Abstract
A fully integrated platform was developed for capturing/fractionating human fucome from disease-free and breast cancer sera. It comprised multicolumn operated by HPLC pumps and switching valves for the simultaneous depletion of high abundance proteins via affinity-based subtraction and the capturing of fucosylated glycoproteins via lectin affinity chromatography followed by the fractionation of the captured glycoproteins by reversed phase chromatography (RPC). Two lectin columns specific to fucose, namely Aleuria aurantia lectin (AAL) and Lotus tetragonolobus agglutinin (LTA) were utilized. The platform allowed the “cascading” of the serum sample from column-to-column in the liquid phase with no sample manipulation between the various steps. This guaranteed no sample loss and no propagation of experimental biases between the various columns. Finally, the fucome was fractionated by RPC yielding desalted fractions in volatile acetonitrile-rich mobile phase, which after vacuum evaporation were subjected to trypsinolysis for LC-MS/MS analysis. This permitted the identification of the differentially expressed proteins (DEP) in breast cancer serum yielding a broad panel of 35 DEP from the combined LTA and AAL captured proteins and a narrower panel of 8 DEP that were commonly differentially expressed in both LTA and AAL fractions, which are considered as more representative of cancer altered fucome.
Keywords: Affinity chromatography, Breast cancer, Glycoproteins, Multidimensional liquid chromatography, Differential protein expression
1. Introduction
Protein glycosylation is one of the most common and complex post-translational modifications of proteins. In fact, glycosylation affects more than half of all the proteins in an eukaryotic cell [1, 2]. Protein glycosylation can occur at more than one position in the amino acid sequence, and the glycans at even a single position may be heterogeneous or may be missing from some protein molecules. This yields the so-called glycoforms, and the relative proportions of the glycoforms are found to be reproducible, not random, and depend mainly on the environment in which the protein is glycosylated [3].
It has been well known for decades that glycoproteins in eukaryotic cells, undergo changes in their glycosylation with the onset of and during cancer and inflammation (for recent reviews, see refs. [4, 5]). For instance, cancer associated changes in glycosylation include both the under- and over-expression of naturally occurring glycans. These structures most often arise from changes in the expression levels of glycosylating enzymes such as glycosyltransferases and glycosidases in cancerous cells versus healthy cells [6]. Therefore, changes in glycoprotein expression are often a hallmark of disease states. Most often, the disease is associated with changes in a set of glycoproteins, or the so-called sub-glycoproteomics. On this basis, profiling the glycoproteins that are most affected in biologically diseased matrices and fluids should in principle be sufficient, and represents a promising strategy for determining glycoprotein biomarkers. This sub-glycoproteomics approach to find glycoprotein biomarkers of carcinoma or chronic inflammation would involve the selective isolation and targeting of the sub-glycoproteomics from serum samples or that are shed or secreted by diseased cell lines. Of interest to the present research report is a major glycan alteration that involves increased Lewis (Le) antigen expression in many epithelial cancers, which are synthesized by fucosyltransferases I-VIII, which add fucose in a (1,2/3/4) or (6)-linkage to galactose (Gal) or N-acetylglucosamine (GlcNAc) residues [7]. Thus, the sub-glycoproteomics involving the selective isolation and targeting of aberrantly fucosylayted proteins (or the so called fucome) is the major objective of this research report in the aim of providing a promising strategy for revealing differentially expressed proteins.
When focusing on profiling specific sub-glycoproteomics, it becomes necessary to remove some of the high abundance proteins such as albumin (HSA), which represents ~50% of the total mass of serum proteins and immunoglobulins (Ig’s) which represent an additional ~20% of the total mass of serum proteins. The removal of this group of high abundance proteins was achieved in the present investigation by immunosubtraction for HSA and microbial protein-based affinity subtraction (e.g., protein A and protein G′) for Ig’s. For a very recent review on depletion of high abundance proteins by a variety of approaches, see Selvaraju and El Rassi [8].
To capture a given sub-glycoproteomics, e.g., the fucome, lectin affinity chromatography (LAC) offers the potential to achieve this goal [9]. In fact, LAC is an important tool to study and unlock glycosylation. LAC generally results in an enrichment of classes of different glycoproteins, possessing similar carbohydrate determinants recognized by the immobilized lectin and called “lectin receptors”. Therefore, LAC should facilitate the profiling of specific sub-glycoproteomics, e.g., the fucome. Indeed, LAC has been used to selectively capture sub-glycoproteomics in various formats including single, serial-LAC, and multi-LAC (M-LAC) [10–17]. Serial LAC refers to using LAC in a sequential or serial fashion where the pass through fraction from one lectin column is collected and further analyzed on a second lectin column and so forth. Serial LAC involves excessive discontinuous sample manipulation (e.g., dialysis, concentration, drying, reconstitution), which causes sample loss and propagation of experimental biases from step-to-step or column-to-column. Multi-LAC (M-LAC) refers to using a mixture of immobilized lectins having complementary specificities in a given column, i.e., mixed bed column. After loading the sample onto the column, the M-LAC column is eluted sequentially using specific displacer (i.e., haptenic sugar) for each lectin. Sequential elution may not be a clear cut, and some glycoproteins captured by another lectin may be concurrently eluted (cross inhibitory reactivity). Also, since most lectins are glycoproteins, M-LAC may introduce a lowering in the capturing capacity of each lectin due to possible bead-to-bead interactions in a mixed bed column.
This research report is aimed at eliminating the above drawbacks by achieving the on line depletion of some high abundance proteins (i.e., HSA and Ig’s) simultaneously with the capturing of specific sub-glycoproteomics (i.e., the fucome) followed by the fractionation of the captured glycoproteins using reversed phase chromatography (RPC). This entailed the design of a platform that operates multicolumn columns via switching valves and incorporates multiple high precision HPLC pumps.
With the intention to capture the fucosylated serum glycoproteins (i.e., the fucome), two fucose specific lectins namely, Aleuria aurantia lectin (AAL) and Lotus tetragonolobus agglutinin (LTA) were immobilized onto the surface of glyceryl methacrylate (GMM)/pentaerythritol triacrylate (PETA) monolith. The GMM-PETA monolith was very recently introduced by Gunasena and El Rassi for performing immuno affinity chromatography at reduced nonspecific interactions [18]. Immobilized AAL has a strong affinity towards core fucosylated glycans (i.e.,) where a fucose residue is attached to the innermost GlcNAc of the N-linked-core structure represented as Fucα1→6GlcNAc→R and has weak binding towards fucose in the outer arm such as Fucα1→2Galβ1→4GlcNAcβ1→R, Galβ1→4(Fucα1→3)GlcNAc→R and Galβ1→3(Fucα1→4)GlcNAc→R, where R = H or sugar [19]. Immobilized LTA can bind to glycans having fucose present in the outer arm including Fucα1→3/1→4GlcNAc and Fucα1→2Gal. LTA also has an affinity for glycans containing the Lex determinant represented as Galβ1→4(Fucα1→3)GlcNAcβ1→R [20]. The haptenic sugar for AAL and LTA is α-L-fucose. Similarly, to deplete HSA and Ig’s anti-HSA antibody and protein A and protein G′ were immobilized on the GMM-PETA monolith.
Disease-free and breast cancer sera were selected as the proteomics samples to challenge the platform and evaluate its effectiveness in facilitating the sample preparation and fractionation as far as the fucome capturing is concerned. The integrated platform introduced in this research report is unique in its design and effectiveness in reproducibly preparing serum samples for further differential quantitative analysis by LC-MS/MS. The platform eliminates all the experimental biases that would occur in the multi step methodologies normally required in proteomics/glycoproteomics. In fact, and as will be shown here, the proposed platform eliminates the need for multi step dialysis, dilution, centrifugation, fractionation, transfer from vessel to vessel, etc. The integrated platform allows the transfer/processing of the glycoproteomics/proteomics sample from column-to-column in the liquid phase in a highly reproducible manner using high precision HPLC pumps and valves, and isolate and concentrate the required information with zero sample loss and zero propagation of experimental errors. The platform is a multistage operation working in “cascade” whereby the needed information is processed in a continuous fashion from the first stage to the last stage in the process. The platform assembles various modes of chromatography that are working in harmony to extract, concentrate and fractionate the sub-glycoproteomics of interest. The platform with its unique design is fully amenable to automation and can be readily scaled up or down to fit various sample sizes making the platform the instrument of future for clinical and biomedical research set ups.
