Abstract
Objective
The aim of this study was to investigate the genetic association of the FAT gene with schizophrenia in the Korean population, as well as analyzing the association of FAT gene with clinical variables.
Methods
Four variants within the FAT gene were investigated in 189 patients with schizophrenia and 119 healthy controls (rs2306987 A/C, rs2306990 T/C, rs2637777 G/T, and rs2304865 G/C).
Results
Significant association at the rs273777 with schizophrenia was observed; however, rs2306987, rs2306990, and rs2304865 were not associated with schizophrenia. Haplotype analyses revealed that the haplotype A/T/T/G was associated with a significantly protective effect. Sliding window analysis (rs2637777 G/T and rs2304865 G/C) revealed the more common T/G haplotype, included in the A/T/T/G protective combination, showed a small protective effect, in particular the effect was due to the rs273777 T variant (minor allele).
Conclusion
The present finding suggests that FAT polymorphism may play a putative role in the susceptibility to schizophrenia in the Korean population. Further studies using a larger number of subjects should be performed to determine whether the FAT gene polymorphism may be truly involved in the development of schizophrenia.
Keywords: Association, Cadherins, Polymorphism, Schizophrenia, Korean
INTRODUCTION
Despite continuing efforts aimed at identifying the etiologies of schizophrenia, little is known about the definite causes of the disorder. Disturbance of prenatal brain development and/or postnatal brain maturation in the context of the pathology of schizophrenia is increasingly recognized as one potential cause of schizophrenia.1,2) In this context, cadherin, which is a cell adhesion molecule, is of critical importance to morphogenesis in the central nervous system during embryonic development. They also play a role in neuronal differentiation and synaptogenesis, processes that are believed to underlie the development of schizophrenia.3-5)
The cadherin family consists of nearly 100 different genes scattered throughout the genome either as separate entities or as members of tandem clusters that arose through gene duplication. Recently a gene that is closely related to the classical cadherin genes family has gained the growing attention. There is the coherent evidence suggesting a possible role of classical cadherin in the development of psychiatric disorders.6)
A cadherin gene, FAT (the homolog of the Drosophila tumor suppressor gene fat) on 4q35.2, has been implicated in the development of bipolar disorder.7-10) Increasing evidence suggests that bipolar disorder and schizophrenia have some etiological overlap, which is at least partially due to genetic factors.11-13) Also notable is the fact that the role of FAT in neuronal migration and positioning during development makes it a good candidate gene for schizophrenia. However, no work has yet examined the association between FAT gene and schizophrenia. In this study, we examined FAT gene in the Korean population to determine the association of the FAT gene and schizophrenia in conjunction with clinical variables.
METHODS
The patient group consisted of 189 inpatients with schizophrenia. Ninety were male and 99 were female and the mean age of the participants was 36.4±8.8 years. The diagnosis was done by the consensus of two board-certified psychiatrists (T.Y.J.; Y.E.J) using a Structured Clinical Interview for the 4th edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) Axis I disorders-Clinical Version.14) Subjects who had neurological illness, endocrine disorders, autoimmune disease, or other DSM-IV Axis I diseases were excluded from this study. Clinical variables such as age, age of onset, duration of illness, and history of suicide attempts were collected. Symptom severity was assessed using the Clinical Global Impression Severity Scale (CGI-S)15) and Positive And Negative Syndrome Scale (PANSS).16)
One hundred and nineteen voluntary controls were recruited from the personnel and medical students of the Yeouido St. Mary's Hospital (male 51, female 68; mean age 36.3±9.8 years). Before blood sample collection, a semi-structured interview was performed to determine whether the control subjects had current psychiatric problems, or had history of psychiatric or neurological illness. There was no difference in the distribution of gender (p=0.41) and age (p=0.90) between patients and controls.
All subjects were biologically unrelated and were native Koreans residing in the Republic of Korea. The objectives and procedures of the study were explained to all subjects and written informed consent was obtained. The Ethics Committee of the Yeouido St. Mary's Hospital, The Catholic University of Korea, approved this study.
