Abstract
Immunity to intracellular pathogens and cancer relies on the generation of robust CD8+ T cell effector responses as well as the establishment of immunological memory. During a primary immune response CD8+ T cells experience diverse extracellular environmental cues and cell-cell interactions that trigger downstream transcriptional programs ultimately guiding a CD8+ T cell to undertake either an effector or a memory cell fate. Here, we discuss our current understanding of the signaling pathways and transcriptional networks that regulate effector and memory commitment in CD8+ T lymphocytes.
Introduction
CD8+ T cells play a critical role in the immune responses to both intracellular pathogens and cancer [1;2]. Upon pathogen-antigen or tumor-antigen stimulation, naïve CD8+ T cells (TN) undergo a massive clonal expansion to generate large numbers of effector T cells capable of eliminating cells bearing the target antigen. At the end of the primary response the majority of responding CD8+ T cells will undergo apoptosis; however, a small fraction of activated cells will persist long-term establishing a memory T cell population [3]. Expression of killer cell lectin-like receptor G1 (KLRG1) and IL-7 receptor-α (IL-7Rα) on responding CD8+ T cells can distinguish cells that are destined to die or survive as long-lived memory cells. Specifically, IL-7Rα+KLRG1− CD8+ T cells have a greater potential to enter into the memory pool, whereas IL-7Rα−KLRG1+ CD8+ T cells represent terminally differentiated, short-lived effector T cells (SLEC) [4]. The transcriptional regulation of these cell-fate decisions has undergone much scrutiny over the past years. Early studies establishing the transcriptional regulators Eomesodermin (EOMES), T-BET (encoded by T-BOX 21), B-cell CLL/lymphoma 6 (BCL-6) and B lymphocyte induced maturation protein 1 (BLIMP-1, encoded by PRDM1) as critical determinants of CD8+ T cell differentiation have been reviewed in detail elsewhere [5;6]. Here, we discuss more recent advances that have shaped our understanding of the signaling pathways and transcriptional programs that regulate the formation of effector and memory CD8+ T cells.
STAT signaling
Signal transducer and activator of transcription (STAT) signaling pathways are central to the differentiation and long-term survival of CD8+ T cells. Seven members of the STAT family have been described in mammals (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6) [7]. While a single cytokine receptor can activate downstream multiple STATs, most receptors function through a dominant STAT protein. For instance, interleukin (IL)-6, IL-10 and IL-21 preferentially act through STAT3 while IL-12 and IL-2 activate STAT4 and STAT5, respectively (Figure 1).
Figure 1. Signaling pathways modulating memory and effector CD8+ T cell fates.
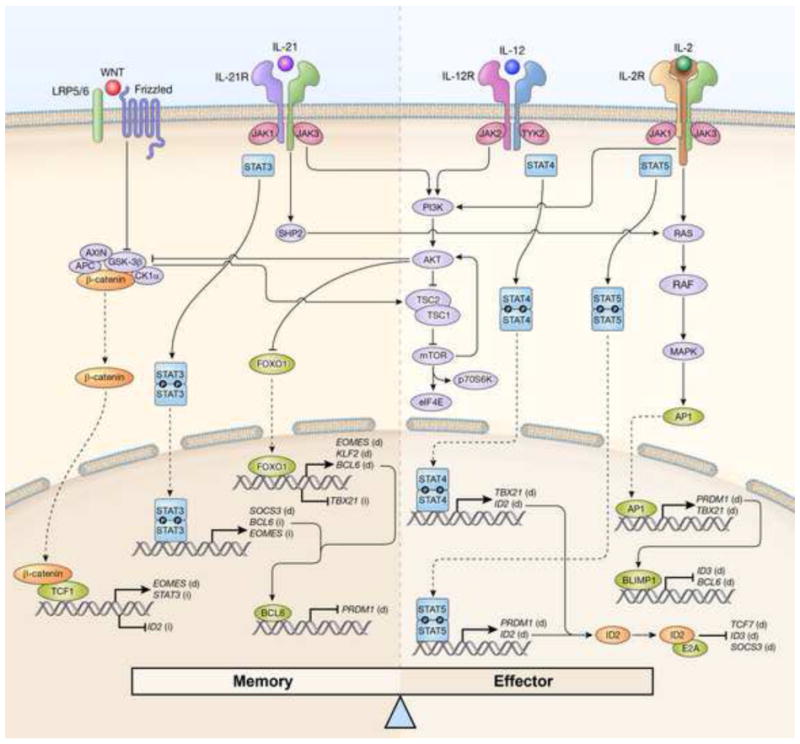
CD8+ T cell memory and effector differentiation are tightly regulated by opposite signals received from inflammatory cytokines and developmental modulators. CD8+ T cell memory is promoted by binding of WNT ligands to Frizzled/Low-density lipoprotein receptor related protein (LRP) 5/6 complexes and by interleukin-21 (IL-21) or other cytokines signaling through signal transducer and activator of transcription 3 (STAT3). Activation of these signaling pathways result in the expression of transcriptional regulators and molecules that favor self-renewal and memory formation, including STAT3, Eomesodermin (EOMES), B-cell CLL/lymphoma 6 (BCL6), Kruppel-like