Abstract
Activation of metabotropic glutamate receptor subtype 4 has been shown to be efficacious in rodent models of Parkinson’s disease. Artificial neural networks were trained based on a recently reported high throughput screen which identified 434 positive allosteric modulators of metabotropic glutamate receptor subtype 4 out of a set of approximately 155,000 compounds. A jury system containing three artificial neural networks achieved a theoretical enrichment of 15.4 when selecting the top 2% compounds of an independent test dataset. The model was used to screen an external commercial database of approximately 450,000 drug-like compounds. 1,100 predicted active small molecules were tested experimentally using two distinct assays of mGlu4 activity. This experiment yielded 67 positive allosteric modulators of metabotropic glutamate receptor subtype 4 that confirmed in both experimental systems. Compared to the 0.3% active compounds in the primary screen, this constituted an enrichment of 22 fold.
Keywords: Metabotropic glutamate receptor subtype 4, Virtual high-throughput screening, Machine learning, Quantitative structure-activity relationship, Enrichment
Introduction
This study implements a machine learning approach (Artificial Neural Networks, ANNs) to virtually screen commercially available compounds for positive allosteric modulators (PAMs) of metabotropic glutamate receptor subtype 4 (mGlu4). Marino and Conn [1, 2] showed that activation of mGlu4 is a viable option in treating Parkinson’s disease (PD), a debilitating movement disorder that afflicts more than 1 million people in North America. In Parkinson’s patients, there is a decrease in GABAergic transmission at the inhibitory striatopallidal synapse within the basal ganglia; this abnormality is thought to contribute to the motor dysfunctions observed in PD patients. Current PD treatments that are focused on dopamine-replacement strategies ultimately fail in most patients because of loss of efficacy and severe adverse effects that worsen as the disease progresses [3, 2]. Selective activation of mGlu4 could provide palliative benefit in PD. Further, selectively targeting mGlu4 avoids the loss of efficacy and severe side-effects of long-term dopamine replacement therapy. In 2003 Maj [4, 2] et al. reported on the discovery of (−)-PHCCC, the first positive allosteric modulator of mGlu4 with demonstrated selectivity for group III mGlus, but also a partial antagonist for mGlu1 (group I). Around the same time, Mathiesen [5] et al. showed that SIB-1893 and MPEP (a known mGlu5 antagonist [6]) are mGlu4 PAMs.
Despite the success of many GPCR-based drug discovery programs, many of the developed ligands lack selectivity. The traditional approach to target the endogenous ligand (orthosteric)-binding site has suffered from a paucity of suitably subtype-selective ligands as orthosteric binding sites are highly conserved between GPCR subtypes. An alternative approach is to target allosteric sites that are topographically distinct from the orthosteric site, either enhancing or inhibiting receptor activation [7]. Discovery and characterization of allosteric modulators of GPCRs has gained significant momentum over the last few years, especially since the clinical validity of GPCR allosteric modulators was demonstrated with two allosteric modulators entering the market [8, 9]. Thus, allosteric modulation represents an exciting novel means of targeting GPCRs particularly for CNS disorders, a therapeutic area with one of the highest rates of attrition in drug discovery [10].
Recently, Niswender [11] et al. reported the discovery of 434 PAMs of mGlu4 from a high-throughput screen of approximately 155,000 compounds. The study highlighted a series of cyclohexyl amides joined to a substituted phenyl ring. The structures of these tested molecules and their experimentally determined EC50 towards mGlu4 potentiation were employed in the ANNs described in this paper. Engers et al. [12] discuss the synthesis and evaluation of a set of heterobiarylamides optimized for penetrating the central nervous system. Around the same time, several pyrazolo[3,4-d]pyrimidines were also described to be novel mGlu4 positive allosteric modulators [13]. Two challenges in further developing PAMs of mGlu4 as a PD treatment strategy are the low hit-rate of 0.3% in the original high-throughput screen resulting in a small number of available ligands and the ‘flat’ structure activity relationship (SAR) around the ‘proof of concept’ compound PHCCC. Even slight structural modifications lead to complete loss of activity for the reported compounds [14]. However, successful modifications of PHCCC were reported later by our group [15]. The present study addressed both challenges by identifying additional PAMs from commercially available compound libraries and exploring the chemical space around the known active compounds.
Quantitative structure activity relationship (QSAR) models describe the often complex, non-linear relation between the chemical and physical properties of molecules and their biological activity; for a review of different methods see Todeschini et al. [16, 17]. Classical QSAR was introduced by Hansch et al. by deriving biological activity from electron density [18]. In 1988 Cramer [19] introduced Comparative Molecular Field Analysis (CoMFA.) CoMFA establishes 3D-QSAR by correlating sterics and electrostatics to the bioactivity data. This approach was expanded into the Comparative Molecular Similarity Index Analysis (CoMSIA) by Klebe [20] in 1994. However, both approaches rely on the spatial alignment of small molecules sharing a common scaffold. The QSAR techniques employed in the present and similar studies utilize 2D molecular fingerprints and 3D molecular descriptors coupled with machine learning [21–23]. These descriptors are independent from the orientation of the small molecule and the existence of a common scaffold. ANNs have been successfully applied in biochemistry to generate QSAR models [23–27]. Our group recently published a theoretical comparison of machine learning techniques for identification of compounds that are predicted allosteric modulators of the mGlu5 glutamate response [28, 29].
In the present study, artificial neural networks (ANNs) were trained on descriptors computed with the software package ADRIANA [30] linking chemical properties of small molecules to their potency as PAMs of mGlu4. Fragment-independent scalar descriptors, 2D and 3D surface and auto-correlation functions, and radial distribution functions are employed to encode a large diversity of chemotypes into comparable mathematical representations [29].