2. Experimental
2.1. Instrumentation
The HPLC setup consisted of a quaternary solvent delivery system Model Q-grad pump from Lab Alliance (State College, PA, USA), a solvent delivery system Model CM4000, and a Model 3100 UV-Vis variable wavelength detector from Milton Roy, LDC division (Riviera Beach, FL, USA) and a Rheodyne injector Model 7010 (Cotati, CA, USA) equipped with a 20-μL loop. All the switching valves were from Rheodyne, while one 3-way valve was from SSI (State College, PA, USA). The Spectra/Chrom CF-1 fraction collector was from spectrum chromatography (Houston, TX, USA). All mass spectra were obtained using a hybrid LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.2. Reagents and materials
The unconjugated lectins namely, AAL and LTA were purchased from Vector Laboratories (Burlingame, CA, USA). Pooled breast cancer serum from five donors and pooled disease-free human serum from five donors (same age group and race as the cancer serum) were purchased from Bioreclamation (Westbury, NY, USA). Stainless steel tubing of 4.6 mm ID was obtained from Alltech Associates (Deerfield, IL, USA). Glycerylmethacrylate (GMM) was purchased from Monomer-polymers & Dajac Labs (Feaster-Ville, PS, USA). Pentaerythritol triacrylate (PETA), 2,2′-azobis(isobutyronitrile) (AIBN) and 1-dodecanol were purchased from Aldrich Chemical Co. (Milwaukee, WI, USA). Sodium periodate, sodium cyanoborohydride, trifluoroacetic acid (TFA), L-(−)-fucose, protein A (from Staphylococcus aureus), protein G′ (from Streptococcus sp.) and IgG fraction of anti-human albumin (rabbit) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cyclohexanol and HPLC grade acetonitrile were obtained from Fisher Scientific (Fair Lawn, NJ, USA). The reversed phase chromatography column (ProSwift™ RP-1S) was purchased from Dionex Corporation (Sunnyvale, CA, USA).
2.3. Monolithic affinity columns
A 20 g polymerization mixture containing 7.6% w/w GMM, 7% w/w PETA, 59.1% w/w cyclohexanol 22.9% w/w dodecanol and 3.4% w/w water containing 1.0% w/w AIBN with respect to the monomers [18] was sonicated for 15 min, purged with nitrogen for 5 min and introduced into three stainless steel columns of dimensions 25 cm × 4.6 mm ID each that function as molds for the monoliths, and were heated at 60 °C for 15 h in a gas chromatography oven. The resulting monolithic columns were washed extensively with acetonitrile followed by water. The monolithic support was transferred from the 25 cm column to a shorter column (5 or 3 cm × 4.6 mm ID) by connecting the two columns with a ¼″-union and running water through the columns at flow rate of 3.0 mL/min until the unmodified monolithic support is transferred. A total of four columns of dimensions 3 cm × 4.6 mm ID and two columns of dimensions 5 cm × 4.6 mm ID were prepared.
The 3 or 5 cm monolithic columns were allowed to react with freshly prepared 0.1 M NaIO4 for 2 h at room temperature to convert the diol groups into aldehyde groups followed by a 5 min water wash. Then, the immobilization of lectin was done on the column by passing a solution of 1 mg of AAL or LTA in 0.5 mL of 0.1 M sodium acetate, pH 6.4, containing 0.1 M fucose and 50 mM sodium cyanoborohydride through the column for overnight at room temperature. For the anti-HSA column, a 5-mL solution containing 2 mg/mL of anti-HSA antibody containing 50 mM sodium cyanoborohydride in 0.1 M sodium acetate, pH 6.4, was passed through the column overnight [21, 22]. Protein A and G′ columns were prepared in the same way as the anti-HSA column except that a 1-mL solution containing 2 mg/mL of Protein A or Protein G′ was used. After passing the immobilization solution for overnight, a solution containing 0.4 M Tris/HCl, pH 7.2, and 50 mM sodium cyanoborohydride was passed through the columns for 3 h at room temperature to scavenge the residual aldehyde groups. The immobilized lectin columns as well as the anti-HSA, Protein A and Protein G′ columns were stored with the mobile phase containing 20 mM Tris-HCl, pH 7.4, containing 0.08% NaN3 at 4 °C until use.
2.4. Chromatographic conditions for the depletion of albumin and Igs followed by enrichment of the fucosylated glycoproteins
The simultaneous depletion of albumin and Igs (i.e., IgG’s, IgA and IgM) followed by enrichment/concentration and RPC separation of the fucosylated glycoproteins was achieved by the following order of tandem affinity columns. Two anti-HSA columns (5 cm × 4.6 ID mm each) → Protein G′ column (3 cm × 4.6 mm ID) → Protein A column (3 cm × 4.6 mm ID) → LTA column (3 cm × 4.6 mm ID)→ AAL column (3 cm × 4.6 mm ID) → RPC column (3 cm × 4.6 mm ID). The chromatographic set-up is shown in Fig. 1. The RPC column, which was initially stored in ACN was washed with 20 column volumes of water to remove the ACN. The binding mobile phase consisted of 20 mM Tris, pH 7.4, containing 0.3 M NaCl. The eluting mobile phase for the depletion column was 50 mM NaH2PO4, pH 2.2. The eluting mobile phase for the lectin columns was 5 mM fucose in the binding mobile phase. The aqueous-rich mobile phase (mobile phase A) for the RPC column consisted of H2O/ACN (95:5 v/v) containing 0.1% TFA and the organic-rich mobile phase (mobile phase B) consisted of ACN/H2O (95:5 v/v) containing 0.1% TFA.
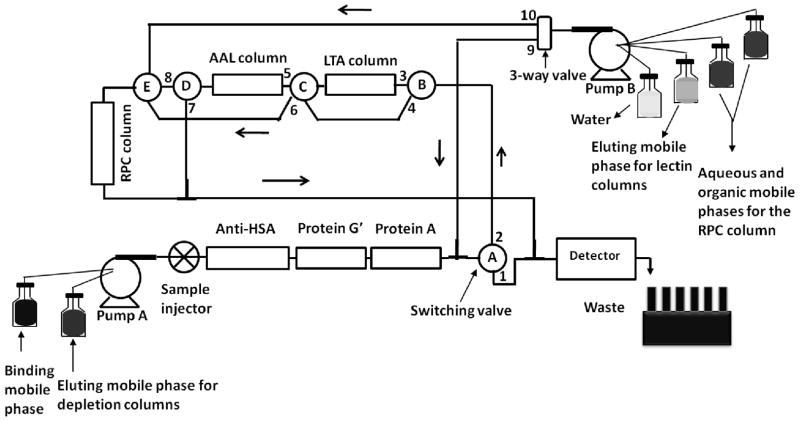
Figure 1.
Schematic representation of the platform for the simultaneous depletion of albumin and Igs, and the enrichment of fucosylated glycoproteins, and the subsequent RPC fractionation of the captured proteins. When the switching valve (SV) A, B, C and D were in 2, 3, 5 and 7 positions, respectively, the 3-fold diluted serum was injected onto the depletion and the lectin columns, followed by washing with the binding mobile phase (BMP) using pump A. Then, the eluting mobile phase for the depletion columns was passed by changing the SV-A position to 1, thus by-passing the lectin and the RPC columns. The depletion columns were re-equilibrated again with the BMP, after which the SV-A was changed back to position 2. Then, the LTA column was eluted using pump B, while the 3-way valve was in position 9, SV-B in position 3 and SV-C in position 6, thus by-passing the AAL column and passing through the RPC column. This was followed by washing, eluting and re-equilibrating the RPC column using the mobile phase from pump B, while the 3-way valve is in position 10. Then, the AAL column was eluted by changing the 3-way valve position back to 9, SV-B position to 4, SV-C position to 5 and SV-D position to 8. This was again followed by washing, eluting and re-equilibrating the RPC column by keeping the 3-way-valve in position 10.