factor 2 (KLF2) and suppressor of cytokine signaling 3 (SOCS3). At the same time, these signaling pathways inhibit the expression of transcription regulators that promote CD8+ T cell effector differentiation and senescence, such as T-BET (encoded by T-box 21 (TBX21)), B-lymphocyte-induced maturation protein 1(BLIMP1, encoded by PR domain containing 1, with ZNF domain (PRDM1)) and inhibitor of DNA-binding 2 (ID2). Conversely, pro-inflammatory cytokines such as IL-2 and IL-12 drive CD8+ T cell differentiation by triggering STAT4 and STAT5 signaling as well as the phosphoinositide-3-kinase (PI3K)/AKT/ mammalian target of rapamycin (mTOR) pathway and the RAS/RAF/ mitogen-activated protein kinase (MAPK) pathway. These signaling pathways induce the expression of the pro-effector molecules T-BET, BLIMP1 and ID2, as well as the inhibition of pro-memory transcription modulators such as forkhead box O1 (FOXO1), BCL6 and ID3. Between these self-renewal and pro-differentiation pathways there is a considerable amount of cross-talk such that the net effect of each pathways is precisely balanced. APC, adenomatous polyposis coli; CK1α, casein kinase 1, alpha 1; GSK-3β, glycogen synthase 3β; TCF, T cell factor; JAK, janus kinase; SHP2, SH2 domain-containing protein tyrosine phosphatase-2; TSC, tuberous sclerosis; eIF4E, eukaryotic translation initiation factor 4E; p70S6K, p70 ribosomal protein S6 kinase 1; TYK2, tyrosine kinase2; AP1, activator protein 1. (d), direct regulation; (i) indirect regulation.
There is now evidence indicating that STAT4 and STAT5 signaling drive T cells towards terminal differentiation, whereas STAT3 withholds differentiation favoring the establishment of CD8+ T cell memory. Increased levels of Stat4 activity resulting from IL-12 signaling promoted the generation of SLEC [4] whereas memory responses were enhanced in mice deficient of IL-12 [8;9]. Sustained Stat5 signaling also favors terminal differentiation as cells perceiving prolonged IL-2 signals exhibited a more pronounced effector phenotype and increased amounts of KLRG1 [10]. By contrast, Stat3 signaling is critical for the generation of memory CD8+ T cells as Stat3-deficient T cells underwent terminal differentiation and failed to form self-renewing TCM [11]. Moreover, disruption of IL-6, IL-10 or IL-21 signaling by genetic depletion of either the cytokine itself or the cytokine receptor resulted in the accumulation of SLEC and impaired memory responses [11–14]. Consistent with these findings, patients with autosomal-dominant hyper-IgE syndrome, a disease often caused by dominant-negative mutations in STAT3, form decreased numbers of TCM and exhibit defective immune responses against viral infections [15]
Mechanistically, the pro-differentiating activity of Stat4 and Stat5 appears to be secondary to the induction of key master regulators of effector differentiation such as T-bet [4;9], Blimp-1 [10;16–18] and, as discussed below, inhibitor of DNA-binding 2 (Id2) [19] (Figure 1). Stat3, instead, was found to control CD8+ T cell differentiation by sustaining the expression of Eomes which is key for the long-term persistence of memory CD8+ T cells as it regulates IL-15-dependent homeostatic turnover via the induction of IL-2Rβ [20], as well as Bcl-6, a transcriptional repressor of Blimp-1 [11;21;22](Figure 1). Additionally, Stat3 can favor memory CD8+ T cell formation by mitigating the activity of IL-12 through the induction of suppressor of cytokine signaling 3 (Socs3) [11] (Figure 1).