Methods
Two independent assays for primary and confirmatory screen
The compounds identified in the virtual screen were ordered from ChemBridge and tested at the Vanderbilt HTS facility. These compounds were screened in single point at a nominal 10 μM concentration employing the human mGlu4/Gqi5 calcium mobilization assay as well as the rat mGlu4 thallium flux assay described in Niswender et al [31, 11]. Compounds exceeding three standard deviations over the control EC20 response were then screened in concentration-response curve format in both assays.
Preparing the input for the ANNs
Only 0.3% (432 molecules) of the whole data set (156,146 molecules) was active. For training, the dataset was oversampled by a factor of 360 (see Fig. 1) [29]. This leads to a data set with 311,234 molecules where approximately half of the data points were active and the other half inactive.
Fig. 1.
Overall model generation workflow: (a) SD files were provided with active and inactive compounds towards mGlu4 determined by HTS and CRC; CORINA and ADRIANA were employed to generate 3D structures and molecular descriptors; (b) active molecules were oversampled 360 times to balance data sets; (c) molecules were randomly distributed between training (80%), monitoring (10%), and independent (10%) datasets; (d) ANNs were trained and (e) low sensitivity descriptors were removed until the quality measures (see Table 2) no longer improved; the best three ANNs were combined into a jury network
Three-dimensional models of all 156,146 molecules from the original HTS were generated using CORINA [32]. These models served as input for the ADRIANA [33] software package. All 35 categories (scalar, 2D/3D auto-correlation, RDF (eight each), surface auto-correlation (three), see Table 1) were computed implementing the default values in each category. Approximately 4% of all molecules were not properly encoded by ADRIANA and removed from the data set. The final data set consisted of 298,914 data points.
Table 1.
Summary of 1,252 molecular descriptors in 35 categories computed with ADRIANA
| Description Method | Description Property | Abbreviation | Number | |
|---|---|---|---|---|
| 1 | Scalar descriptors | Molecular weight of compound | Weight | 1 |
| 2 | Number of hydrogen bonding acceptors | HDon | 1 | |
| 3 | Number of hydrogen bonding donors | HAcc | 1 | |
| 4 | Octanol/water partition coefficient in [log units] | XlogP | 1 | |
| 5 | Topological polar surface area in [Å2] | TPSA | 1 | |
| 6 | Mean molecular polarizability in [Å3] | Polariz | 1 | |
| 7 | Dipole moment in [Debye] | Dipol | 1 | |
| 8 | Solubility of the molecule in water in [log units] | LogS | 1 | |
|
| ||||
| 9 | 2D Autocorrelation | atom identities | 2DA_Ident | 11 |
| 10 | σ atom charges | 2DA_SigChg | 11 | |
| 11 | π atom charges | 2DA_PiChg | 11 | |
| 12 | total charges | 2DA_TotChg | 11 | |
| 13 | σ atom electronegativities | 2DA_SigEN | 11 | |
| 14 | π atom electronegativities | 2DA_PiEN | 11 | |
| 15 | lone pair electronegativities | 2DA_LpEN | 11 | |
| 16 | effective atom polarizabilities | 2DA_Polariz | 11 | |
|
| ||||
| 17 | 3D Autocorrelation | atom identities | 3DA_Ident | 12 |
| 18 | σ atom charges | 3DA_SigChg | 12 | |
| 19 | π atom charges | 3DA_PiChg | 12 | |
| 20 | total charges | 3DA_TotChg | 12 | |
| 21 | σ atom electronegativities | 3DA_SigEN | 12 | |
| 22 | π atom electronegativities | 3DA_PiEN | 12 | |
| 23 | lone pair electronegativities | 3DA_LpEN | 12 | |
| 24 | effective atom polarizabilities | 3DA_Polariz | 12 | |
|
| ||||
| 25 | Radial Distribution Function | atom identities | RDF_Ident | 128 |
| 26 | σ atom charges | RDF_SigChg | 128 | |
| 27 | π atom charges | RDF_PiChg | 128 | |
| 28 | total charges | RDF_TotChg | 128 | |
| 29 | σ atom electronegativities | RDF_SigEN | 128 | |
| 30 | π atom electronegativities | RDF_PiEN | 128 | |
| 31 | lone pair electronegativities | RDF_LpEN | 128 | |
| 32 | effective atom polarizabilities | RDF_Polariz | 128 | |
|
| ||||
| 33 | Surface Autocorrelation | molecular electrostatic potential | Surf_ESP | 12 |
| 34 | hydrogen bonding potential | Surf_HBP | 12 | |
| 35 | hydrophobicity potential | Surf_HPP | 12 | |
|
| ||||
| Total | 1252 | |||
Training the ANN on the logarithm of the half maximal effective concentration EC50
The experimentally determined EC50 values of the active compounds ranged from 0.4 μM to 15.8 μM. To distinguish between active and inactive compounds, all inactive compounds were set to an arbitrary potency of 1 mM. The output for training the ANN consisted of the natural logarithm of the ln(EC50) values ranging from −14.7 (most active) over −11.1 (least active) to −6.9 (inactive). The root mean square deviation (rmsd) between experimental and predicted EC50 values was employed as objective function in training the ANNs:
| (1) |
where expi is the experimentally determined ln(EC50) value and predi the predicted ln(EC50) value.
From the 298,914 data points in the oversampled data set, 239,132 (80%) were employed in the actual training of the ANN. The monitoring data set consisted of 29,891 data points (10%). The rmsd between experimental and predicted ln(EC50) was computed for the monitoring data set after each iteration over the full training data set. Once the rmsd was minimized, the training was terminated, and the rmsd of the remaining 10% (independent data set) computed (see Table 2). Care was taken to exclude overlap between training, monitor, and independent data set.
