Abstract
Recent studies suggested that miRNAs are involved in the development of the pathogenesis of HIV-associated nephropathy (HIVAN). Rapamycin, a widely used mTOR inhibitor, has been demonstrated to slow down the progression of HIVAN. However, the role of miRNA in the regulation of these processes has not been investigated so far. In the current study, we have used a microarray-based approach in combination with real-time PCR to profile the miRNA expression patterns in rapamycin-treated HIVAN mice (Tg26). Our results demonstrated that 19 miRNAs belonging to 13 different families expressed differentially in renal tissues of rapamycin-receiving Tg26 mice when compared to Tg26 mice-receiving saline only. The patterns of miRNAs expression in rapamycin-receiving Tg26 mice took a reverse turn. These miRNAs were classified into 8 functional categories. In in vitro studies, we examined the expression of specific miRNAs in HIV-1 transduced human podocytes (HIV/HPs). HIV/HPs displayed attenuation of expression of miR-99a, -100a, -199a and miR-200, whereas, rapamycin inhibited this effect of HIV. These findings suggest that rapamycin-mediated up-regulation of specific miRNAs could contribute to amelioration of renal lesions in HIVAN mice.
Keywords: MicroRNA, HIV-associated nephropathy, Epithelial mesanchymal transition, Apoptosis, Mammalian target of rapamycin
Introduction
MicroRNAs (miRNAs) are a family of small, 20- to 22-nucleotide (nt)-long noncoding RNAs, which have been implicated in the regulation of multiple biological processes including developmental timing, signal transduction, and cell maintenance and differentiation [17,32]. Recent studies indicated that miRNAs play important roles in the maintenance of glomerular homeostasis and progression of renal disease [2,4]. Profiles of miRNAs expression in kidney diseases, i.e., polycystic kidney disease [26], diabetic nephropathy [18], ischemia reperfusion injury [9], renal cancer [10], hypertensive nephrosclerosis [37] and kidney fibrosis [6,38] have been investigated. Recently, we reported the profile of miRNAs expression in HIV-1 transgenic mice (Tg26) [5], the most frequently used mouse model of HIV-associated nephropathy (HIVAN) [15]. This report suggested that miRNAs had potential to play a role in the pathogenesis of HIVAN.
HIVAN is the third leading cause of end-stage renal disease in the HIV-1-seropositive African American males, and is character-ized by collapsing focal segmental glomerulosclerosis and micro-cystic dilatation of tubules [1,19]. However, the exact mechanism which contributes to the pathogenesis of HIVAN is not clear. Recently, the role of mammalian target of rapamycin (mTOR) pathway has been demonstrated in the progression of in HIVAN [16]. Rapamycin, an inhibitor of mTOR pathway, not only inhibited the activation of the mTOR pathway but also attenuated the development of proliferative phenotype in HIVAN [35].
Rapamycin is a macrolide which has been implicated in repressing HIV-1 replication in both T cells and peripheral blood leukocytes [31]. The inhibitory effect of rapamycin on HIV-1 replication was associated with diminished basal HIV-1 long-terminal repeat (LTR) gene expression [31]. Rapamycin has been demonstrated to decrease transcription of LTR both in renal tissues of HIVAN mice in in vivo studies and HIV-infected podocytes in in vitro studies [28]. However, the role of miRNA in the regulation of these processes has not been reported so far.
The potential role of miRNA in mTOR regulation has previously been described in human cancer cells [8,25]. MiR-99a induced down regulation of mTOR and suppressed tumor growth in certain types of human cancer cells [23,25]. MiR-199 has been demonstrated to target mTOR in human hepatocellular carcinoma (HCC): over expression miR-199 led to G1-phase cell cycle arrest and reduced invasive capability in cancer cells [8]. In cardiac cells, miR-199a was found to be a master regulator of a hypoxia-triggered pathway and can be exploited for preconditioning of cells against hypoxic damage [29]. These results suggested that miRNAs had potential to play a role in regulation of mTOR pathway in HIVAN.