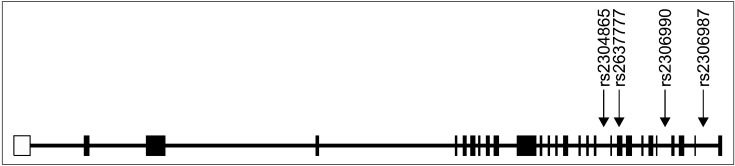
Genomic DNA was extracted from blood by standard methods and quantified. The high-throughput genotyping method using pyrosequencer (Biotage AB, Uppsala, Sweden) was used for genotyping four single nucleotide polymorphisms (SNPs) (rs2306987, rs2306990, rs2637777 and rs2304865) within the FAT gene, which were selected based on public database (National Center for Biotechnology Information, dbSNP, http://www.ncbi.nlm.nih.gov/SNP/) and data from previous studies.7,8) Polymerase chain reactions (PCR) primers (Bioneer, Daejeon, Korea) and sequencing primers (Bioneer) used for the pyrosequencing assay were designed using the Pyrosequencing Assay Design Software (ver 1.0; Biotage AB, Upssala, Sweden) and one primer of each primer set was biotinylated. A map of the positions of the four SNPs in FAT gene is shown in Fig. 1.
Fig. 1.
Gene map and location of single nucleotide polymorphisms (SNPs) in FAT gene.
Haploview 4.2 (Daly Lab, Broad Institute, Cambridge, MA, USA) was used to generate a linkage disequilibrium (LD) map and to test for Hardy-Weinberg equilibrium (HWE). Association for single markers and clinical variables were performed using the chi-square, t-test and the analysis of variance. Tests for associations using multimarker haplotypes were performed using the statistics software "R" (http://www.R-project.org), package 'haplo.stat'. Permutations (n=1,000) were performed to estimate the global significance of the results for all haplotype analyses and to validate the expectation maximization values. Rare haplotypes (<1%) were excluded from the analysis. All p-values were two-tailed, and in accordance with Bonferroni correction for multiple testing (four tests), statistical significance was conservatively set at 0.0125 to reduce the likelihood of false-positive results.
RESULTS
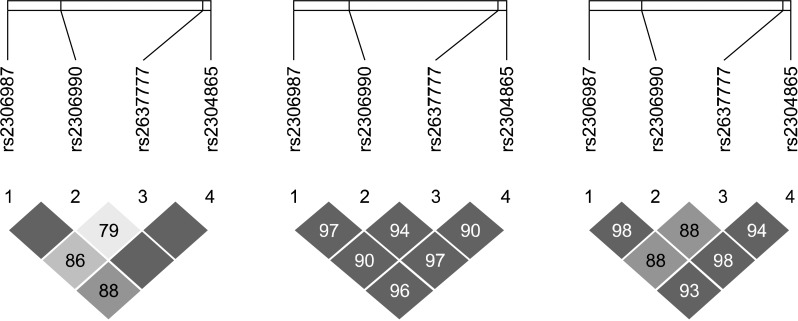
Genotypes of all markers were in HWE in both patients with schizophrenia and the controls. All markers were in HWE: rs2306987(A/T), p=0.9040; rs2306990(T/C), p=0.6254; rs2637777(G/T), p=0.0705; rs2304865(G/C), p=0.2780. All investigated loci were in strong LD in the controls and in patients as well as in whole subjects (Fig. 2).
Fig. 2.
Linkage disequilibrium (LD) mapping in FAT gene. All investigated loci were in strong LD in the control (left) and in the patients (middle) as well as in whole samples (right).
Single marker analyses are presented in Table 1. Schizophrenia was not associated with rs2306987, rs2306990, and rs2304865. A significant difference in genotypic distributions was observed between schizophrenia and controls in rs2637777, where the rs2737777 G/T genotype was more represented in the control than in patients with schizophrenia (p=0.006). Haplotype analyses revealed that the haplotype A/T/T/G was associated with a significant protective effect (see Table 2 for each haplotype's details). Sliding window analysis of two SNPs (rs2637777 G/T, rs2304865 G/C) revealed the more common T/G haplotype, included in the A/T/T/G protective combination, showed a small protective effect, in particular the effect was due to the rs273777 T variant (minor allele) (Table 3).
Table 1.