While by passing the RPC column, 20 μL of 3-fold diluted serum were injected onto the depletion and the lectin columns that were previously equilibrated with the binding mobile phase. After 20 min of washing with the binding mobile phase, the depletion columns were eluted at a flow rate of 0.8 mL/min, followed by re-equilibration of the depletion columns. Another 20 μL of the same 3-fold diluted serum was injected onto the depletion and the lectin columns. Thus, serum proteins from a total of 40 μL (20 + 20 μL) of 3-fold diluted serum were accumulated onto the lectin columns. Using the eluting mobile phase, the proteins captured by the LTA column were transferred to the RPC column, which was in water. The RPC column was washed with mobile phase A to remove the salts for 20 min. This was followed by a linear gradient elution of the RPC column at increasing ACN concentration in the mobile phase. The linear gradient consisted of increasing the % of mobile phase B from 0% to 75% v/v in mobile phase A for 12 min. This was followed by 2 min at 75% mobile phase B, and then returning to initial elution conditions in 1 min. Before the next gradient run, the RPC column was washed with water for 15 min, and then for 20 min with 100% mobile phase A. Now, the accumulated proteins on the AAL lectin column were eluted and transferred to the RPC column. The elution of the RPC column was performed in the same way as described above. The protein fractions from the RPC column were collected every 24 s and evaporated to dryness in a SpeedVac from Savant Instruments, Inc. (Holbrook, NY) and stored at -20 °C until further use.
3. Results and Discussion
3.1. Some operational aspects and basic components of the integrated platform
3.1.1. Overall strategy: Simultaneous depletion of albumin and Igs followed by selective enrichment/concentration of proteins
The integrated platform for achieving the simultaneous depletion of albumin and Ig’s as well as the selective capturing and enrichment of proteins is shown in Fig. 1. The operational steps of the platform are summarized in the legend of Fig. 1 while the chromatographic conditions are detailed in the experimental section above.
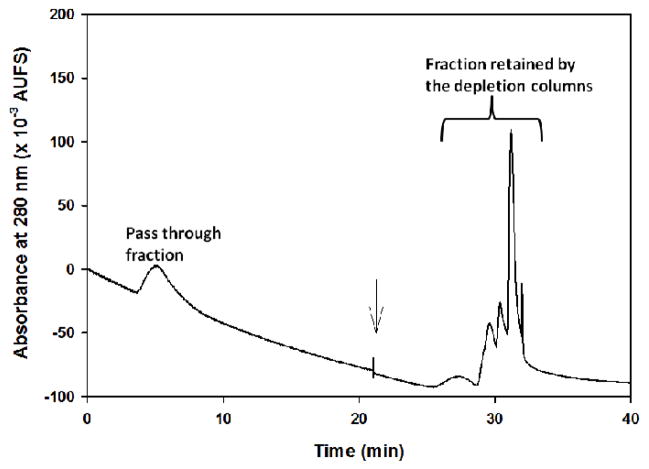
The GMM-PETA monolith, which has been shown previously to exhibit minimal amount of non-specific hydrophobic interactions with serum proteins [18], was selected as an ideal monolith for the preparation of the affinity columns used in the current study. As mentioned in the experimental section, 20 μL of 3-fold diluted serum were injected into the depletion and the lectin columns. This was followed by elution and re-equilibration of the depletion columns. The pass-through fraction from the depletion and lectin columns as well as the eluted albumin and Igs fractions from the depletion columns are shown in Fig. 2. The combined areas of the retained peaks by the depletion columns were almost 4 times larger than the area of the non-retained peak (i.e., pass through) by the depletion and the lectin columns. This is not surprising since albumin and Ig’s correspond up to 70% of the total mass of serum proteins. The pass through fraction, which contains in principle the non-glycosylated proteins and the glycoproteins lacking affinity to LTA and AAL columns (e.g., non-fucosylated glycoproteins), is quite large signaling the relatively low amount of the particular glycoproteins (e.g., fucosylated proteins) retained by the lectin columns.
Figure 2.
Chromatogram of the pass-through fraction and the eluted fraction from the anti-albumin and protein A and protein G′ columns when 20 μL of a 3-fold diluted serum was injected into the tandem affinity columns (the depletion and the lectin columns). The binding mobile phase consisted of 20 mM Tris, pH 7.4, containing 0.3 M NaCl and the eluting mobile phase for the depletion columns was made up of 50 mM NaH2PO4, pH 2.2. Flow rate, 0.8 mL/min.
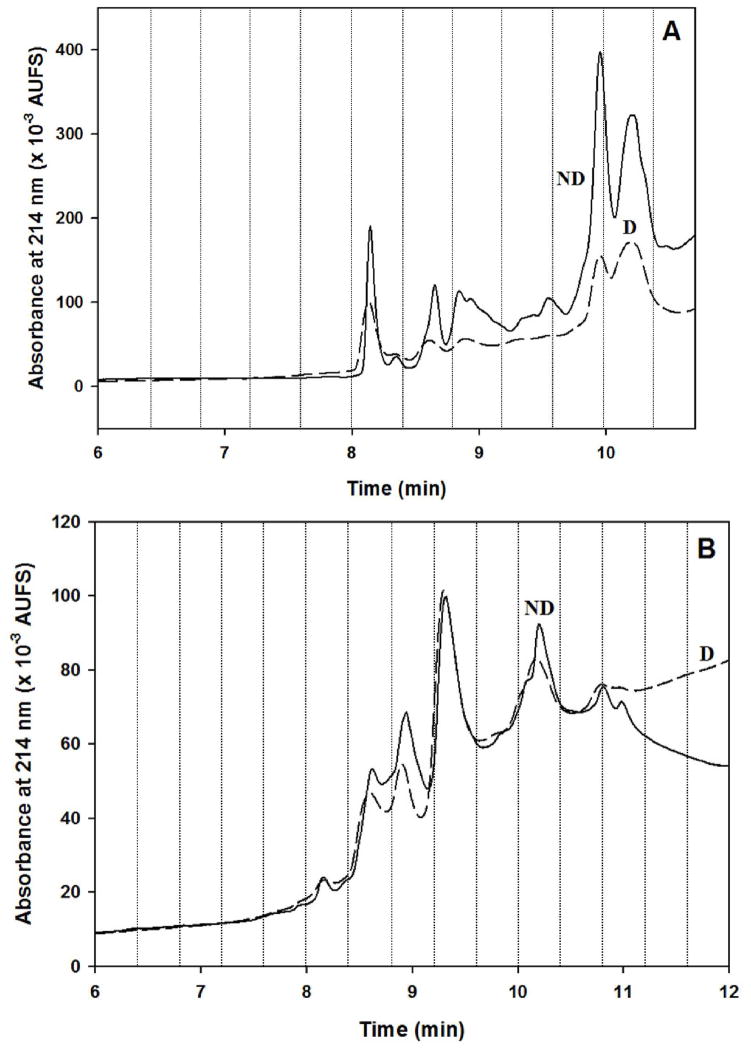
As expected, it was observed that when the same amount of serum was injected onto the lectin columns in the absence of the depletion columns, the lectin columns captured higher amount of proteins. This is clearly seen in Fig. 3 that shows the RPC chromatograms of the LTA and AAL captured proteins with and without depletion. This might be due to the fact that the lectin columns captured some of the glycosylated Igs. The most noticeable difference was observed with the RPC fractionation of the proteins captured by the LTA column, probably due to the fact that the LTA column is the first column in the tandem column format used. A detailed discussion is given below in section 3.1.3. A salient feature of the integrated platform that should be mentioned here is the reproducibility of the elution profile from the RPC column as far as the retention time of the major peaks in the RPC chromatograms portrayed in Figs 5 and 6 despite the fact that these chromatograms were generated on two different days and replicate preparation of the lectin columns. This is discussed further in section 3.1.4.
Figure 3.
Chromatograms of (A) RPC fractionation of the LTA captured proteins and (B) RPC fractionation of the AAL captured proteins with depletion (D), and without depletion (ND) of albumin and Igs. The RPC fractionation was carried out by a linear gradient elution at increasing ACN concentration in the mobile phase by going from 0 % to 75 % of mobile phase B in 12 min. Mobile phase A consisted of H2O/ACN (95:5 v/v) containing 0.1% TFA and mobile phase B consisted of ACN/H2O (95:5 v/v) containing 0.1% TFA. Flow rate, 1 mL/min; UV detection, 214 nm.