Table 2.
QSAR: rmsd, auc, and enrichment values per model
| Iteration | Number and type of descriptors | train | rmsd monitor | independent | auc | enrichment at 2% | |
|---|---|---|---|---|---|---|---|
| all | 1252 | 1–35 | 0.204 | 0.232 | 0.234 | 0.708 | 7.3 |
| scalar | 8 | 1–8 | 0.232 | 0.236 | 0.239 | 0.631 | 1.2 |
| 1 | 741 | 1–8, 14–16, 21–23, 25, 29–33, 35 | 0.224 | 0.224 | 0.227 | 0.703 | 7.1 |
| 2 | 578 | 1–8, 15–16, 23, 25, 30–33, 35 | 0.187 | 0.212 | 0.229 | 0.706 | 13.0 |
| 3 | 415 | 1–8, 15, 25, 30–31, 35 | 0.192 | 0.211 | 0.222 | 0.804 | 10.7 |
| jury | - | - | 0.159 | 0.214 | 0.207 | 0.732 | 15.4 |
Overall structure of the ANNs and the jury system
The trained ANNs consisted of the input layer with up to 1,252 chemical descriptors, the hidden layer consisting of eight neurons, and one neuron in the output layer predicting the ln(EC50) of the described molecule. The sigmoid function S(x) = 1/(1+exp(−x)) served as activation function of the neurons. The ANNs were trained by implementing resilient back-propagation of errors [34], a supervised learning approach. The training was terminated after up to 40,000 iterations when the monitoring dataset achieved its minimum rmsd. It took up to 13 hours per network using eight cores of a core2 quad 2.33GHz Intel Xeon microprocessor in parallel on the 64-bit version of Red Hat Enterprise Linux 5.2.
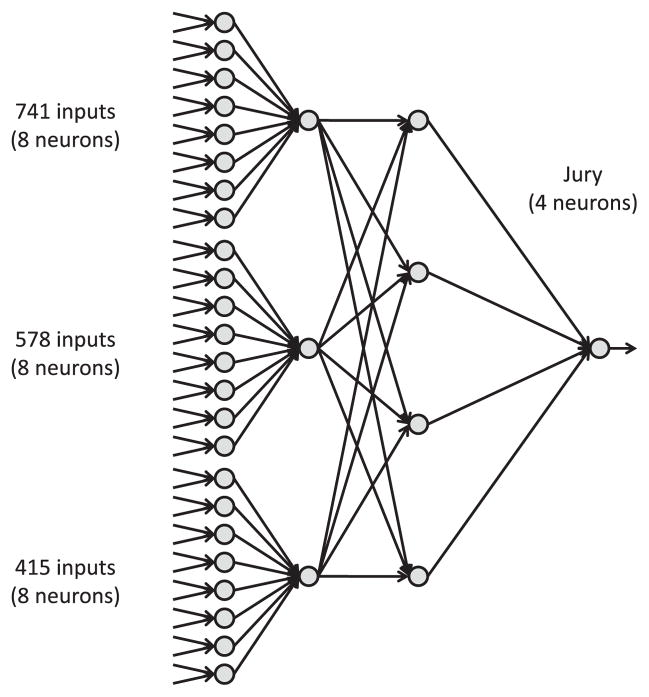
The outputs of the three best ANNs were used as input for a jury ANN that consisted of three inputs, four hidden neurons, and one output (Fig. 2). The training of the jury ANN terminated after 290 steps.
Fig. 2.
Schematic view of the jury system: The output of the best three ANNs according to the quality measures reported in Table 2 were employed as inputs for a jury ANN with four hidden neurons
Selection of the optimal set of descriptors of chemical structure
It is crucial to select the optimal set of descriptors from the 35 available categories. In a top-down approach, the least significant categories for predicting ln(EC50) were successively removed to increase the predictive power of the according ANNs. The advantage lies in removing degrees of freedom from the ANN by reducing the number of inputs. Since the number of data points stays the same, the signal-to-noise ratio improves. This procedure is described in detail elsewhere [29].
The input sensitivity of each of the 27 non-scalar descriptor categories was determined as norm over the individual input sensitivity values within this category. The descriptor categories were sorted by input sensitivity. In each step, categories comprising the least 10% of input sensitivity were removed. This process was repeated until the quality measures were optimized (see Table 2).
Enrichment and area under the curve complement rmsd as quality measures
Analysis of the rmsd proved to be a poor indicator for model quality (see Table 2). Hence, all models were also assessed in terms of their binary classification power using enrichment and area under the curve (auc) quality measures. Receiver operating characteristic (ROC) curves were generated as a measure to evaluate predictive power of the machine learning approaches. ROC curves plot the rate of true positives TP/P versus the rate of false positives FP/N of a binary classifier, here biological activity (see Fig. 3 and Fig. 4). TP and FP represent the numbers of known active and inactive compounds within a given subset of small molecules ordered by their predicted biological activity. Similarly, P and N represent the according numbers for the whole HTS dataset. The diagonal represents the performance expected from a random predictor. A higher auc of a ROC curve represents a better predictive power of the according model.
