzgibbon M, Pereira-Faça S, Pitteri S, Green A, Ireton R, Zhang Q, Wang H, O'briant K, Drescher C, Schummer M, Mcintosh M, Knudsen B, Hanash S, Imhof A. Proteomic Analysis of Ovarian Cancer Cells Reveals Dynamic Processes of Protein Secretion and Shedding of Extra-Cellular Domains. PLoS ONE. 2008;3:e2425. doi: 10.1371/journal.pone.0002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gunawardana CG, Kuk C, Smith CR, Batruch I, Soosaipillai A, Diamandis EP. Comprehensive analysis of conditioned media from ovarian cancer cell lines identifies novel candidate markers of epithelial ovarian cancer. Journal of Proteome Research. 2009;8:4705–4713. doi: 10.1021/pr900411g. [DOI] [PubMed] [Google Scholar]
- 80.Waldemarson S, Krogh M, Alaiya A, Kirik U, Schedvins K, Auer G, Hansson KM, Ossola R, Aebersold R, Lee H, Malmström J, James P. Protein Expression Changes in Ovarian Cancer during the Transition from Benign to Malignant. Journal of Proteome Research. 2012 doi: 10.1021/pr201258q. pr201258q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.