WNT–β-catenin signaling
WNT–β-catenin signaling has recently emerged as a critical determinant of CD8+ T cell differentiation. This signaling pathway revolves around β-catenin which in the absence of WNT signals is targeted for proteasomal degradation by a ‘destruction complex’ consisting of Axin, Adenomatosis Polyposis Coli (APC), and the serine/threonine kinases Casein Kinase 1 (CK1) and Glycogen-Synthase Kinase 3β (GSK-3β) [23]. Binding of WNT to the Frizzled receptor and LRP5 or 6 co-receptors triggers a signaling cascade resulting in the disruption of the ‘destruction complex’ and subsequent accumulation and nuclear translocation of β-catenin which allows β-catenin to interact with T cell factor (TCF) and lymphoid enhancer-binding factor (LEF) family members to promote specific gene expression [23] (Figure 1).
Studies employing a physiological WNT ligand, WNT3A, or inhibitors of GSK-3β to activate WNT–β-catenin signaling revealed that this pathway withholds the differentiation of TN into SLEC while promoting the generation of long-lived memory stem cells (TSCM) and central memory T cells (TCM) [24–27]. These observations have recently been corroborated using gain- and loss-of function genetic approaches. Overexpression of a stabilized form of β-catenin in CD8+ T cells hampered cell proliferation and the acquisition of effector functions [28]. Furthermore, constitutive activation of WNT signaling by transgenic expression of Tcf-1 and stabilized β-catenin resulted in reduced expansion of antigen-specific CD8+ T cells and enhanced memory T cell formation [29]. Conversely, deletion of Tcf1 promoted CD8+ T cell differentiation into SLEC and impaired the maintenance of IL-7R+KLRG1− memory precursors resulting in decreased TCM cells and impaired immune responses to pathogen re-challenge [30;31]. Lef-1 deficient CD8+ T cells exhibit only minor alterations in the development of effector and memory responses, indicating functional redundancy in Lef-1 activity through Tcf-1 dependent mechanism[32]. Indeed, deletion of both Tcf-1 and Lef-1 caused virtually a complete loss of memory precursors and failure to mount recall responses [32]. Although deletion of β-catenin alone was insufficient to quench WNT signaling in CD8+ T cells as indicated by WNT reporter activity [33] nor did it alter CD8+ T cell effector and memory responses [34], several lines of evidence indicate that the phenotype of Tcf-1 and Lef-1 knockout cells is dependent on WNT–β-catenin signaling. For instance, memory recall responses were impaired in CD8+ T cells lacking both β-catenin and its homolog γ-catenin [30]. Perhaps more compelling, genetic complementation of Tcf-1 knockouts with Tcf1 p45, but not the p33 isoform which lacks the catenin-binding domain, rescued defective T cell memory responses [30].
How WNT–β-catenin signaling affects CD8+ T cell differentiation has just begun to be elucidated. Tcf-1 was found to partially act through the induction of Eomes [31]. Additional mechanisms might involve an indirect regulation of the pro-memory transcription STAT3 [35] and ID2 as discussed below [36] (Figure 1).