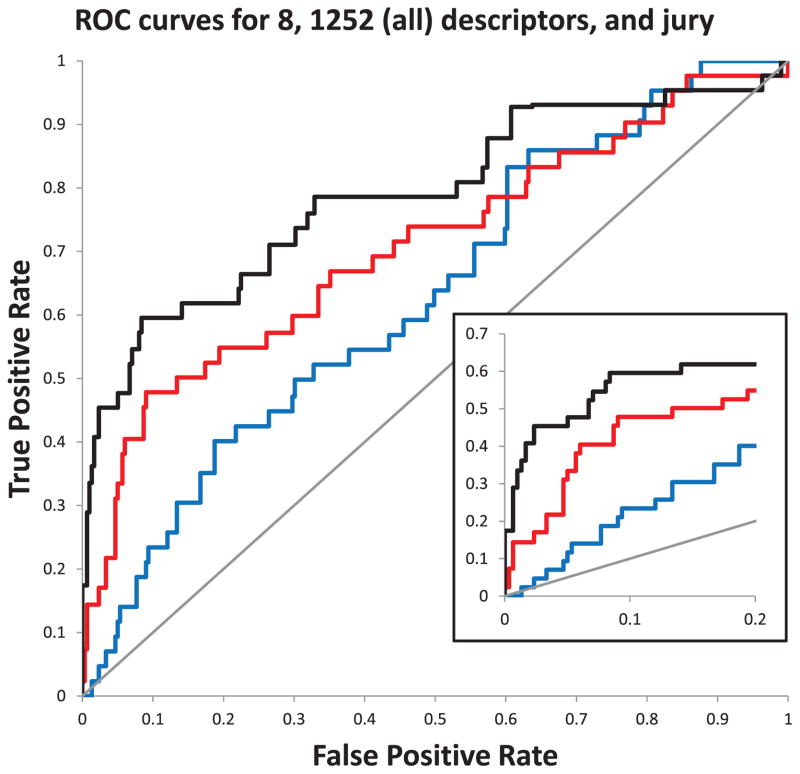
Fig. 3.
Receiver Operating Characteristic (ROC) curve plot for classical (blue), all (red), and jury (black) approach: This plot compares classical QSAR (eight scalar descriptors) with utilizing all (1,252) available ADRIANA descriptors and a jury approach. It demonstrates that ADRIANA descriptors add to the classical approach and that a jury approach improves performance even further. The inset shows the first 20% of the False Positive rate
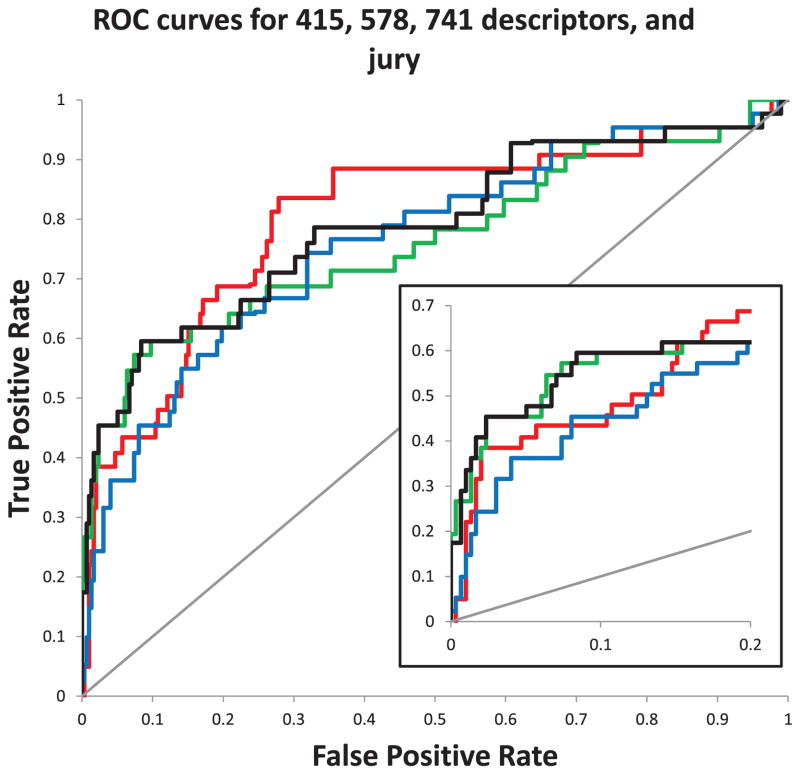
Fig. 4.
Receiver Operating Characteristic (ROC) curve plot for 415 (red), 578 (green), 741 (blue), and jury (black) approach: The three optimized descriptor sets (415, 578, 741) perform similarly well as the jury approach. However, the jury approach is more stable compared to the three other ANNs, as can be seen in Table 2. The inset shows the first 20% of the False Positive rate
The enrichment represents the expected factor by which the fraction of active compounds is increased in a small set of compounds resulting from an in silico virtual screen compared to the same fraction in the original HTS dataset (0.28%).
| (2) |
where P represents the total number of active compounds (positives) in the training dataset and N the total number of inactive compounds (negatives). TP stands for true positives – active compounds found in a compound library from virtual screening. FP are false positives – compounds predicted to be active that turn out to be inactive. Enrichments can be determined when experimentally testing a compound library obtained from a virtual screen or for an independent dataset set aside during training of the QSAR method. The enrichments reported in Table 2 were determined when selecting 2% of the compounds in the independent dataset predicted to be most active.
Implementation
The ANN and the Resilient Propagation training algorithm [34] were implemented in the BioChemistryLibrary (BCL). The BCL is an in house developed, object-oriented library written in the C++ programming language currently consisting of approximately 400 classes and 300,000 lines of code. Chemical descriptors were computed with ADRIANA [30, 33] based on three-dimensional structures generated by CORINA [32].
Results
Artificial Neural Networks (ANNs) were trained to predict the capability of drug-like molecules for allosteric potentiation of the metabotropic glutamate receptor subtype 4 (mGlu4) based on a High Throughput Screen (HTS) as reported by Niswender et al [11]. Commercially available databases of small molecules were virtually screened for novel PAMs of mGlu4. Hit compounds were verified experimentally in a human mGlu4 Gqi5-mediated calcium assay and a rat mGlu4 thallium flux assay.