Genotype and alleles distribution in 4 FAT SNPs in patients (n=189) and the controls (n=119)
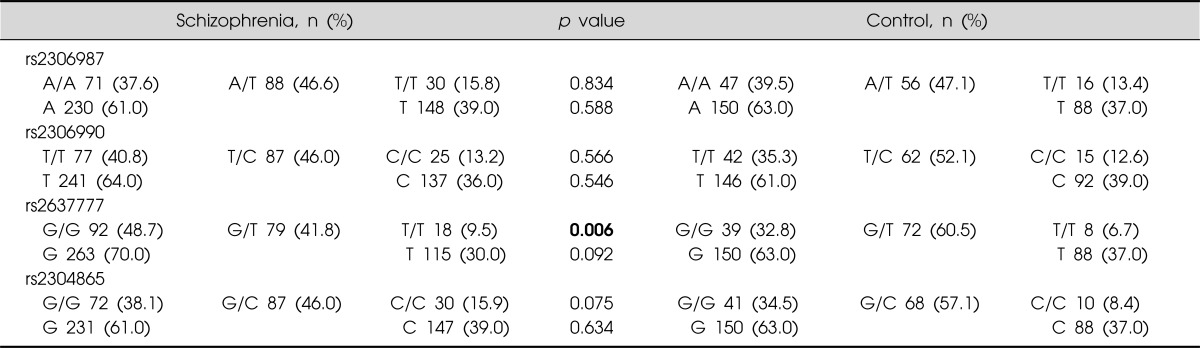
SNP, single nucleotide polymorphisms.
Bold number indicates statistically significant.
Table 2.
Haplotype analysis in patients and the controls
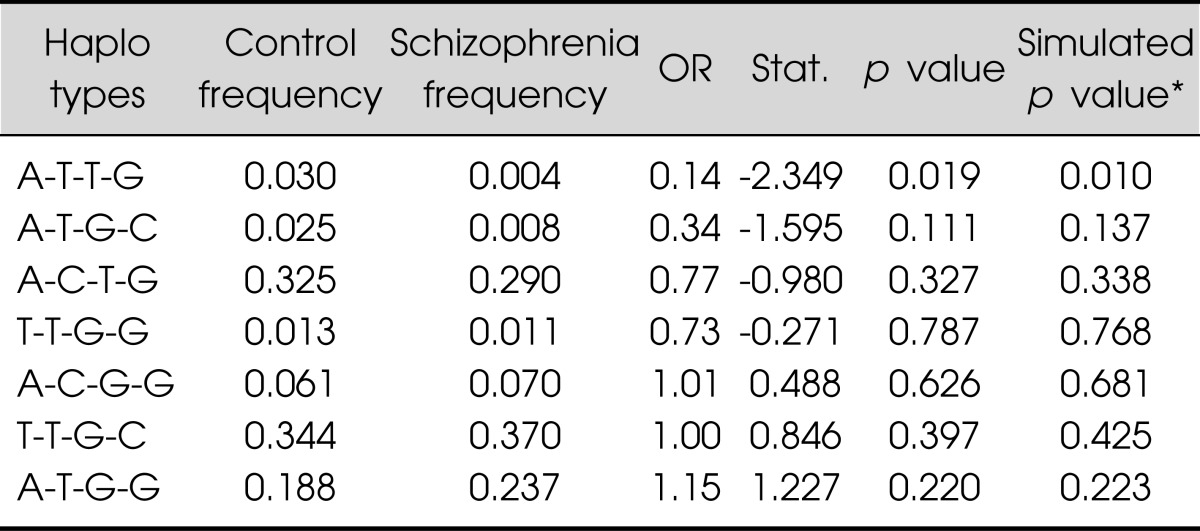
Haplotype analysis in patients and the controlsHaplotypes comprised of rs2306987, rs2306990, rs2637777 and rs2304865. Haplotypes whose frequencies were estimated >1% were described. Bold number indicates significant association.
*1,000 simultations.
OR, odds ratio.
Table 3.