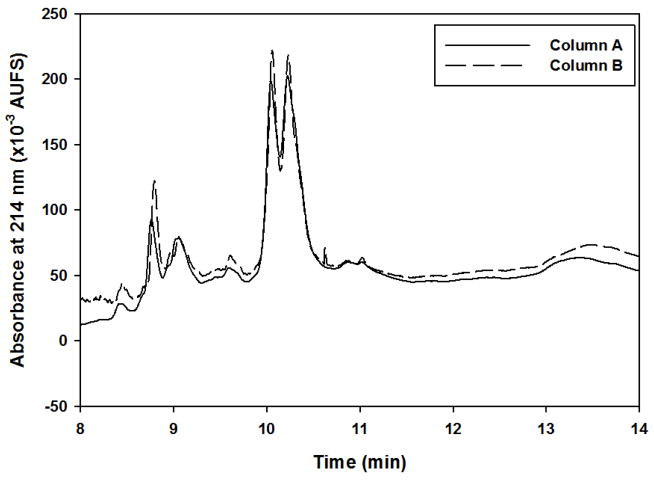
Figure 5.
Representative RPC fractionation chromatograms of proteins captured by the LTA column showing the reproducibility from lectin-to-lectin column. Experimental conditions for RPC fractionation are the same as in Fig. 3.
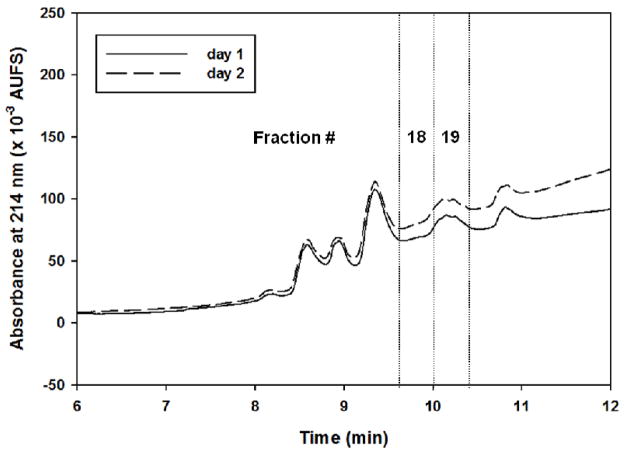
Figure 6.
Representative RPC fractionation chromatograms of proteins captured by the AAL column showing the reproducibility from day-to-day. Experimental conditions for RPC fractionation are the same as in Fig. 3.
Furthermore, the platform is fully integrated comprising multi-affinity columns operating in cascade in the sense that there is no sample handling between the various steps, which guarantee no sample loss and no propagation of experimental biases from step-to-step or column-to-column. In fact, the sample is moved from column-to-column in the liquid phase and requiring no sample manipulation (e.g., collecting, desalting, dialysis) between columns. Finally, the sample is fractionated by RPC yielding desalted fractions in volatile ACN – rich mobile phase, which after evaporation in a speed vacuum are subjected to LC-MS/MS analysis. Thus, the “cascading” concept involving the continuous transferring and processing of the proteomic sample from column to column has been put to work in a multi-column liquid phase separation platform yielding a highly reproducible depletion/capturing/fractionation unit that is readily amenable to automation.
3.1.2. Loading capacity of the anti-HSA column
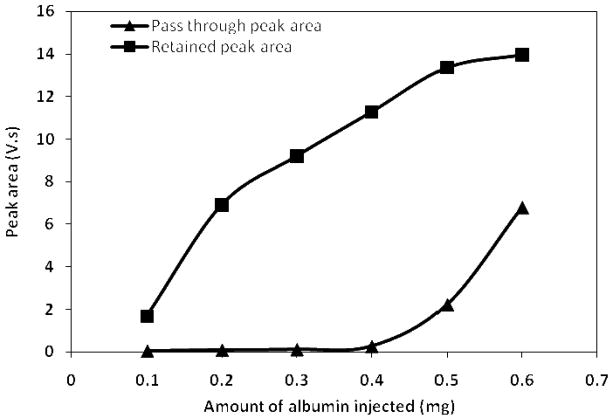
In a recent study from our laboratories, Jmeian and El Rassi reported the sample loading capacity of the protein A and protein G′ columns to be ~ 7–25 fold higher than that of the antibody affinity columns [21]. In the current study, it was therefore very important to evaluate the sample loading capacity of the anti-HSA column since it is the “weak link” column in the tandem depletion column format composed of the anti-HSA column → protein G′ column → protein A column. Standard solutions of human serum albumin of varying concentration were injected onto the anti-HSA column (5 cm × 4.6 mm ID) and the peak areas of the retained and the pass through peaks were plotted against the amount of injected albumin (see Fig. 4). It can be seen from Fig. 4 that one single anti-HSA column of the dimensions 5 cm × 4.6 mm ID can handle a maximum of 0.4 mg of albumin before an overflow peak becomes noticeable and its area can be evaluated. Considering the fact that human serum contains 35–50 mg albumin/mL of serum [23], the injection of 20 μL of 3-fold diluted serum into the depletion columns corresponds to injecting 0.24 – 0.34 mg of albumin into the integrated platform, which in its actual operation includes two of the 5 cm × 4.6 mm ID anti-HSA columns. The 0.24 – 0.34 mg quantity is therefore well below the 0.4 mg maximum capacity of one single anti-HSA column.
Figure 4.
Plot of peak area versus the amount of standard human serum albumin protein injected on the anti-HSA column (5 cm × 4.6 mm ID). Flow rate 1 mL/min, other conditions same as Fig. 2.
3.1.3. Assessing the need for depleting albumin and Igs
In order to assess the virtue of depleting albumin and Igs on the results of the integrated platform, 3 fold diluted disease-free human serum was processed by the platform in the presence and absence of the 3 depletion columns under otherwise the same operating conditions, see Fig. 3. As one would expect, besides the commonly captured proteins, the two lectin columns of the integrated platform captured some different proteins or the so-called unique proteins in the presence than in the absence of the depletion columns. It is the nature of the unique proteins as well as their level of abundance, which should favor one approach over another (i.e., depletion vs. no depletion).
For the LTA column, 31 unique proteins were captured by this column in the presence of depletion columns versus 30 unique proteins in the absence of the depletion columns (Table S-1, Supplementary Information) as was measured in the RPC collected fractions by LC-MS/MS. In the case of depletion, the 31 unique proteins listed in Table S-1 comprised 17 low abundance proteins and only one Ig chain, whereas in the case of absence of depletion columns, the 30 unique proteins captured by the LTA column included 10 different Ig chains and 12 low abundance proteins that are listed in Table S-1. In the case of online depletion, and to enumerate, the 17 low abundance proteins were cartilage acidic protein 1, cartilage oligomeric matrix protein, cathelicidin antimicrobial peptide, cholesteryl ester transfer protein, dermcidin, desmoplakin, extracellular matrix protein 1, filaggrin-2, histone H4, hornerin, keratin type I cytoskeletal 16, keratin type I cytoskeletal 17, protein S100-A8, secreted phosphoprotein 24, semenogelin-1, semenogelin-2 and titin. In the case of absence of online depletion columns, the 12 low abundance proteins found in the LTA fractions included 8 proteins of known concentrations such as adipocyte plasma membrane-associated protein, apolipoprotein (a), cadherin-5, complement C1r subcomponent-like protein, EGF-containing fibulin-like extracellular matrix protein 1, ficolin-2, mannose-binding protein C, and serum paraoxonase/lactonase 3. The 4 remaining proteins have unknown concentrations including c4b-binding protein beta chain, collectin-11, keratinocyte proline-rich protein and solute carrier family 2 facilitated glucose transporter member 2.
The protein fractions captured by the AAL column in the presence or absence of the online depletion columns contained 32 and 28 unique proteins (Table S-2), respectively, as measured in the RPC fractions by LC-MS/MS. In the case of online depletion, the 32 unique proteins comprised 6 Ig chains and 12 low abundance proteins. To enumerate, the 12 low abundance proteins included annexin A2, beta-Ala-His dipeptidase, cholinesterase, dermcidin, desmoglein-1, extracellular matrix protein 1, filaggrin, histone H4, low affinity immunoglobulin gamma Fc region receptor III-A, phosphatidylinositol-glycan-specific phospholipase D, protein S100-A8 and vitamin K-dependent protein C. Oppositely, in the absence of online depletion, the 28 unique proteins captured by the AAL column comprised 10 low abundance proteins whereby the concentration of the C4b-binding protein beta chain is not known while the following 9 low abundance proteins have known concentrations: apolipoprotein(a), CD44 antigen, complement factor H-related protein 3, filaggrin-2, hepatocyte growth factor activator, low affinity immunoglobulin gamma Fc region receptor III-B, L-selectin, mannan-binding lectin serine protease 1, scavenger receptor cysteine-rich type 1 protein M130 and vascular cell adhesion protein 1. Furthermore, in the absence of online depletion, the AAL columns captured 10 Ig chains (see Table S-2).