ID proteins
ID proteins are a key family of transcription regulators that control effector and memory CD8+ T cell development. These proteins, which lack a basic DNA-binding region, mainly function as negative regulators of E protein transcriptional activity by forming heterodimer through a helix–loop–helix (HLH) domain and preventing E proteins from binding to DNA [37]. Four ID proteins (ID1–ID4) have been described, but only the function of ID2 and ID3 has been investigated in the differentiation of mature CD8+ T cells. Both Id2 and Id3 are found in naïve CD8+ T cells, but following T cell activation their expression is dichotomously regulated [19;38]. While Id3 expression is actively repressed by Blimp-1 [38], Id2 is upregulated as cells differentiate into SLEC as the result of Stat4/Stat5 signaling [19] and possibly the Tcf-1 down-regulation that accompanies this process [36] (Figure 1). Id2 is required to support the survival of expanding effector CD8+ T cells [39] and to induce sufficient levels of T-bet to sustain the accumulation of SLEC [40;41]. By contrast, Id3 is maintained in memory precursors and is critical for the maintenance of long-lived memory CD8+ T cells [19;38]. Enforced expression of Id3 in SLEC was sufficient to confer long-term survival to these cells otherwise destined to die [38]. Consistently, deletion of the E proteins, E2a and HEB, led to long-term accumulation of KLRG1+ T cells [42]. Both Id2 and Id3 partly operate through inhibition of E2A, but they clearly exert different functions in effector and memory T cells, perhaps as results of diverse binding affinities for E protein members or context-dependent interactions with additional molecules. For instance, Id2 restrained the expression of E2A-target genes Tcf7, Socs3 and Id3, which are key for memory T cell development [41] (Figure 1), and altered the expression of pro- and anti-apoptotic genes such as Bcl2l1, Bcl2 and Serpin peptidase inhibitor, clade B, member 9 (Serpinb9) [39]. Id3, instead, was found to affect a set of genes that control genome stability including forkhead box M1 (Foxm1), NIMA- related kinase 2 (Nek2) and members of the minichromosome maintenance and kinesin complexes [38]. Nevertheless, the activity of Id2 and Id3 appear to extend beyond the mere regulation of E proteins as T cells deficient of both E2A and HEB display a more subtle phenotype compared to those observed in the absence of Id2 and Id3 [42].
PI3K–AKT–mTOR signaling pathway
The nutrient-sensing serine/threonine protein kinase mammalian target of rapamycin (mTOR) is a well-established regulator of cell growth and metabolism, but more recently has emerged as a pivotal modulator of CD8+ T cell fate decisions. mTOR integrates signals from pro-inflammatory cytokines such as IL-2 and IL-12 through phosphoinositol 3-kinase (PI3K)–AKT signaling as well as Wnt proteins via GSK-3β (Figure 1). Sustained activation of AKT/mTOR activity by IL-12 [43], expression of a constitutively active form of AKT [44;45], and deletion of Tuberous sclerosis 1 (Tsc1) [46] all drive naive T cells towards a terminally differentiated effector state. Remarkably, modulation of mTOR activity with low doses of the mTOR inhibitor, rapamycin, increases the numbers of memory T cells as well as promotes the preferential formation of TCM [47–49]. Similarly, pharmacological blockade of AKT can augment CD8+ memory T cells although through a different mechanism involving the rescue of SLEC survival [45].
The transcriptional mechanisms by which AKT/mTOR signaling favor CD8+ T cell effector differentiation have recently been resolved and implicate Forkhead Box O1 (FOXO1) at the center stage [50]. Phosphorylation of FOXO1 by AKT facilitates its binding to cytosolic 14-3-3 scaffold proteins which prevents translocation of FOXO1 into the nucleus. The consequent loss of Foxo1 transcriptional activity resulted in reduced mRNA levels of the pro-memory transcription factors Eomes [43] and Bcl-6 [51] whereas indirectly augmented the expression T-bet [4;50] (Figure 1). Additionally, inhibition of Foxo1 causes down-regulation of Kruppel-like factor 2 (Klf-2), a key regulator of lymph node homing molecules which are critical for TCM and TSCM cell function [52].