We recently reported the miRNAs’ expression, including miR-200 family, were down regulated in HIVAN mice and HIV-transduced podocytes [5]. MiR-200 has been revealed to inhibit EMT in cancer cells as well as in renal tubular cells by targeting Zeb1 and Zeb2, the negative transcription factors of E-cadherin [11,27]. We have demonstrated that epithelial–mesenchymal transition (EMT) in renal epithelial cells could contribute to the development of proliferative phenotype in HIVAN [16]. These results indicate that miR-200 can be involved in the progression of HIVAN. In addition, rapamycin has been demonstrated to modulate HIV transcription as well as attenuation of the proliferative phenotype of HIVAN through inhibition of epithelial mesenchymal transition (EMT) in renal cells [35]. However, the contribution of rapamycin to miRNA expression in HIVAN and their potential role in the pathological processes has not been investigated to date.
In the present study, we evaluated the effect of rapamycin on the miRNA expression patterns in HIVAN (Tg26) mice. We used a microarray-based approach to examine the miRNA expression in rapamycin-treated HIVAN mice. The miRNA expression was further validated and confirmed by quantitative PCR in renal tissues and HIV-transduced human podocytes. Our results showed that selective miRNAs were down regulated in HIVAN mice, and the patterns of miRNA expression could be modulated by rapamycin. These studies suggested that rapamycin-induced attenuation of pathological processes in HIVAN was probably mediated through reversible regulation of miRNA expression.
Materials and methods
Animals
The Ethics Review Committee for Animal Experimentation of Feinstein Institute for Medical Research-North Shore LIJ Health System approved the study protocols. FVB/N background of HIV transgenic mice (Tg26), which have the proviral transgene, pNL4-3:d1443, encoding all the HIV-1 genes except gag and pol, were kindly gifted by Dr. Paul E. Klotman (Mount Sinai Medical Center, New York, NY). They were bred on FVBN background and maintained in a laminar-flow facility (Animal Facility of Feinstein Institute). Breeding pairs of FVB/N mice were originally purchased from Jackson Labs (Bar Harbor, ME). Genotyping for Tg26 was performed by DNA amplification for HIV-1 gene (forward primer: 5′-ACATGAGCAGTCAGTTCTGCCGCAGAC-3′; reverse primer: 5′-GTGTGGACGCGTAGTCTCAGGAAC-3′). These mice developed proteinuria at approximately 24 days of age, progressed into nephrotic syndrome and renal failure, and were used as mouse model of HIVAN.
Experimental protocol
Tg26 mice (aged 3 weeks, n=6) were administered 5 mg/kg rapamycin (Sigma-Aldrich) or normal saline by intraperitoneal (i.p.) injection every other day for 4 weeks. Age- and sex-matched control (FVBN) mice (n=6) were also administered normal saline for the same duration. At the end of scheduled periods, the animals were anesthetized and sacrificed. Kidneys were removed, one kidney was stored for RNA and microRNA analysis at −80 °C and the other was fixed in 10% buffered formalin for 24 h, for renal histology.
Grading of renal lesions
Tissues were embedded in paraffin and 5 μm thick sections were cut and stained with hematoxylin–eosin and Periodic Acid Schiff (PAS). Renal histology was scored for both glomerular and tubular injury. Twenty random microscopic fields per tissue section were reviewed under ×200 magnification with blinded observers. Glomerular lesions were classified as segmental glomerulosclerosis (SGS), global glomerulosclerosis (GGS), and collapsing glomerulosclerosis (CGS). Tubular injury was scored in the form of tubular dilatation (% of dilated tubules/section) and size of microcysts (1+ to 4+).
RNA extraction and miRNA microarray
Total RNA was extracted with Trizol reagent (Invitrogen) from kidney tissues of FVB/N, Tg26 and rapamycin-treated Tg26 mice (n=3 each group). MicroRNA expression analysis was performed by LC Sciences Microarray Service (Houston, TX). The standard mouse chip MRA-1002 (μParaflo® microfluidic chip platform) was used. The chips layout included probes (in triplicates) against all unique mature miRNAs from miRBase version 17. The samples were run on chips with three probe repeats for each miRNA. The signals were normalized using a LOWESS (Locally-weighted Regression) method. The average of these three probe repeats was used. The ratios of Tg26 vs. control and rapamycin-treated Tg26 vs. control for differentially expressed miRNAs were log2 transformed and plotted. The differentially expressed miRNAs were further analyzed with assistance of Database (TargetScan 6.2 and miRDB) for miRNA target prediction and functional annotation. The biological association networks and specific pathways were analyzed using PANTHER classification system (version 7.2).