On the basis of the number of the low abundance proteins that were unique to the lectin columns in the presence or absence of online depletion columns (17 vs. 12 proteins, respectively) and also considering the amount of Igs captured under both conditions (7 unique Igs with depletion vs. 20 unique Igs without depletion), the depletion method is considered superior. This finding has justified the implementation of online depletion in the integrated platform.
On the other hand and despite the fact that the two 5 cm × 4.6 mm ID anti-HSA columns were used below their maximum capacity, there is still some serum albumin captured by the LTA and the AAL columns. However, the combined average spectral count of serum albumin for both lectin columns was 39 in the presence vs. 313 in the absence of the anti-HSA column. This represents 8-fold reduction in the serum albumin binding to the lectin columns. Also, there were an overall 15 different Ig chains identified in the LTA fractions in the presence of depletion columns whereas without the depletion column 25 different Ig chains were identified. The Igs and HSA found in the presence of depletion columns might be due in part to protein-protein interactions that are often observed in complex proteomics samples and protein-based affinity assays [24, 25].
3.1.4. Reproducibility of the platform as manifested by its major output, the RPC fractionation of proteins captured by the lectin columns
In proteomic analysis, the fractionation of the complex protein mixture is an important step in the overall workflow and any inconsistency in this step directly affects the identification of the differentially expressed proteins in the diseased serum when compared to the disease-free serum. On this basis, the integrated platform was evaluated for its reproducibility in the RPC profiling of the proteins captured by the lectin columns, which is the final output of the platform.
The reproducibility from run-to-run was evaluated in terms of the RPC fractionation by gradient elution of the LTA and AAL captured proteins. The visual examination of the RPC chromatograms readily showed that the platform generated reproducible RPC profiles. Furthermore, the reproducibility was assessed by calculating the percent relative standard deviation (% RSD) of the retention time and peak area. The average % RSD (n = 2) for the retention time and peak areas of the peaks in the RPC chromatogram of LTA captured proteins were 0.24 % and 11.2 %, respectively. For the RPC chromatograms obtained for the AAL captured proteins, the respective average % RSD (n = 2) for the retention time and peak areas were 0.13 % and 4.8 %. It was observed that not only the retention time/peak areas were very reproducible in terms of %RSD but also the spectral count for most of the proteins were similar in both runs. For example, it was seen that the spectral count was the same (12 vs. 12) in the LTA fraction from run 1 and run 2 for the low abundance glycoprotein kallistatin, which has a concentration of 22.1 ± 3.5 μg/mL in plasma [26].
The reproducibility of the platform as manifested by the RPC fractionation of the proteins captured by two lectin columns prepared on two different days (i.e., lectin column-to-lectin column reproducibility) was also assessed. For instance, the RPC chromatograms obtained for the proteins captured by two different LTA column preparations without the depletion columns were very much reproducible as is readily revealed in the chromatograms shown in Fig. 5. The % RSD (n=2) for retention time and peak area was 0.13 % and 7.43 %, respectively.
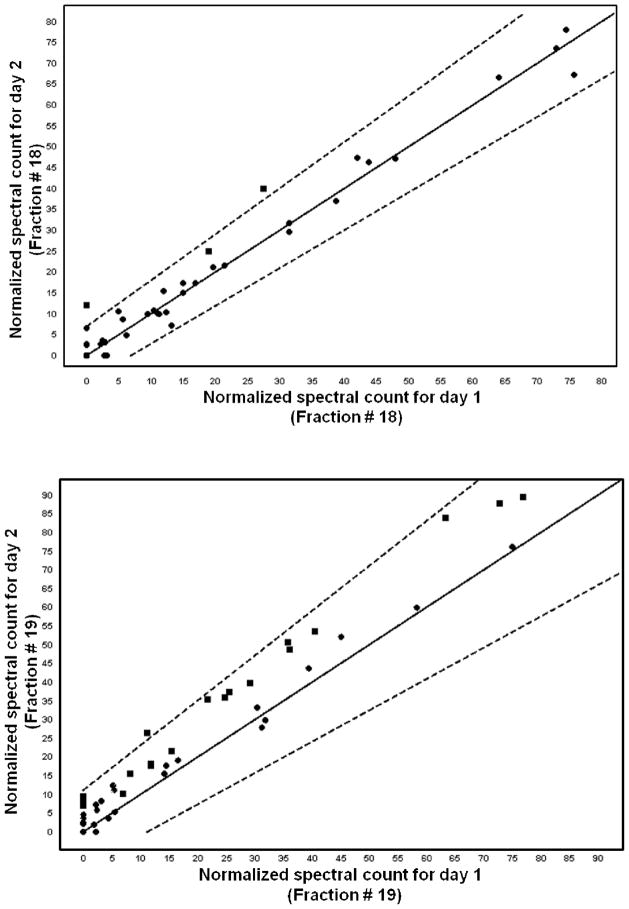
To evaluate the day-to-day reproducibility of the platform, some selected fractions of the AAL captured proteins that were fractionated by RPC were subjected to mass spectrometric analysis. The stringent conditions that were used to identify the differentially expressed proteins in cancer serum with respect to disease-free serum were also used to make the comparison of the mass spectral counts for the proteins that were identified from fractions collected on different days (i.e., the day-today reproducibility analysis). The spectral counts were strictly those of the proteins identified on the basis of 99.9% protein identification probability, 95% peptide identification probability with minimum 5 unique peptides. Also, the keratins that had grouping ambiguity in the sense that they shared some of the peptides with one or more proteins were not considered for the reproducibility or differential analysis. Figure 6 shows representative chromatograms of RPC gradients of the AAL captured proteins for the day-to-day reproducibility. It was observed that not only retention time and peak areas were very reproducible (%RSD for retention time was on the average 0.57% and %RSD for peak areas was on the average 7.8%) but also most of the proteins identified from the two different days did have spectral counts that were less than two standard deviations away from each other as determined by the scatterplots or Q-Q plots, which plot the normalized spectral counts for each protein found in a given fraction on a certain day versus the normalized spectral count of that same protein found in that same fraction on another day. For example, with the above said conditions, and using Q-Q plots for fraction # 18 from day 1 and day 2, two proteins only (Ig mu chain C region and fibronectin) were two standard deviations away. The protein Ig mu chain C region was found on both days whereas fibronectin was found only on day 2. In the same way, in fraction #19 from day 1 and day 2, only the protein fibronectin that was found on both days was different and all other proteins were less than two standard deviations away from each other (see the Q-Q plots in Fig. 7).
Figure 7.
Representative Q-Q scatterplots for the to day-to-day reproducibility of MS spectral counts. The squares that plot outside the dotted lines are more than two standard deviations away from being the same in both categories. The dotted lines delimit the upper and lower error bars.