Optimization of molecular descriptor set improves prediction results
An ANN was trained using only the scalar descriptors 1–8 to report a baseline performance using only naïve descriptors (Fig. 3 and Table 2) yielding an auc value of 0.631. The enrichment equals 1.2 at a compound cutoff of 2%. The relative rmsd value for the independent data set is 0.238. ‘Total Polarizable Surface Area’ was the input with the highest sensitivity (0.87) in this model with the other descriptor sensitivities ranging from 0.07 (‘Dipole Moment’) to 0.48 (‘Hydrogen Bond Acceptors’). The second baseline model involved all 1,252 descriptors as inputs. The auc and enrichment values improved to 0.708 and 7.3, respectively, while the rmsd dropped to 0.234.
In a 1st round of descriptor optimization, one third of the descriptor categories with the lowest input sensitivity were removed. Note that the scalar descriptors were kept in all models to facilitate comparison of input sensitivities with the baseline. This procedure leads to a final model containing 741 descriptors in 21 categories (see Table 2) without a significant change in model quality (auc: 0.703, rmsd: 0.227, enrichment: 7.1). The 2nd round yielded a model with 578 descriptors in 17 categories. The quality measures were better than the model with all descriptors with an auc of 0.706, rmsd of 0.229, and an improved enrichment of 13.0. The last iteration left 415 descriptors in 13 categories. While the auc value (0.804) and rmsd value (0.222) improved, the enrichment (10.7) dropped.
Jury model combines favorable features of all previous models
As these QSAR models have a comparable quality and enrichment values are affiliated with high uncertainties, a jury approach was tested to combine models. An ANN was trained on the output of the three ANNs with the reduced descriptor sets (see Fig. 2). This procedure improved the critical enrichment value to 15.4 and reduced the rmsd to 0.207. The auc value is with 0.732, a value lower than the 0.804 value reported for the ANN model with 415 descriptors. However, the reduced auc value this model results from the second half of the ROC curve, which is not employed when predicting molecules with high activities (Fig. 4).
Virtual screening of ChemBridge compound library
The ANN QSAR model was applied in a virtual screen of the ChemBridge EXPRESS-Pick collection of commercially available compounds. In silico screening of the entire library of ~450,000 compounds took approximately ten hours on a regular personal computer. A total of 1,108 compounds with predicted EC50 values below 3 μM for mGlu4 PAM activity were selected.
Screening these compounds in the human mGlu4/Gqi5 calcium mobilization assay as well as the rat mGlu4 thallium flux assay described in Niswender et al [31, 11] identified 168 primary hits which were then moved to screening in concentration-response curve format in both assays. 67 compounds were confirmed as potentiators in both assays, representing an enrichment of 67/1,108 × 156,184/434 = 22 relative to the initial experimental HTS hit rate. The experimentally observed enrichment is consistent with the enrichment values predicted from analysis of an independent dataset during development of the QSAR model, given the large uncertainty of these values (Table 2).
The EC50 of the 67 confirmed potentiators ranged from 0.4 μM to 15.8 μM with three compounds below 1 μM in the human Gqi5 assay and from 0.4 μM to >10 μM with three compounds below 1 μM in the rat thallium flux assay.
Simple similarity search fails to identify 54 out of 67 confirmed potentiators
To compare the ANN virtual screen with a more naïve similarity search, we selected 1,116 small molecules from the ChemBridge database based on MACCS structural key fingerprint similarity to known actives. The number of compounds was chosen to create a dataset of similar size to the one selected through the ANN. The artificially high Tanimoto coefficient cutoff of 96.7% indicates that a similarity search with a reasonable Tanimoto coefficient would create a dataset much too large for experimental verification. The intersection between this set of 1,116 compounds and the 67 potentiators identified by the ANN contains 13 compounds (19%) missing 54 compounds identified by the ANN virtual HTS.
Enrichment of 22 in known benzo-oxazoles scaffold
The rate of PAMs from the benzo-oxazole scaffold in the original HTS constituted 88 out of 26,180 (0.34%, MOE [35] similarity search employing MACCS fingerprints at 85% Tanimoto Superset/Subset.) A simple similarity search for benzo-oxazoles would preserve this rate while the virtual HTS enriched benzo-oxazoles by a factor of 8/133*156,146/432=22.
Discussion
Radial Distribution Functions (RDFs) are most important descriptors for predicting mGlu4 PAM activity
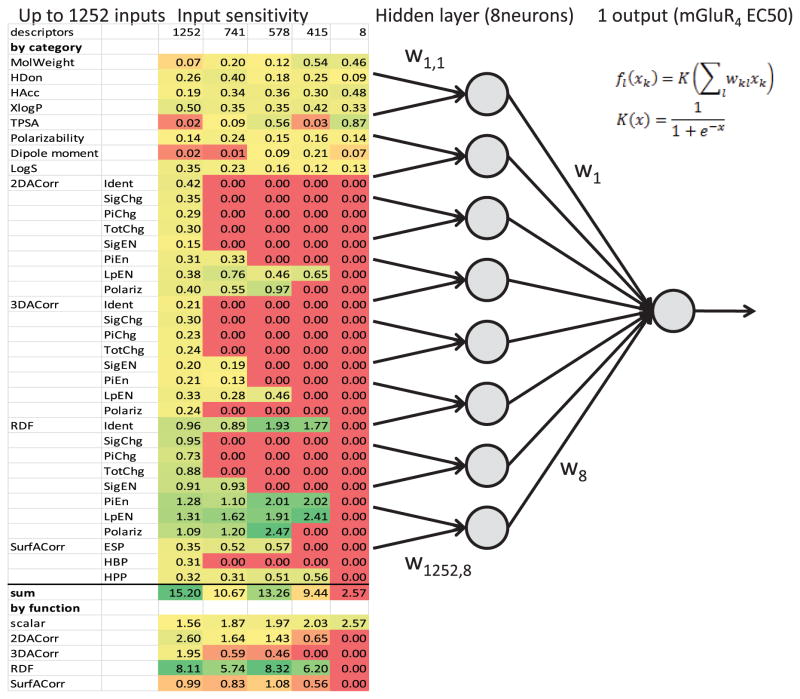
Several of the descriptor categories (see Table 1) employ the same chemical property but different encoding functions (2D vs. 3D auto-correlation and Radial Distribution Functions). Therefore, a descriptor optimization strategy was critical to identify the smallest set of descriptors needed for optimal QSAR models. Using this technique, the number of parameters (weights) in the ANNs is reduced, improving the signal-to-noise ratio for the trained models. To determine the ‘least necessary’ descriptor categories, the input sensitivity (see Methods) of each input with respect to the output of the ANN, i.e. biological activity prediction, was determined.