Pseudotyped retrovirus production
The replication defective HIV-1 virus stocks were obtained by cotransfected pNL4-3:ΔG/P-GFP construct and packaging constructs pCMV R8.91 and pMD.G plasmids into 293 T cells as previously described [5]. Briefly, pNL4-3:ΔG/P-GFP construct was produced by reconstruction of pNL4-3 clone, the same proviral construct used to generate Tg26 transgenic mice, by substituting gag/pol genes with GFP reporter gene. The HIV-1 gag/pol and VSV.G envelope were provided by pCMV R8.91 and pMD.G plasmids, respectively. The negative control virus was generated from pHR-CMV-IRES2-GFP-ΔB construction, which carried HIV-1 long term repeats and GFP reporter. The viral concentration was titrated by infecting Hela cells with 10-fold serial dilution of virus stocks and analyzed by FACS for GFP. Viral stocks ranging from 106 to 108 GEU (titration units) per ml were obtained.
Human podocytes transduction
Immortalized human podocytes (HPs) were kindly gifted from Dr. Moin A. Saleem (Children’s Renal Unit and Academic Renal Unit, University of Bristol, South Mead Hospital, Bristol, UK). The cells were conditionally immortalized by transfection of human telomerase gene and additionally introducing a temperature-sensitive SV40-T antigen, which allowed the cells to proliferate at a permissive temperature of 33 °C and enter growth arrest at a nonpermissive temperature of 37 °C. Cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mm l-glutamine and 1% insulin–transferrin–selenium solution (ITS, Invitrogen). HPs were incubated with 1 × 106 GEU of HIV-1 pseudotyped virus per 1 × 106 cells at 37 °C and replaced with fresh medium after 2 h. The control vector was used as a negative control. GFP expression was examined under fluorescence microscopy, 24 h after virus infection. The cells were harvested after 72 h of infection. Total cellular RNA was isolated (Trizol) and subjected to qPCR analysis.
Quantitative reverse transcription PCR analysis
Quantitative RT-PCR (qPCR) reactions were performed to validate and confirm miRNAs expression in renal tissues of FVB/N, saline-receiving Tg26, and rapamycin-receiving Tg26 mice, and HIV-1 transduced human podocytes using ABI Prism 7900HT sequence detection system. cDNA was synthesized using NCode™ EXPRESS SYBR® GreenER™ miRNA qRT-PCR Kit (Invitrogen) according to the manufacturer’s instructions. SYBR Green assays were performed using forward primers from sense strands of specific mature miRNAs and universal reverse primer (Invitrogen). U6 small nuclear RNA (snRNA) was used as endogenous control to normalize the respective miRNA cycle thresholds (Ct) values. The relative expression level of miRNA was calculated using modified 2−ΔΔCt method [36].
Results
Rapamycin attenuates renal lesions in HIVAN mice
Periodic acid-Schiff (PAS) stained renal cortical sections of 8 weeks old FVB/N mice, saline- receiving Tg26 mice, and rapamycin-receiving Tg26 mice were coded and evaluated for severity of renal lesion by two investigators unaware of the experimental conditions. Tg26 mice displayed both sclerosed glomeruli, including focal segmental glomerulosclerosis (FSGS) not otherwise specified, tip variant, hilar variant and global sclerosis, and collapsed glomeruli along with tubular dilatation and micro cyst formation. Rapamycin-receiving Tg26 mice displayed only occasional glomerular lesions with minimal tubular dilatation and micro cyst formation (Fig. 1).
Fig. 1.