The Hippo signaling pathway
The Hippo pathway is an evoultionarily conserved intracellular signal transduction cascade that transduces cell-cell contact signals to trigger differentiation [53]. Recently this signaling pathway has also been demonstrated to affect the transcriptional regulation of differentiation of CD8+ T cells. The core components of this serine/threonine kinase pathway consists of Mammalian sterile-20-like kinase (MST), MOB kinase activator (MOB), Salvador homolog 1 (SAV1) and Large tumor suppressor homolog (LATS) (only MST and LATS are kinases). In the original description of the pathway, it was noted that following cell-cell contact a wave of phosphorylation is initiated leading to activation of the serine kinase, LATS, which in turn phosphorylates the tran- scriptional co-activator Yes-associated protein (Yap) [53]. Phosphorylated Yap is excluded from the nucleus and is degraded, preventing Yap-dependent transcription. In CD8+ T cells the Hippo pathway is assembled in response to the signals necessary for CD8+ T cell differentiation, IL-2, and antigen, but in the absence of contact between activated CD8+ T cells, Hippo signaling is prevented (Figure 2A) [54]. However, in conditions in which antigen/IL-2 activated CD8+ T cells come into contact with each other, triggering of the Hippo pathway leads to the degradation of Yap, suggesting that a receptor/ligand pair is present on activated CD8+ T cells that is absent on TN (Figure 2B) [54]. CTLA-4 and its ligand CD80, which are both induced upon antigen/IL-2 stimulation of TN were found to be the triggering receptor/ligand pair activating the Hippo pathway (Figure 2B) [54]. The recent in vivo demonstration that, following vaccination, activated CD8+ cells in lymph nodes form aggregated synapses with each other, provides visually striking evidence of how this signalling may occur [55]. Hippo activation increases the generation of SLEC in response to infectious challenge [54]. The pro-differentiating effects of Hippo siganling appear to be dependent on suppression of Eomes, and induction of Blimp-1. Indeed, overexpression of a Yap isoform not subject to Hippo-mediated negative regulation enhanced expression of Eomes, suppressed the induction of Blimp-1 and promoted the maintenance of IL-7Rα+ and KLRG1− memory precursors [54].
Figure 2. Differentiation of clonally expanding CD8+ T cells is triggered by cell-cell contact between activated cells.
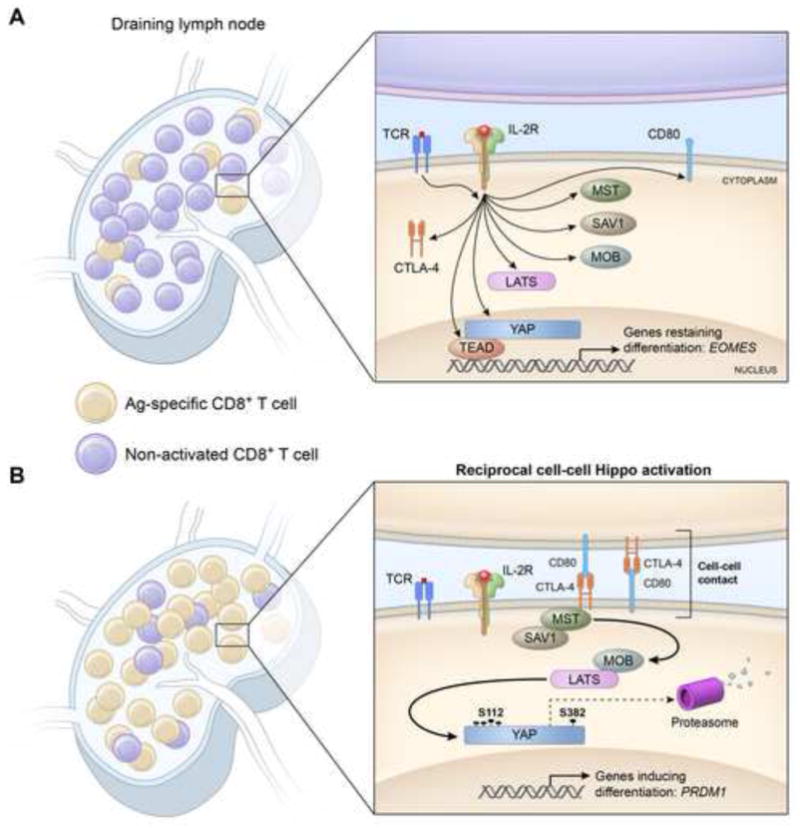
(A) In the initial stages of the primary response, stimulation of CD8+ T cells by antigen and interleukin-2 (IL-2) leads to transcription and translation of the components of the Hippo pathway as well as cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), and its ligand, CD80. However, contact between antigen-specific cells is unlikely, and intra-nuclear Yes-associated protein (Yap) is maintained because the Hippo pathway is not activated. This circumstance enables Yap-dependent transcription to suppress a commitment to terminal differentiation, which may be mediated by the ‘memory’ associated transcription factor, Eomesodermin (EOMES). (B) As clonal expansion increases the frequency of activated antigen-specific T cells, contact between these cells is more likely to occur, leading to surface expression of CTLA-4 and its ligation by CD80, thereby triggering the Hippo signaling cascade. Ultimately, this activates the serine/threonine kinase, Large Tumor Suppressor Homolog (LATS), which phosphorylates Yap at 5 serine residues, one of which, pS112, leads to Yap being trapped in the cytosol, and another, pS382, causes its ubiquitinylation and proteasomal degradation. This loss of Yap is associated with the expression of PR domain containing 1, with ZNF domain (PRDM1) and terminal differentiation. SAV1, salvador homolog 1; MST, mammalian sterile-20-like kinases MOB, MOB kinase activator; TEAD, TEA domain family member.