As more descriptors are removed from the inputs, the input sensitivity values increase for RDFs (see Fig. 5). Specifically, RDFs for π- and lone pair electronegativity play an important role. RDFs for identity and polarizability are featured most prominently in the model with 578 descriptors which is the best non-jury network (see Table 2). The importance of these descriptors immediately makes sense, since the active compounds of the original High Throughput Screening often feature phenyl rings and amide substructures that are well described by such RDFs.
Fig. 5.
Schematic view of an ANN: The input to the ANN consists of up to 1,252 descriptors in 35 categories. The weighted sum of the inputs is propagated through the activation function and fed into the hidden layer (8 neurons). The output is the predicted value of the logarithm of the EC50 of the small molecule towards potentiation of glutamate response at the metabotropic glutamate receptor 4. The heat map shows the input sensitivity of each category from lowest (red) to highest (green)
Virtual screening yields active compounds similar to known hits
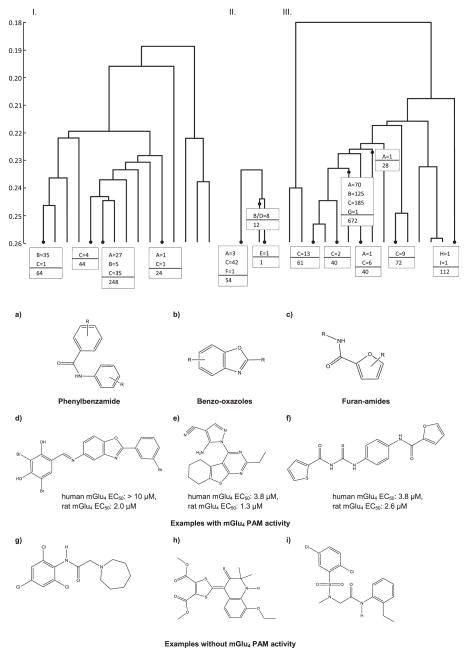
The 67 newly identified mGlu4 PAM compounds contained eight benzo-oxazoles, 42 furan-amides including 22 thioureas, and three phenylbenzamides. All three compound classes were represented in the original HTS hits [36, 37, 11] and featured modifications in R-groups (Fig. 6). Experimentally inactive compounds from these classes included 72 phenylbenzamides, 125 benzo-oxazoles, and 215 furanamides out of 1,041 inactive compounds.
Fig. 6.
Scaffold category analysis: (I) Scaffold composition of 432 mGlu4 PAMs from HTS. mGlu4 PAMs were clustered with the Mathematica package using the Tanimoto coefficient of the largest common substructure as distance measure. Three major scaffolds are constituted by 28 phenylbenzamides (6.5%, a), 40 benzo-oxazoles (9.3%, b), and 41 (9.5%, c) furan-amides. (II) Scaffold composition of 67 active compounds in the postscreen. (III) Scaffold composition of inactive compounds in the postscreen. Compounds d, e, and f are examples for active compounds identified by the virtual HTS, where g, h, and i were found to be inactive
Conformational ensembles could improve activity prediction from 3D molecular descriptors
CORINA provides one low energy conformation per small molecule. For flexible molecules multiple conformations of similar energy can exist. The conformation of the small molecule binding allosterically to the mGlu4 is then unknown. This shortcoming of the present approach could be addressed by generating an ensemble of low energy conformations for each small molecule and using the lowest predicted EC50 value in virtual screening. The molecular libraries employed in the present study are dominated by rather rigid molecules with few rotatable bonds. Therefore we speculate that the impact of additional conformational sampling is small.
Conclusions
Artificial Neural Networks were trained to generate QSAR models from an HTS experimental dataset of compounds with hmGlu4 Gqi5 assay activity. A jury system, based on the three ANN models, generated improved enrichments when compared to each individual model. The enrichment factor of 22 determined from biological testing of 1,100 compounds prioritized from a commercial library of ~450,000 substances demonstrates the predictive power of the method.