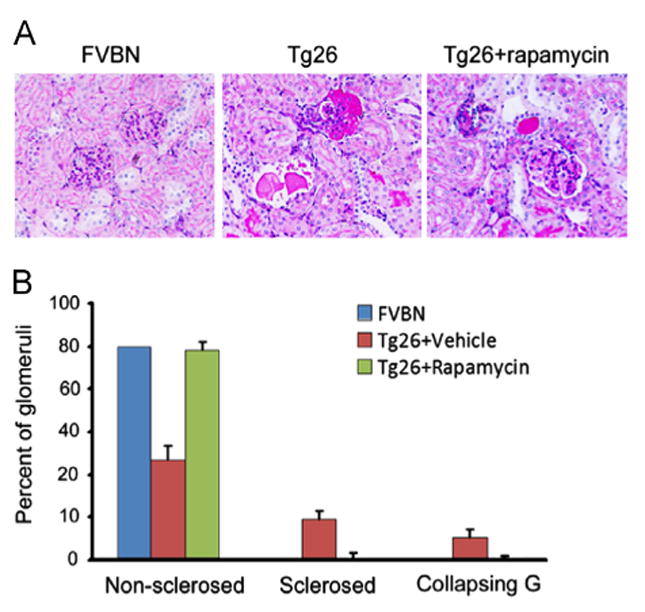
Rapamycin attenuates renal lesions in HIVAN mice. (A) Representative microphotographs of FVB/N, Tg26 and rapamycin-treated Tg26 mice. Tg26 mice-receiving normal saline showed sclerosed glomerulus and tubular cyst filled with proteinaceous cast. Rapamycin-treated Tg26 mice showed minimal tubular dilatation without glomerulosclerosis. (B) Cumulative data showing percentage of normal glomeruli, sclerosed glomeruli and collapsing glomeruli.
Differential miRNAs expression
A global miRNA expression profile (total number of 1096 miRNAs analyzed) in renal tissues of saline-receiving Tg26 and rapamycin-receiving Tg26 was developed by microarray. MiRNAs expression levels in rapamycin-receiving Tg26 differed considerably from saline-receiving Tg26 mice. Rapamycin led to reverse expression patterns of miRNAs in Tg26 mice. After excluding miRNAs, either expressed at extremely low levels (<500) or statistically not significant (p>0.05), 19 miRNAs belonging to 13 different families were differentially expressed in renal tissues of rapamycin-receiving Tg26 mice when compared to renal tissues of saline-receiving Tg26 mice (p<0.05). Of these miRNAs, the down regulated mRNAs in Tg26 mice were as follows: miR-497, miR-140, miR-145, miR-331, miR-16, miR-30a, miR-22, miR-30c, miR-378, miR-99a, miR-100, miR-199a, miR-200a, miR-200b, miR-200c and miR-429; whereas miR-466i, miR-467f, and miR-669a were upregulated in rapamycin-receiving mice (Fig. 2).
Fig. 2.
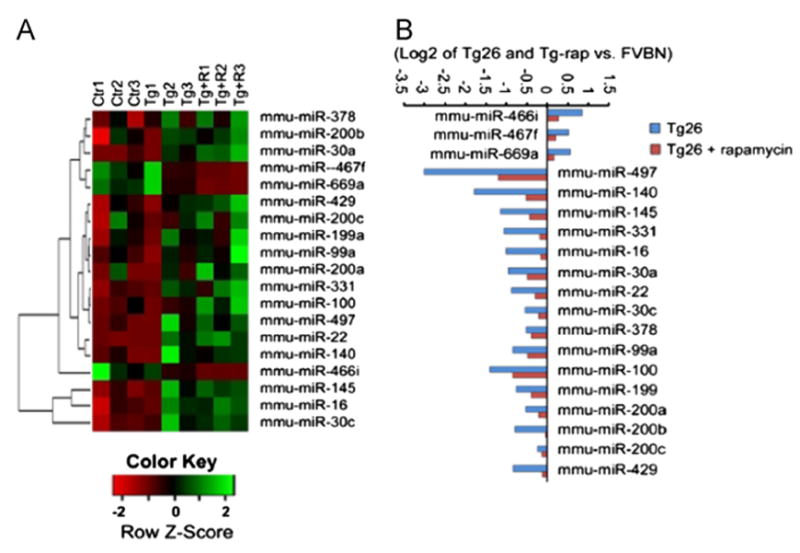
Differential expression of miRNAs in kidneys of Tg26 and rapamycin-treated Tg26 mice. (A) Heatmap depicts triplicate microarray hybridizations, revealing a subset of miRNAs that are differentially expressed in rapamycin-treated Tg26 mice compared with Tg26 and control FVB/N mice. The color scale shown at the bottom: green denotes expression >40 and red denotes an expression <0. (B) The relative expression of miRNAs level is present in log2 transformation. Total 3 miRNAs were upregulated (right) and 16 miRNAs were downregulated (left) in rapamycin-treated Tg26 mice (P<0.05).