The realization that CD8+ T cell differentiation could be influenced not only by the inflammatory environment but also by the relative density of responding CD8+ T cells has provided new insights to resolve seemingly ambiguous observations in the field such as the relationship between IL-2 and terminal differentiation. The role of IL-2 in CD8+ terminal differentiation has remained controversial. Studies on its transcriptional effects have demonstrated that it both leads to expression of senescence-associated Blimp-1 [56] and memory-associated Eomes [57]. Furthermore, separate in vivo studies have revealed that antigen-specific cells expressing IL-2 receptor-α, the high affinity IL-2 receptor, have a continued proliferative advantage [58] and are more prone to terminally differentiate [10]. Confusingly, in certain infections, IL-2 signaling appears to be essential for maintenance of a normal proliferative secondary responses [59–61], while for other infection models, IL-2 signaling correlates with loss of secondary responses [62]. A way to resolve these conflicting results is offered by the recent investigations into the Hippo pathway detailed above. IL-2 signaling, in the context of an inactive Hippo pathway, promotes Yap dependent expression of Eomes, which prevents differentiation (Figure 2A). However, in the context of Hippo activation, Yap dependent transcription of Eomes is terminated and IL-2 signaling leads to Blimp-1 expression and terminal differentiation (Figure 2B). Kinetic consideration of the primary immune response suggests when these two different contexts occur. At initiation of the CD8+ T cell response, antigen-specific frequency is low and the chance of activated cell-cell contact is correspondingly low (Figure 2A). However, after clonal expansion, activated cell frequency increases and, with this, there is an increase in the likelihood of activated cell-cell contact triggering the Hippo pathway and so of terminal differentiation (Figure 2B). These findings could provide a mechanism for the important question of how the CD8+ T cell links terminal differentiation to the size of clonal expansion that is reminiscent of quorum sensing in bacteria and yeast, in which population size is sensed by the detection of soluble factors. The advantage of the Hippo pathway is that activation by cell-cell contact is a more direct means for sensing population size than is the concentration of a soluble surrogate of clonal expansion.
Concluding remarks
Over the past decade our understanding of the signaling pathways and transcriptional progams that control effector and memory CD8+ T cell fates has advanced considerably. It is becoming increasingly appreciated that between pathways regualting self-renewal and effector differentiation exist a significant amount of cross-talk such that the net influence of each pathways is finely balanced and tuned. For example, observations in other systems have demonstrated links between WNT and mTOR [63], Hippo and WNT [64;65], STAT3 and WNT [35], and Hippo and mTOR signaling [66]. Moreover, additional complexity is present at the transcriptional level whereby transcription factors that favor memory formation appear to be mutally enhancing and at the same time reciprocally antagonistic to the transcriptional network regulating effector differentiation, and vice versa. In summary, investigations of each pathway individually has provided great insight into what extracellular cues regulate each independently. Future work needs to address which extracellular context selects for the particular pathway that will exert dominant control over the transcriptional regulation of CD8+ T cell differentiation.
Review Highlights.
Evolutionarily conserved signaling pathways such as WNT, STAT3, mTOR and Hippo regulate effector and memory T cell fate decisions.
Cross-talk exists between signaling pathways governing CD8+ T cell differentiation.
Transcription factors controlling memory T cell fates are often self-reinforcing and antagonistic to the transcriptional program regulating effector differentiation.