References
- 1.Marino MJ, Conn PJ. Glutamate-based therapeutic approaches: allosteric modulators of metabotropic glutamate receptors. Curr Opin Pharmacol. 2006;6 (1):98–102. doi: 10.1016/j.coph.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Marino MJ, Williams DL, O’Brien JA, Valenti O, McDonald TP, Clements MK, Wang R, DiLella AG, Hess JF, Kinney GG, Conn PJ. Allosteric modulation of group III metabotropic glutamate receptor 4: A potential approach to Parkinson’s disease treatment. Proc Natl Acad Sci U S A. 2003;100 (23):13668–13673. doi: 10.1073/pnas.1835724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marino M, Valenti O, Conn PJ. Glutamate receptors and Parkinson’s disease: opportunities for intervention. Drugs Aging. 2003;20 (5):377–397. doi: 10.2165/00002512-200320050-00006. [DOI] [PubMed] [Google Scholar]
- 4.Maj M, Bruno V, Dragic Z, Yamamoto R, Battaglia G, Inderbitzin W, Stoehr N, Stein T, Gasparini F, Vranesic I, Kuhn R, Nicoletti F, Flor PJ. (−)-PHCCC, a positive allosteric modulator of mGluR4: characterization, mechanism of action, and neuroprotection. Neuropharmacology. 2003;45 (7):895–906. doi: 10.1016/s0028-3908(03)00271-5. [DOI] [PubMed] [Google Scholar]
- 5.Mathiesen JM, Svendsen N, Bräuner-Osborne H, Thomsen C, Ramirez MT. Positive allosteric modulation of the human metabotropic glutamate receptor 4 (hmGluR4) by SIB-1893 and MPEP. Br J Pharmacol. 2003;138 (6):1026–1030. doi: 10.1038/sj.bjp.0705159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasparini F, Lingenhöhl K, Stoehr N, Flor PJ, Heinrich M, Vranesic I, Biollaz M, Allgeier H, Heckendorn R, Urwyler S, Varney MA, Johnson EC, Hess SD, Rao SP, Sacaan AI, Santori EM, Veliçelebi G, Kuhn R. 2-Methyl-6-(phenylethynyl)-pyridine (MPEP), a potent, selective and systemically active mGlu5 receptor antagonist. Neuropharmacology. 1999;38 (10):1493–1503. doi: 10.1016/s0028-3908(99)00082-9. [DOI] [PubMed] [Google Scholar]
- 7.Gregory KJ, Dong EN, Meiler J, Conn PJ. Allosteric Modulation of Metabotropic Glutamate Receptors: Structural Insights and Therapeutic Potential. Neuropharmacology. 2010 doi: 10.1016/j.neuropharm.2010.07.007. S0028-3908(10)00168-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindberg JS, Culleton B, Wong G, Borah MF, Clark RV, Shapiro WB, Roger SD, Husserl FE, Klassen PS, Guo MD, Albizem MB, Coburn JW. Cinacalcet HCl, an Oral Calcimimetic Agent for the Treatment of Secondary Hyperparathyroidism in Hemodialysis and Peritoneal Dialysis: A Randomized, Double-Blind, Multicenter Study. J Am Soc Nephrol. 2005;16 (3):800–807. doi: 10.1681/asn.2004060512. [DOI] [PubMed] [Google Scholar]
- 9.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Maraviroc (UK-427,857), a Potent, Orally Bioavailable, and Selective Small-Molecule Inhibitor of Chemokine Receptor CCR5 with Broad-Spectrum Anti-Human Immunodeficiency Virus Type 1 Activity. Antimicrob Agents Chemother. 2005;49 (11):4721–4732. doi: 10.1128/aac.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3 (8):711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 11.Niswender CM, Johnson KA, Weaver CD, Jones CK, Xiang Z, Luo Q, Rodriguez AL, Marlo JE, de Paulis T, Thompson AD, Days EL, Nalywajko T, Austin CA, Williams MB, Ayala JE, Williams R, Lindsley CW, Conn PJ. Discovery, Characterization, and Antiparkinsonian Effect of Novel Positive Allosteric Modulators of Metabotropic Glutamate Receptor 4. Mol Pharmacol. 2008;74 (5):1345–1358. doi: 10.1124/mol.108.049551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engers DW, Niswender CM, Weaver CD, Jadhav S, Menon UN, Zamorano R, Conn PJ, Lindsley CW, Hopkins CR. Synthesis and Evaluation of a Series of Heterobiarylamides That Are Centrally Penetrant Metabotropic Glutamate Receptor 4 (mGluR4) Positive Allosteric Modulators (PAMs) J Med Chem. 2009;52 (14):4115–4118. doi: 10.1021/jm9005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niswender CM, Lebois EP, Luo Q, Kim K, Muchalski H, Yin H, Conn PJ, Lindsley CW. Positive allosteric modulators of the metabotropic glutamate receptor subtype 4 (mGluR4): Part I. Discovery of pyrazolo[3,4-d]pyrimidines as novel mGluR4 positive allosteric modulators. Bioorg Med Chem Lett. 2008;18 (20):5626–5630. doi: 10.1016/j.bmcl.2008.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams R, Niswender CM, Luo Q, Le U, Conn PJ, Lindsley CW. Positive allosteric modulators of the metabotropic glutamate receptor subtype 4 (mGluR4). Part II: Challenges in hit-to-lead. Bioorg Med Chem Lett. 2009;19 (3):962–966. doi: 10.1016/j.bmcl.2008.11.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams R, Zhou Y, Niswender CM, Luo Q, Conn PJ, Lindsley CW, Hopkins CR. Re-exploration of the PHCCC Scaffold: Discovery of Improved Positive Allosteric Modulators of mGluR4. ACS Chem Neurosci. 2010;1 (6):411–419. doi: 10.1021/cn9000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todeschini R, Consonni V. Methods Princ Med Chem. Vol. 11. Wiley - VCH; 2000. Handbook of Molecular Descriptors. [Google Scholar]