Results of 2 haplotypes analysis with rs2637777 and rs2304865
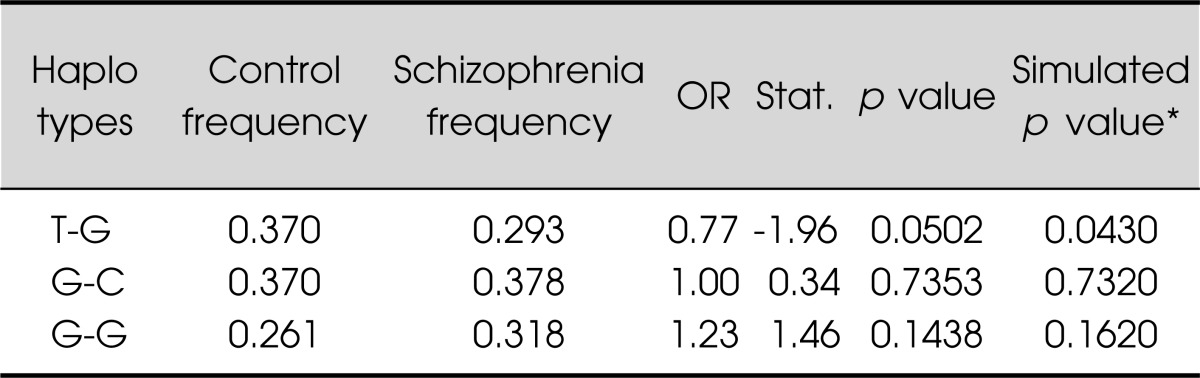
Haplotypes whose frequencies were estimated >1% were described.
*1,000 simultations.
OR, odds ratio.
No association was observed with the other clinical variables, such as age of onset, history of suicide attempts, diagnostic subtype (paranoid subtype vs. non-paranoid subtype), duration of illness, and symptom severity score (Table 4).
Table 4.
FAT SNPs stratified for demographic and clinical data in patients with schizophrenia (n=189)
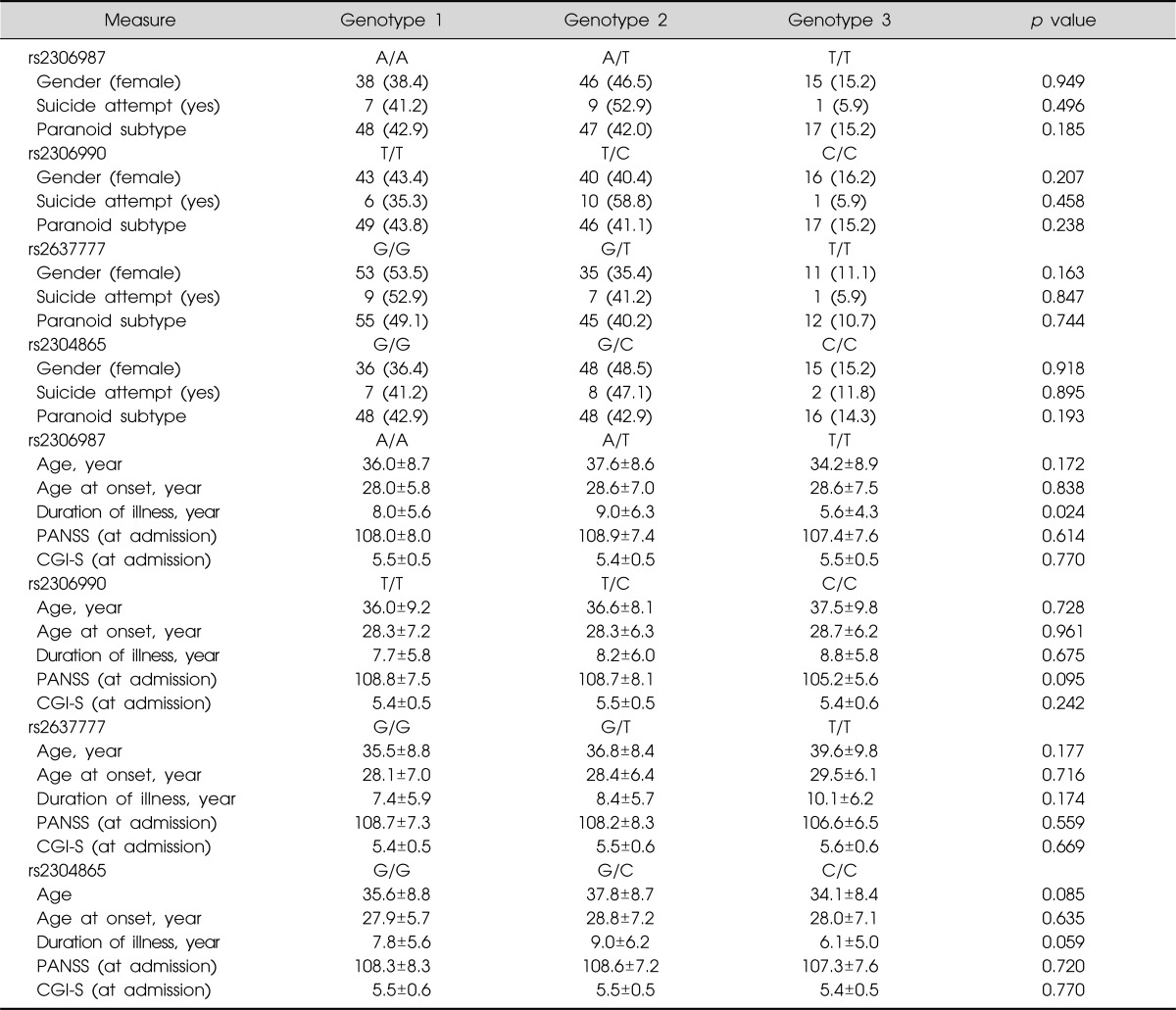
Values are presented as number (%) or mean±standard deviation.
SNP, single nucleotide polymorphisms; PANSS, Positive And Negative Syndrome Scale; CGI-S, Clinical Global Impression Severity.
DISCUSSION
This study was the first attempt to investigate the association of FAT gene polymorphism with schizophrenia. In the present study, we found significant association at the rs273777, and the rs273777 T variant (minor allele) was found to be protective against schizophrenia. On the other hand, rs2306987, rs2306990, and rs2304865 were not found to be associated with schizophrenia in the Korean population sample examined in this study.
We tested the four markers (rs2306987, rs2306990, rs2637777 and rs2304865) that showed the strongest association in previous studies. Blair et al.8) performed systematic LD studies in the candidate region and produced the evidence that the susceptibility to bipolar disorder was conferred by a haplotype block at the 3' end of the cadherin gene FAT (rs2306987, rs1298865, rs2306990, rs2637777 and rs2304865). Abou Jamra et al.7) recently reported that bipolar disorder had significantly relevance to nine SNPs in FAT (rs4862718, rs7683023, rs2306987, rs1298865, rs2306990, rs10009030, rs2637777, rs2304865 and