3.2. LC-MS/MS identification of proteins captured by the lectin columns
3.2.1. Identification of the proteins captured by the LTA column
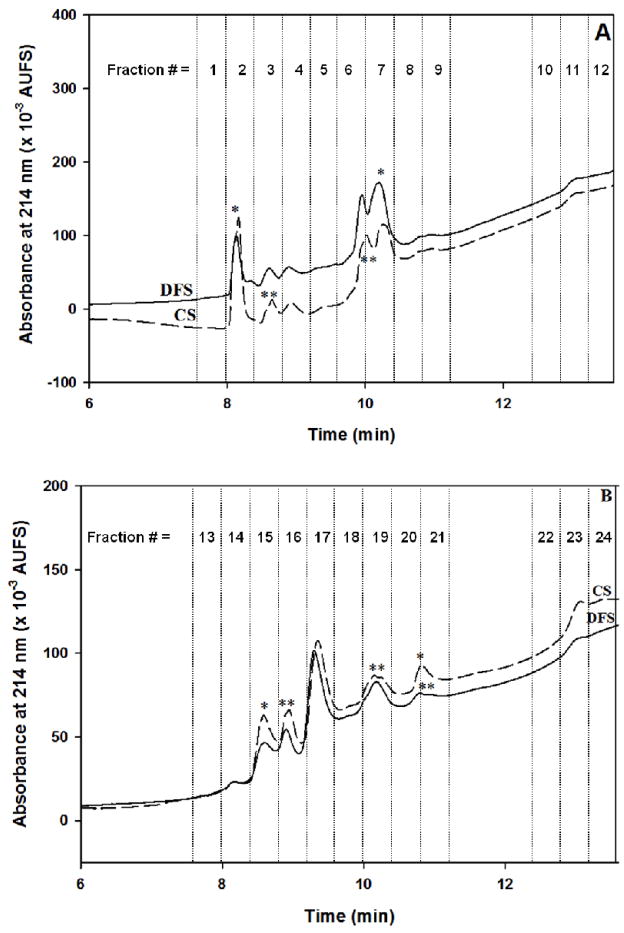
The captured proteins by the LTA column were eluted stepwise with the haptenic sugar fucose and subsequently trapped on the RPC column, which were then fractionated by a linear gradient elution of the RPC column at increasing acetonitrile concentration in the mobile phase. This led to various collected RPC fractions (see Fig. 8A) that were subjected to LC-MS/MS analysis. Only proteins that exhibited protein identification probability greater than 99 % with peptide identification probability greater than 95 % containing at least two unique peptides were considered and are listed in Table S-3. The false rate discovery for both protein and peptide identification was 0.0% for both the LTA and AAL captured proteins. The numbers of the identified proteins from the disease-free serum and cancer serum were 148 and 138 non-redundant proteins, respectively. In the LTA fractions, the identified proteins that were unique to the disease-free and the cancer sera were 22 and 12 proteins, respectively, while 126 proteins were common to both sera (Table S-3). Some of the identified proteins were previously reported as cancer biomarker candidates, namely apolipoprotein C-I, neutrophil defensin1 and serum paraoxonase/arylesterase 1 [27]. Furthermore, some other proteins that were identified in the LTA fractions such as plasminogen, coagulation factor XII, complement C3, kininogen-1, Ig mu chain C region, Ig α-2 chain C, apolipoprotein A-I, apolipoprotein E, apolipoprotein C-III, apolipoprotein D, fibronectin, protein AMBP, vitronectin, histidine-rich glycoprotein, complement factor H, clusterin, galectin-3-binding protein, inter αtrypsin inhibitor heavy chain H4 and the fibrinogen chains were reported to have glycans containing the Lex determinant [28], for which LTA exhibits distinct and strong affinity.
Figure 8.
Comparison of chromatograms of the RPC fractionation of proteins captured by the LTA column (in A) and AAL column (in B) from disease-free (DFS) and breast cancer sera (CS). Experimental conditions for RPC fractionation are the same as in Fig. 3.
It should be noted that the depletion/enrichment/fractionation achieved by the integrated platform allowed the detection and identification of 45 low abundance proteins in the LTA captured protein fractions. Also, proteins with estimated concentrations in the range between ng level and = 1 ≤ 1 μg level were considered as low abundance proteins. Typical examples of these low abundance proteins are filaggrin (0.82 ng/mL of plasma), titin (4.1 ng/mL), filaggrin-2 (4.6 ng/mL), semenogelin-1 and 2 (5.9 ng/mL), and N-acetylglucosamine-1-phosphotransferase subunit gamma (6.8 ng/mL) [29]. The concentrations of some of the other low abundance proteins such as actin cytoplasmic 1, complement C5 and complement C1r subcomponent are not known and these proteins are not listed in the human plasma proteome reference set that has non-redundant set of 1929 protein sequences compiled by Farah et al [29].
3.2.2. Identification of the proteins captured by the AAL column
Using the same approach as in the above section, 141 and 184 non-redundant proteins were identified in the proteins captured by the AAL column from the disease-free and breast cancer sera, respectively, that were further fractionated on the RPC column using a linear gradient elution at increasing acetonitrile concentration in the mobile phase (see Fig. 8B). Again, only proteins that exhibited protein identification probability greater than 99 % with peptide identification probability greater than 95 % containing at least two unique peptides were considered and are listed in Table S-4. In the AAL captured proteins, there were 6 and 49 proteins unique to the disease-free and the cancer sera, respectively, with 135 proteins common to both sera. Some of the identified proteins such as afamin, attractin, complement C1r subcomponent-like protein, cholinesterase, Ig alpha-2 chain C region, kallistatin, sulfhydryl oxidase 1, vitronectin, and scavenger receptor cysteine-rich type 1 protein M130 have been identified to have core fucosylation [30].
Similarly to what is reported in the above section, numerous proteins that belonged to the low abundance category (54 proteins) were identified. Typical low abundance proteins could be cited such as filaggrin (0.82 ng/mL of plasma), transferrin receptor protein 1 (2.9 ng/mL), long palate, lung and nasal epithelium carcinoma-associated protein 1 (3.3 ng/mL), follistatin-related protein 1 (3.6 ng/mL), plakophilin-1 (3.6 ng/mL), filaggrin-2 (4.6 ng/mL), galectin-7 (6.9 ng/mL), angiotensin-converting enzyme (7.4 ng/mL) and calmodulin-like protein 5 (8.1 ng/mL) [29]. The concentration of some of the low abundance proteins such as arginase-1, deleted in malignant brain tumors 1 protein, mucin-5B, neutrophil elastase and premature ovarian failure protein 1B (protein POF1B) are not known [29]. Furthermore, the concentrations of some of the other low abundance proteins such as actin aortic smooth muscle, actin cytoplasmic 1, complement C5, complement C1r subcomponent, keratinocyte proline-rich protein, histone H2B type 1-C/E/F/G/I and putative lipocalin 1-like protein1 are not known and these proteins are not listed in the human plasma proteome reference set that has non-redundant set of 1929 protein sequences compiled by Farah et al [29].
Overall, and as determined from SWISSPROT database searching and using NetNGly –predictor, which can predict the N-glycosylation sites in proteins, there were a certain number of non glycoproteins in both the AAL and LTA captured proteins from disease free and breast cancer sera. In fact, searching the LTA captured proteins from the disease free serum in the SWISSPROT database, and using the NetNGly – predictor revealed that 79% of the identified proteins were glycoproteins, and 21% were non-glycoproteins. Similarly, in the cancer serum there were 83% glycoproteins and 17% non-glycoproteins. Also, searching the AAL captured proteins from the disease free serum in the SWISSPROT database and using NetNGly – predictor revealed that 75% of the identified proteins were glycoproteins and 25% were non-glycoproteins. In the case of AAL captured proteins from cancer serum, the percentage of glycoproteins was 77% and that of non-glycoproteins was 23%. As mentioned above in section 3.1.3, the capturing of non glycoproteins by the two lectin columns might be due in part to protein-protein interactions that are often observed in complex serum samples and protein-based affinity assays [24, 25].
3.2.3. Differentially expressed proteins in the LTA and AAL fractions
The RPC chromatograms of the LTA and AAL captured proteins from disease-free and cancer sera are shown in Figs 8A and 8B, respectively. In both figures, one can readily see the differences between the two serum profiles, which are reflected by peak intensity (noted by *) and shouldering (noted by **) on the chromatograms. In both cases (i.e., LTA and AAL fractions), for the identification of the differentially expressed proteins in the cancer serum relative to the disease-free serum, only proteins with 99.9% protein identification probability and 95% peptide identification probability with a minimum of 5 unique peptides were considered (this is the same norm used in section 3.1.4). The differentially expressed proteins between breast cancer and disease-free sera were revealed using the quantitative Q-Q scatterplot which plots the normalized spectral count for each protein found in the breast cancer serum versus the normalized spectral count of that same protein found in the disease-free serum. The proteins that are more than two standard deviations away from being the same in both categories are considered as differentially expressed proteins. This scatterplot approach was used very recently by Selvaraju and El Rassi [9], and proved effective in revealing candidate biomarkers.