CD8+ T cell fate is influenced by both the inflammatory environment and the relative density of responding cells
Acknowledgments
This work was supported by the Intramural Research Programs of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research, the Ludwig Institute for Cancer Research, the NIHR Cambridge Biomedical Research Centre and the Wellcome Trust. The authors would like to thank Y. Ji, C.A. Klebanoff and J. Crompton for helpful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Klenerman P, Hill A. T cells and viral persistence: lessons from diverse infections. Nat Immunol. 2005;6:873–879. doi: 10.1038/ni1241. [DOI] [PubMed] [Google Scholar]
- 2.Gattinoni L, Klebanoff CA, Restifo NP. Paths to stemness: building the ultimate antitumour T cell. Nat Rev Cancer. 2012;12:671–684. doi: 10.1038/nrc3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 4.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutishauser RL, Kaech SM. Generating diversity: transcriptional regulation of effector and memory CD8 T-cell differentiation. Immunol Rev. 2010;235:219–233. doi: 10.1111/j.0105-2896.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- 6.Belz GT, Kallies A. Effector and memory CD8+ T cell differentiation: toward a molecular understanding of fate determination. Curr Opin Immunol. 2010;22:279–285. doi: 10.1016/j.coi.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 7.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce EL, Shen H. Generation of CD8 T cell memory is regulated by IL-12. J Immunol. 2007;179:2074–2081. doi: 10.4049/jimmunol.179.4.2074. [DOI] [PubMed] [Google Scholar]
- 9.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- 10.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 11**.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. This study identifies Stat3 as a pivotal transcription factor required for the formation and maintenance of memory T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellino F, Germain RN. Chemokine-guided CD4+ T cell help enhances generation of IL-6RalphahighIL-7Ralpha high prememory CD8+ T cells. J Immunol. 2007;178:778–787. doi: 10.4049/jimmunol.178.2.778. [DOI] [PubMed] [Google Scholar]
- 13.Yi JS, Ingram JT, Zajac AJ. IL-21 deficiency influences CD8 T cell quality and recall responses following an acute viral infection. J Immunol. 2010;185:4835–4845. doi: 10.4049/jimmunol.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572–1576. doi: 10.1126/science.1175194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, Milner JD. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 17.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. This study together with reference 38 demonstrates that the DNA-binding inhibitor Id3 is critical for the maintenance of long-lived memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol. 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 22.Ichii H, Sakamoto A, Hatano M, Okada S, Toyama H, Taki S, Arima M, Kuroda Y, Tokuhisa T. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 23.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, Paulos CM, Muranski P, Restifo NP. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gattinoni L, Ji Y, Restifo NP. beta-catenin does not regulate memory T cell phenotype Reply. Nature Medicine. 2010;16:514–515. doi: 10.1038/nm0510-513. [DOI] [PubMed] [Google Scholar]
- 26.Muralidharan S, Hanley PJ, Liu E, Chakraborty R, Bollard C, Shpall E, Rooney C, Savoldo B, Rodgers J, Dotti G. Activation of Wnt signaling arrests effector differentiation in human peripheral and cord blood-derived T lymphocytes. J Immunol. 2011;187:5221–5232. doi: 10.4049/jimmunol.1101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forget MA, Huon Y, Reuben A, Grange C, Liberman M, Martin J, Mes-Masson AM, Arbour N, Lapointe R. Stimulation of Wnt/ss-catenin pathway in human CD8+ T lymphocytes from blood and lung tumors leads to a shared young/memory phenotype. PLoS One. 2012;7:e41074. doi: 10.1371/journal.pone.0041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Driessens G, Gajewski T. Beta-catenin negatively regulates peripheral T cell activation. J Immunol. 2009;182:35.36. [Google Scholar]