rs32843. However, between the studies, the tendencies of risk allele frequencies, genetic distribution, or haplotypes were different. More recently, Pae et al.17) reported that, in schizophrenia patients, the FAT gene did not play a role in the response to aripiprazole. The most common haplotype of this study consisted of the common alleles of each SNP (T/T/G/C), which was similar pattern of allele and genotype frequencies with Pae et al.'s data. They suggested that their work should be considered preliminary, because no work has yet confirmed either an association between the FAT gene and response to mood stabilizing agents or an association between FAT and schizophrenia.
Our results raise the possibility that FAT gene variants might be involved in the development of schizophrenia. However, several issues should be noted in the present study. The major limitations of the present study were the relatively small sample size and the lack of genomic control, which is liable to result in a stratification bias. Further, the significant haplotype (A/T/T/G) is one of rare haplotype (3.0% in controls, 0.4% in patients), therefore the variance explained by the present association is very low. Positive associations sometimes could be due to genetic heterogeneity, and protective haplotypes could vary according to ethnic differences. Thus, further studies using a larger number of subjects in different ethnic groups should be performed to determine whether the FAT gene polymorphism may be truly involved in the development of schizophrenia. Also, the choice of SNPs was based on previous research and focused only on the 3' of the FAT gene. Accordingly it is necessary that further research provide more complete coverage of the FAT gene and investigate other variants that may have an effect on the expression of the gene.