Using the Q-Q scatterplot just mentioned, 17 identified proteins were found to be either up or down regulated in the LTA captured proteins (Table S-5). The proteins α-1-antitrypsin, α-2-HS-glycoprotein and serotransferrin, which were found to be differentially expressed in the present study have been reported to possess fucosylation in their glycan structures [31]. Some of the other differentially expressed proteins such as apolipoprotein B-100, α-1-antichymotrypsin, inter-α-trypsin inhibitor heavy chain H4, α-2-HS-glycoprotein, CD5 antigen-like and prothrombin have been reported to have altered fucosylation in hepatocellular carcinoma (HCC) [32]. The observed elevated level of the protein complement C4-B in breast cancer serum is in accordance with another report that used L-phytohemagglutinin (L-PHA) lectin affinity to identify differentially expressed glycoproteins in breast cancer tissue [33]. Serum amyloid A-4 protein was up regulated which is in accordance with that reported in ref. [34], where it was found to be at elevated level in stage 2 breast cancer patients. The protein α-2-HS-glycoprotein that was down regulated in the LTA fractions has been reported to be a potential cancer biomarker [29].
Similarly to the LTA captured proteins, using Q-Q scatterplots, 26 proteins in the AAL fractions were found to either be up or down regulated in the cancer serum relative to the disease-free serum fractions (Table S-6). The differentially expressed proteins such as afamin, α-1-antitrypsin, α-2-macroglobulin, ceruloplasmin, hemopexin, inter-α-trypsin inhibitor heavy chain H1 and serotransferrin have been reported to have core fucosylated glycopeptides [30]. With the exception of serotransferrin, all these core fucosylated proteins were identified in HCC serum [32]whereby agarose-bound AAL was used to capture glycoproteins from serum. Also, the differentially expressed proteins found in the AAL fractions, namely α-1-antitrypsin, α-1-antichymotrypsin, α-2-macroglobulin, ceruloplasmin, hemopexin and serotransferrin were reported to possess altered fucosylation by Comunale et al [35] in HCC serum. Furthermore, afamin was found to be at elevated level in the AAL fraction, which agrees with a recent finding in the sense that this protein was reported to be significantly up-regulated in pre-diagnostic breast serum [36]. In the same study, it was also reported that the proteins α-2-macroglobulin and ceruloplasmin [36] were significantly down regulated in breast cancer serum. The same was observed in the current study for α-2-macroglobulin but in the case of ceruloplasmin it was up- and down-regulated in the AAL fractions. In another study, α-2-macroglobulin and serotransferrin were reported to be at elevated levels in breast cancer plasma [37]. The up-regulated protein serotransferrin has been listed as candidate cancer biomarker with more than 500 citations in a compilation of proteins that are differentially expressed in human cancer [27]. The protein complement C3 was elevated in some of the AAL fractions and down regulated in one fraction. This finding corroborates with that observed in another study [38] as far as the elevated level of fucosylation of complement C3 is concerned. Also, complement C3 was identified as a potential marker of colorectal cancer [38] and a potential candidate to study changes in breast cancer serum [39]. The proteins α-2-macroglobulin, apolipoprotein A-I, ceruloplasmin, lactotransferrin and serum albumin have been reported to be potential cancer biomarkers [29]. The proteins apolipoprotein A-I and hemopexin were reported to be at elevated level in breast cancer tissues [33] and in this study it was the same in the case of hemopexin, whereas apolipoprotein A-I was down regulated. The observed difference might be due to the fact that the lectins L-PHA and AAL have different affinity toward glycoproteins, that is, L-PHA has affinity towards β(1,6)-branched N-linked glycans whereas AAL has affinity towards the fucosylated glycans. In a recent study from our laboratory involving the use of broad selectivity lectins namely, concanavalin A, wheat germ agglutinin and ricinus communis agglutinin-I to identify the proteins that were differentially expressed in breast cancer serum [9], the proteins afamin and hemopexin were found to be up-regulated and in the current study also elevated levels were observed for these two proteins. In the same study, the proteins α-2-antiplasmin and inter-α-trypsin inhibitor heavy chain H1 were down regulated whereas in the current study these two proteins were up regulated. This might be due to the difference in the lectin specificities. As discussed above, the protein plasma kallikrein was up regulated in one of the RPC fraction of LTA captured proteins, whereas in the AAL captured proteins kallikrein was found to be down-regulated in breast cancer serum. This might be due to the partial loss of the core fucosylation in the protein for which AAL has affinity. Finally, the two differentially expressed proteins desmoplakin and phosphatidylinositol-glycan-specific phospholipase D in the AAL fractions have been previously predicted to be at 20 ng/mL and 0.46 μg/mL concentration levels in plasma, respectively [29].
Overall, among the 43 DEP in the AAL and LTA fractions (26 proteins from AAL + 17 proteins from LTA), 8 DEP were found in common to both LTA and AAL fractions, see Table 1. On the other hand, 9 proteins were found to be unique to the LTA column and 18 other proteins were unique to the AAL column fractions. Therefore, a combined total of 35 proteins are in fact differentially expressed in the LTA and AAL captured proteins when taking into consideration the 8 common differentially expressed proteins in the fractions of LTA and AAL columns.
Table 1.
Differentially expressed proteins in the LTA and AAL fractions. la, low abundance (few ng/mL to ≤1 μg/mL level); CF, core fucosylated; F, fucosylated
| Differentially expressed proteins in the LTA fractions | Differentially expressed proteins common to both LTA and AAL fractions | Differentially expressed proteins in the AAL fractions |
|---|---|---|
| Alpha-2-HS-glycoprotein (CF, F) | Alpha-1-antichymotrypsin (F) | Afamin (CF, F) |
| Apolipoprotein A-IV | Alpha-1-antitrypsin (CF, F) | Alpha-2-antiplasmin (F) |
| C4b-binding protein alpha chain (F) | Apolipoprotein B-100 (F) | Alpha-2-macroglobulin (CF, F) |
| CD5 antigen-like | Inter-alpha-trypsin inhibitor heavy chain H2 (F) | Apolipoprotein A-I (F) |
| Complement C1r subcomponent (F) | Inter-alpha-trypsin inhibitor heavy chain H4 (F) | Ceruloplasmin (CF, F) |
| Complement C4-B (F) | Plasma kallikrein (F) | Complement C1s subcomponent (F) |
| Ig lambda-2 chain C regions | Pregnancy zone protein la | Complement C3 (F) |
| Prothrombin (CF, F) | Serotransferrin (CF, F) | Complement C5 (F) |
| Serum amyloid A-4 protein (F) | Desmoplakin la | |
| Haptoglobin-related protein (F) | ||
| Hemopexin (CF) | ||
| Inter-alpha-trypsin inhibitor heavy chain H1 (CF, F) | ||
| Lactotransferrin | ||
| Phosphatidylinositol-glycan-specific phospholipase D la | ||
| Plasminogen (F) | ||
| Protein AMBP | ||
| Serum albumin | ||
| Vitamin K-dependent protein S (F) |
The combined 35 DEP constitute a broad panel of DEP while the 8 common DEP may be viewed as a narrower panel of DEP (see Table 1). The narrower panel may be more representative of what is happening at the fucome level in breast cancer since this panel is common to both lectins (i.e., LTA and AAL) that have complementary affinity towards fucosylated glycoproteins. The fucosylation of the protein panels have been discussed in the above section and the fucosylated proteins are also indicated in Table 1.
It is interesting to note that among the combined 35 DEP in the LTA and AAL fractions, 28 proteins were found in the regions marked by * and ** in Figs 8A and 8B, which indicate difference in peak intensity and the presence of shouldering. This quick visual finding of the difference between disease-free and breast cancer sera by a simple visual inspection of the corresponding RPC chromatograms of targeted captured proteins may prove useful in expediting the screening of a large number of biological samples in clinical and medical setups. Also, this quick visual inspection should in principle reduce the exuberant cost and length of the LC-MS/MS analysis in the sense that only those fractions corresponding to the RPC peaks with apparent difference would be analyzed by LC-MS/MS rather than analyzing all the collected fractions.
4. Concluding remarks
The developed and tested multicolumn liquid phase sample preparation, separation and fractionation platform has been demonstrated to be very suitable for the reproducible processing and efficient capturing and concentration of the human fucome from disease-free and breast cancer serum. The platform allowed the convenient comparison of the fucosylated proteins in breast cancer serum to those in disease-free serum yielding a broad panel of 35 DEP from the combined LTA and AAL captured proteins and a narrower panel of 8 DEP that were commonly differentially expressed in both LTA and AAL captured proteins, which are considered as more representative of the altered fucome in breast cancer due the complementary specificity of both lectins. This was accomplished with virtually no sample loss and dilution or experimental biases when comparing the diseased serum fucome to the disease-free fucome by LC-MS/MS due to the cascading nature of the platform.