- 17.Hansch C, Hoekman D, Leo A, Weininger D, Selassie CD. Chem-bioinformatics: comparative QSAR at the interface between chemistry and biology. Chem Rev. 2002;102(3):783–812. doi: 10.1021/cr0102009. cr0102009 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Hansch CM, Peyton P, Fujita Toshio, Muir Robert M. Correlation of biological activity of phenoxyacetic acids with Hammett substituent constants and partition coefficients. Nature. 1962;(194):178–180. [Google Scholar]
- 19.Cramer RD, Patterson DE, Bunce JD. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc. 1988;110 (18):5959–5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 20.Klebe G, Abraham U, Mietzner T. Molecular Similarity Indices in a Comparative Analysis (CoMSIA) of Drug Molecules to Correlate and Predict Their Biological Activity. J Med Chem. 1994;37 (24):4130–4146. doi: 10.1021/jm00050a010. [DOI] [PubMed] [Google Scholar]
- 21.Bleckmann A, Meiler J. Epothilones: Quantitative Structure Activity Relations Studied by Support Vector Machines and Artificial Neural Networks. QSAR Comb Sci. 2003;22 (7):719–721. [Google Scholar]
- 22.Tetko IV, Sushko I, Pandey AK, Zhu H, Tropsha A, Papa E, Oberg T, Todeschini R, Fourches D, Varnek A. Critical assessment of QSAR models of environmental toxicity against Tetrahymena pyriformis: focusing on applicability domain and overfitting by variable selection. J Chem Inf Model. 2008;48 (9):1733–1746. doi: 10.1021/ci800151m. [DOI] [PubMed] [Google Scholar]
- 23.Winkler D. Neural networks as robust tools in drug lead discovery and development. Mol Biotechnol. 2004;27 (2):139–167. doi: 10.1385/MB:27:2:139. [DOI] [PubMed] [Google Scholar]
- 24.Zupan J, Gasteiger J. Neural Networks for Chemists. VCH Verlagsgesellschaft mbH; Weinheim: 1993. [Google Scholar]
- 25.Burton J, Ijjaali I, Barberan O, Petitet F, Vercauteren DP, Michel A. Recursive Partitioning for the Prediction of Cytochromes P450 2D6 and 1A2 Inhibition: Importance of the Quality of the Dataset. J Med Chem. 2006;49 (21):6231–6240. doi: 10.1021/jm060267u. [DOI] [PubMed] [Google Scholar]
- 26.Hecht D, Cheung M, Fogel GB. QSAR using evolved neural networks for the inhibition of mutant PfDHFR by pyrimethamine derivatives. BioSystems. 2008;92 (1):10–15. doi: 10.1016/j.biosystems.2007.10.005. S0303-2647(07)00164-5 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Hecht D, Fogel GB. High-throughput ligand screening via preclustering and evolved neural networks. IEEE/ACM Trans Comput Biol Bioinform. 2007;4 (3):476–484. doi: 10.1109/tcbb.2007.1038. [DOI] [PubMed] [Google Scholar]
- 28.Butkiewicz M, Mueller R, Selic D, Dawson E, Meiler J. Application of Machine Learning Approaches on Quantitative Structure Activity Relationships. Paper presented at the IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology; Nashville. 2009. [Google Scholar]
- 29.Mueller R, Rodriguez AL, Dawson ES, Butkiewicz M, Nguyen TT, Oleszkiewicz S, Bleckmann A, Weaver CD, Lindsley CW, Conn PJ, Meiler J. Identification of Metabotropic Glutamate Receptor Subtype 5 Potentiators Using Virtual High-Throughput Screening. ACS Chem Neurosci. 2010;1 (4):288–305. doi: 10.1021/cn9000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwab CH, Gasteiger J. Computerchemie MNG, Program Description. 2006. ADRIANA.Code, Algorithms for the Encoding of Molecular Structures, Version 2.0. [Google Scholar]
- 31.Niswender CM, Johnson KA, Luo Q, Ayala JE, Kim C, Conn PJ, Weaver CD. A Novel Assay of Gi/o-Linked G Protein-Coupled Receptor Coupling to Potassium Channels Provides New Insights into the Pharmacology of the Group III Metabotropic Glutamate Receptors. Mol Pharmacol. 2008;73 (4):1213–1224. doi: 10.1124/mol.107.041053. [DOI] [PubMed] [Google Scholar]
- 32.Gasteiger J, Rudolph C, Sadowski J. Automatic generation of 3D atomic coordinates for organic molecules. Tetrahedron Comput Methodol. 1990;3 (6c):537–547. [Google Scholar]
- 33.Schwab CH. ADRIANA.Code. 2.0. Molecular Networks GmbH; Erlangen, Germany: 2007. [Google Scholar]
- 34.Riedmiller M, Rprop Braun H. A Fast Adaptive Learning Algorithm. International Symposium on Computer and Information Science VII.1992. [Google Scholar]
- 35.Molecular Operating Environment (MOE) 2010.10. Chemical Computing Group Inc; 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: 2010. [Google Scholar]
- 36.Engers DW, Field JR, Le U, Zhou Y, Bolinger JD, Zamorano R, Blobaum AL, Jones CK, Jadhav S, Weaver CD, Conn PJ, Lindsley CW, Niswender CM, Hopkins CR. Discovery, Synthesis, and Structure–Activity Relationship Development of a Series of N-(4-Acetamido)phenylpicolinamides as Positive Allosteric Modulators of Metabotropic Glutamate Receptor 4 (mGlu4) with CNS Exposure in Rats. J Med Chem. 2011;54 (4):1106–1110. doi: 10.1021/jm101271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engers DW, Gentry PR, Williams R, Bolinger JD, Weaver CD, Menon UN, Conn PJ, Lindsley CW, Niswender CM, Hopkins CR. Synthesis and SAR of novel, 4-(phenylsulfamoyl)phenylacetamide mGlu4 positive allosteric modulators (PAMs) identified by functional high-throughput screening (HTS) Bioorg Med Chem Lett. 2010;20 (17):5175–5178. doi: 10.1016/j.bmcl.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]