Acknowledgments
This study was supported by a grant of the Korean Healthcare technology R & D Project, Ministry of Health and Welfare, Republic of Korea (A102065).
References
- 1.Halliday GM. A review of the neuropathology of schizophrenia. Clin Exp Pharmacol Physiol. 2001;28:64–65. doi: 10.1046/j.1440-1681.2001.03398.x. [DOI] [PubMed] [Google Scholar]
- 2.Weinberger DR. From neuropathology to neurodevelopment. Lancet. 1995;346:552–557. doi: 10.1016/s0140-6736(95)91386-6. [DOI] [PubMed] [Google Scholar]
- 3.Brusés JL. Cadherin-mediated adhesion at the interneuronal synapse. Curr Opin Cell Biol. 2000;12:593–597. doi: 10.1016/s0955-0674(00)00137-x. [DOI] [PubMed] [Google Scholar]
- 4.Huntley GW, Gil O, Bozdagi O. The cadherin family of cell adhesion molecules: multiple roles in synaptic plasticity. Neuroscientist. 2002;8:221–233. doi: 10.1177/1073858402008003008. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 6.Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 7.Abou Jamra R, Becker T, Georgi A, Feulner T, Schumacher J, Stromaier J, et al. Genetic variation of the FAT gene at 4q35 is associated with bipolar affective disorder. Mol Psychiatry. 2008;13:277–284. doi: 10.1038/sj.mp.4002111. [DOI] [PubMed] [Google Scholar]
- 8.Blair IP, Chetcuti AF, Badenhop RF, Scimone A, Moses MJ, Adams LJ, et al. Positional cloning, association analysis and expression studies provide convergent evidence that the cadherin gene FAT contains a bipolar disorder susceptibility allele. Mol Psychiatry. 2006;11:372–383. doi: 10.1038/sj.mp.4001784. [DOI] [PubMed] [Google Scholar]
- 9.McInnis MG, Lan TH, Willour VL, McMahon FJ, Simpson SG, Addington AM, et al. Genome-wide scan of bipolar disorder in 65 pedigrees: supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol Psychiatry. 2003;8:288–298. doi: 10.1038/sj.mp.4001277. [DOI] [PubMed] [Google Scholar]
- 10.Willour VL, Zandi PP, Huo Y, Diggs TL, Chellis JL, MacKinnon DF, et al. Genome scan of the fifty-six bipolar pedigrees from the NIMH genetics initiative replication sample: chromosomes 4, 7, 9, 18. Am J Med Genet B Neuropsychiatr Genet. 2003;121B:21, 27. doi: 10.1002/ajmg.b.20051. [DOI] [PubMed] [Google Scholar]
- 11.Bramon E, Sham PC. The common genetic liability between schizophrenia and bipolar disorder: a review. Curr Psychiatry Rep. 2001;3:332–337. doi: 10.1007/s11920-001-0030-1. [DOI] [PubMed] [Google Scholar]
- 12.Ivleva E, Thaker G, Tamminga CA. Comparing genes and phenomenology in the major psychoses: schizophrenia and bipolar 1 disorder. Schizophr Bull. 2008;34:734–742. doi: 10.1093/schbul/sbn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier W, Höfgen B, Zobel A, Rietschel M. Genetic models of schizophrenia and bipolar disorder: overlapping inheritance or discrete genotypes? Eur Arch Psychiatry Clin Neurosci. 2005;255:159–166. doi: 10.1007/s00406-005-0583-9. [DOI] [PubMed] [Google Scholar]
- 14.First MB, Spitzer RL, Gibbon M, Williams JB. Structured clinical interview for DSM-IV axis I disorders-clinician version (SCID-CV) Washington, DC: American Psychiatric Press; 1997. [Google Scholar]
- 15.Guy W. Clinical global impression. Assessment manual for psychopharmacology. Washington, DC: Department of Health Education and Welfare; 1976. [Google Scholar]
- 16.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13:261–276. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 17.Pae CU, Chiesa A, Mandelli L, De Ronchi D, Serretti A. No influence of FAT polymorphisms in response to aripiprazole. J Hum Genet. 2010;55:32–36. doi: 10.1038/jhg.2009.117. [DOI] [PubMed] [Google Scholar]