- 29.Zhao DM, Yu S, Zhou X, Haring JS, Held W, Badovinac VP, Harty JT, Xue HH. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J Immunol. 2010;184:1191–1199. doi: 10.4049/jimmunol.0901199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci USA. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. This study identifies Tcf1–β-catenin signaling as critical pathway regulating the formation and maintenance of functional memory CD8+ T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Xue HH. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol. 2012;189:2722–2726. doi: 10.4049/jimmunol.1201150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, Huelsken J, Held W. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 34.Prlic M, Bevan MJ. Cutting edge: beta-catenin is dispensable for T cell effector differentiation, memory formation, and recall responses. J Immunol. 2011;187:1542–1546. doi: 10.4049/jimmunol.1100907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 38.Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, Reger RN, Palmer DC, Borman ZA, Muranski P, Wang E, Schrump DS, Marincola FM, Restifo NP, Gattinoni L. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12:1230–1237. doi: 10.1038/ni.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 40.Knell J, Best JA, Lind NA, Yang E, D’Cruz LM, Goldrath AW. Id2 Influences Differentiation of Killer Cell Lectin-like Receptor G1hi Short-Lived CD8+ Effector T Cells. J Immunol. 2013 doi: 10.4049/jimmunol.1200750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, Speed TP, Busslinger M, Nutt SL, Belz GT. Id2-Mediated Inhibition of E2A Represses Memory CD8+ T Cell Differentiation. J Immunol. 2013;190:4585–4594. doi: 10.4049/jimmunol.1300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Cruz LM, Lind KC, Wu BB, Fujimoto JK, Goldrath AW. Loss of E protein transcription factors E2A and HEB delays memory-precursor formation during the CD8(+) T-cell immune response. Eur J Immunol. 2012 doi: 10.1002/eji.201242497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hand TW, Cui W, Jung YW, Sefik E, Joshi NS, Chandele A, Liu Y, Kaech SM. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proc Natl Acad Sci USA. 2010;107:16601–16606. doi: 10.1073/pnas.1003457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim EH, Sullivan JA, Plisch EH, Tejera MM, Jatzek A, Choi KY, Suresh M. Signal Integration by Akt Regulates CD8 T Cell Effector and Memory Differentiation. J Immunol. 2012 doi: 10.4049/jimmunol.1103568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. This study together with reference 48 shows that CD8+ T cell effector and memory fates are controlled by pharmacologically targetable metabolic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, Rao RR, Araki K, Pollizzi K, Odunsi K, Powell JD, Shrikant PA. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–553. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA. Transcription Factor Foxo1 Represses T-bet-Mediated Effector Functions and Promotes Memory CD8(+) T Cell Differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, Tamas P, Feijoo C, Okkenhaug K, Cantrell DA. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Thaventhiran JE, Hoffmann A, Magiera L, de la Roche M, Lingel H, Brunner-Weinzierl M, Fearon DT. Activation of the Hippo pathway by CTLA-4 regulates the expression of Blimp-1 in the CD8+ T cell. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1209115109. This study highlights the importance of the Hippo signaling pathway in linking the magnitude of clonal expansion to CD8+ T cell terminal differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerard A, Khan O, Beemiller P, Oswald E, Hu J, Matloubian M, Krummel MF. Secondary T cell-T cell synaptic interactions drive the differentiation of protective CD8 T cells. Nat Immunol. 2013;14:356–363. doi: 10.1038/ni.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 57.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505–509. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441:890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feau S, Arens R, Togher S, Schoenberger SP. Autocrine IL-2 is required for secondary population expansion of CD8(+) memory T cells. Nat Immunol. 2011;12:908–913. doi: 10.1038/ni.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bachmann MF, Wolint P, Walton S, Schwarz K, Oxenius A. Differential role of IL-2R signaling for CD8+ T cell responses in acute and chronic viral infections. Eur J Immunol. 2007;37:1502–1512. doi: 10.1002/eji.200637023. [DOI] [PubMed] [Google Scholar]
- 62.Obar JJ, Molloy MJ, Jellison ER, Stoklasek TA, Zhang W, Usherwood EJ, Lefrancois L. CD4+ T cell regulation of CD25 expression controls development of short-lived effector CD8+ T cells in primary and secondary responses. Proc Natl Acad Sci USA. 2010;107:193–198. doi: 10.1073/pnas.0909945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 64.Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. Role of TAZ as Mediator of Wnt Signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 65.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, Shao DD, Schumacher SE, Weir BA, Vazquez F, Cowley GS, Root DE, Mesirov JP, Beroukhim R, Kuo CJ, Goessling W, Hahn WC. beta-Catenin-Driven Cancers Require a Yap1 Transcriptional Complex for Survival and Tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD, Guan KL. Yap mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]