Supplementary Material
Acknowledgments
The financial support of this research by a Grant No 1R15GM096286-01 from the National Institutes of Health is greatly appreciated.
Nonstandard abbreviations
- AAL
Aleuria aurantia lectin
- DEP
differentially expressed protein
- GlcNAc
N-acetylglucosamine
- AIBN
2,2′-azobis(isobutyronitrile)
- GMM
glycerylmethacrylate
- HSA
human serum albumin
- Ig’s
immunoglobulins
- LAC
lectin affinity chromatography
- LTA
Lotus tetragonolobus agglutinin
- PETA
pentaerythritol triacrylate
- L-PHA
L-phytohemagglutinin
- RPC
reversed phase chromatography
- TFA
trifluoroacetic acid
References
- 1.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 2.van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 3.Rademacher TW, Parekh RB, Dwek RA. Glycobiology. Ann Rev Biochem. 1988;57:785–838. doi: 10.1146/annurev.bi.57.070188.004033. [DOI] [PubMed] [Google Scholar]
- 4.Dube DH, Bertozzi CR. Glycans in cancer and inflamation - Potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 5.Fuster MM, Esko JD. The sweet and sour cancer: Glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 6.Drake PM, Cho W, Li B, Prakobphol A, et al. Sweetening the pot: Adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block TM, Comunale MA, Lowman M, Steel LF, et al. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci, USA. 2005;102:779–784. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selvaraju S, El Rassi Z. Liquid-phase-based separation systems for depletion, prefractionation and enrichment of proteins in biological fluids and matrices for in-depth proteomics analysis -An update covering the period 2008 - 2011. Electrophoresis. 2012;33:74–88. doi: 10.1002/elps.201100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selvaraju S, El Rassi Z. Tandem lectin affinity chromatography monolithic columns with surface immobilised concanavalin A, wheat germ agglutinin and Ricinus communis agglutinin-I for capturing sub-glycoproteomics from breast cancer and disease-free human sera. J Sep Sci. 2012;35:1785–1795. doi: 10.1002/jssc.201200230. [DOI] [PubMed] [Google Scholar]
- 10.Durham M, Regnier FE. Targeted glycoproteomics: Serial lectin affinity chromatoraphy in the selection of O-glycosylation sites on proteins from human blood proteome. J Chromatogr A. 2006;1132:165–173. doi: 10.1016/j.chroma.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 11.Madera M, Mann B, Mechref Y, Novotny MV. Efficacy of glycoproteins enrichment by microscale lectin affinity chromatography. J Sep Sc. 2008;31:2722–2732. doi: 10.1002/jssc.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann B, Madera M, Klouckova I, Mechref Y, et al. A quantitative investigation of fucosylated serum glycoproteins with application to esophageal adenocarcinoma. Electrophoresis. 2010;31:1833–1841. doi: 10.1002/elps.201000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu R, Regnier FE. Comparative glycoproteomics of N-linked complex-type glycoforms containing sialic acid in human serum. Anal Chem. 2005;77:7225–7231. doi: 10.1021/ac050554q. [DOI] [PubMed] [Google Scholar]
- 14.Schwientek T, Mandel U, Roth U, Müller S, Hanisch FG. A serial lectin approach to the mucin-type O-glycoproteome of Drosophila melanogaster S2 cells. Proteomics. 2007;7:3264–3277. doi: 10.1002/pmic.200600793. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J Chromatogr A. 2004;1053:79–88. [PubMed] [Google Scholar]
- 16.Yang Z, Hancock WS. Monitoring glycosylation pattern changes of glycoproteins using multi-lectin affinity chromatography. J Chromatogr A. 2005;1070:57–64. doi: 10.1016/j.chroma.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromtography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52:1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 18.Gunasena D, El Rassi Z. Hydrophilic diol monolith for the preparation of immuno-sorbents at reduced nonspecific interactions. J Sep Sc. 2011;34:2097–2105. doi: 10.1002/jssc.201100353. [DOI] [PubMed] [Google Scholar]
- 19.Kobata A, Yamashita K. In: Glycobiology A Practical Approach. Fukuda M, Kobata A, editors. IRL Press; Oxford: 1993. pp. 103–125. [Google Scholar]
- 20.Yan L, Wilkins PP, Alvarez-Manilla G, Do SI, Smith DF. Immobilized Lotus tetragonolobus agglutinin binds oligosaccharides containing the Lex determinant. Glycoconjugate J. 1997;14:45–55. doi: 10.1023/a:1018508914551. [DOI] [PubMed] [Google Scholar]
- 21.Jmeian Y, El Rassi Z. Tandem affinity monolithic microcolumns with immobilized protein A, protein G′ and antibodies for depletion of high abundance proteins from serum samples: Integrated micro columns-based fluidic system for simultaneous depletion and tryptic digestion. J Proteome Res. 2007;6:947–954. doi: 10.1021/pr060660o. [DOI] [PubMed] [Google Scholar]
- 22.Jmeian Y, El Rassi Z. Multicolumn Separation Platform for Simultaneous Depletion and Prefractionation Prior to 2-DE for Facilitating In-Depth Serum Proteomics Profiling. J Proteome Res. 2009;8:4592–4603. doi: 10.1021/pr900399q. [DOI] [PubMed] [Google Scholar]
- 23.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.Stelzl U, Worm U, Lalowski M, Haenig C, et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 25.Stumpf MPH, Throne T, de Silva E, Stewart R, et al. Estimating the size of the human interactome. Proc Natl Acad Sci, USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao J, Schmaier A, Chen LM, Yang Z, Chao L. Kallistatin, a novel human tissue kallikrein inhibitor: Levels in body fluids, blood cells, and tissues in health and disease. Journal of Laboratory and Clinical Medicine. 1996;127:612–620. doi: 10.1016/s0022-2143(96)90152-3. [DOI] [PubMed] [Google Scholar]
- 27.Polanski M, Anderson NL. A List of Candidate Cancer Biomarkers for Targeted Proteomics. Biomarker Insights. 2006;2:1–48. [PMC free article] [PubMed] [Google Scholar]
- 28.Cho W, Jung K, Regnier FE. Sialylated Lewis x antigen bearing glycoproteins in human plasma. J Proteome Res. 2010;9:5960–5968. doi: 10.1021/pr100747p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrah T, Deutsch EW, Omenn GS, Campbell DS, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Molecular & Cellular Proteomics. 2011;10:1–14. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia W, Lu Z, Fu Y, Wang HP, et al. A strategy for precise and large scale identification of core fucosylated glycoproteins. Mol Cell Proteomics. 2009;8:913–923. doi: 10.1074/mcp.M800504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson J, Ruetschi U, Halim A, Hesse C, et al. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Meth. 2009;6:809–811. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- 32.Comunale MA, Wang M, Hafner J, Krakover J, et al. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abbott KL, Aoki K, Lim JM, Porterfield M, et al. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J Proteome Res. 2008;7:1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho W, Jung K, Regnier FE. Use of glycan targeting antibodies to identify cancer-associated glycoproteins in plasma of breast cancer patients. Anal Chem. 2008;80:5286–5292. doi: 10.1021/ac8008675. [DOI] [PubMed] [Google Scholar]
- 35.Comunale MA, Lowman M, Long RE, Krakover J, et al. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res. 2006;5:308–315. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 36.Opstal-van Winden AW, Krop EJ, Karedal MH, Gast MC, et al. Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study. BMC Cancer. 2011;11:381. doi: 10.1186/1471-2407-11-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung K, Cho W, Regnier FE. Glycoproteomics of plasma based on narrow selectivity lectin affinity chromatography. J Proteome Res. 2009;8:643–650. doi: 10.1021/pr8007495. [DOI] [PubMed] [Google Scholar]
- 38.Qiu Y, Patwa TH, Xu L, Shedden K, et al. Plasma Glycoprotein Profiling for Colorectal Cancer Biomarker Identification by Lectin Glycoarray and Lectin Blot. J Proteome Res. 2008;7:1693–1703. doi: 10.1021/pr700706s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng Z, Hincapie M, Haab BB, Hanash S, et al. The development of an integrated platform to identify breast cancer glycoproteome changes in human serum. J Chromatogr A. 2010;1217:3307–3315. doi: 10.1016/j.chroma.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.