We confirmed the reversal of miRNA expressions in renal tissues of rapamycin-receiving Tg26 kidneys using quantitative real-time PCR (qPCR). Consistent with the microarray analysis, qPCR demonstrated that expression of miR-466i, miR-466f and miR-699a was upregulated, whereas expression of miR-497, miR-140, miR-145, miR-331, miR-16, miR-30a, miR-22, miR-30c, miR-378, miR-99a, miR-100, miR-199a, miR-200a, miR-200b, miR-200c, miR-141 and miR-429 was diminished in renal tissues of saline-receiving Tg26 mice. The changes ranged from 0.5 to 3.5 folds, as shown in Fig. 3. Rapamycin-receiving Tg26 mice displayed an inverse miRNA expression profile when compared to saline-receiving Tg26 mice.
Fig. 3.
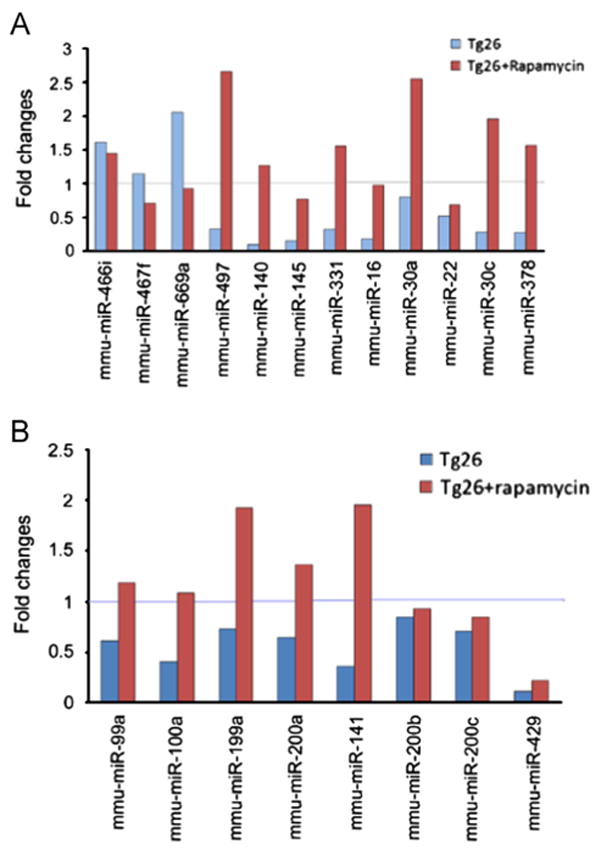
Validation and confirmation of miRNAs expression by real-time quantitative PCR. qPCR demonstrated that rapamycin led to inverse expression patterns of miRNAs in Tg26 mice. Of these, miR-466i, miR-467f and miR-669a were up regulated and miR-497, miR-140, miR-145, miR-331, miR-16, miR-30a, miR-22, miR-30c, miR-378, miR-99a, miR-100a, miR-199a, miR-200a, miR-141, miR-200b, miR-200c and miR-429 were down regulated at 8 wks in HIVAN mice. U6 snRNA was used as an internal control. The relative expression level of specific miRNA was calculated using modified 2−ΔΔCt method.
Microarray analysis of genes involved in HIVAN
In order to understand the functional aspects of the differentially expressed miRNAs in rapamycin-receiving mice, we carried out the biological pathway analysis of the predicted target genes of these miRNAs. We mapped the differentially regulated genes from miRNA target prediction and functional annotations using Tar-getScan 6.2 (http://www.targetscan.org) and miRDB (http://mirdb.org). Only commonly predicted targets for different miR NAs were taken for further analysis. Altogether, from both TargetScan and miRDB genes were identified as miRNA targets. The biological functions of these genes were further categorized into 8 functional groups (Fig. 4A), based on the PANTHER classification system (version 7.2) analyses (http://www.pantherdb.org/panther).
Fig. 4.
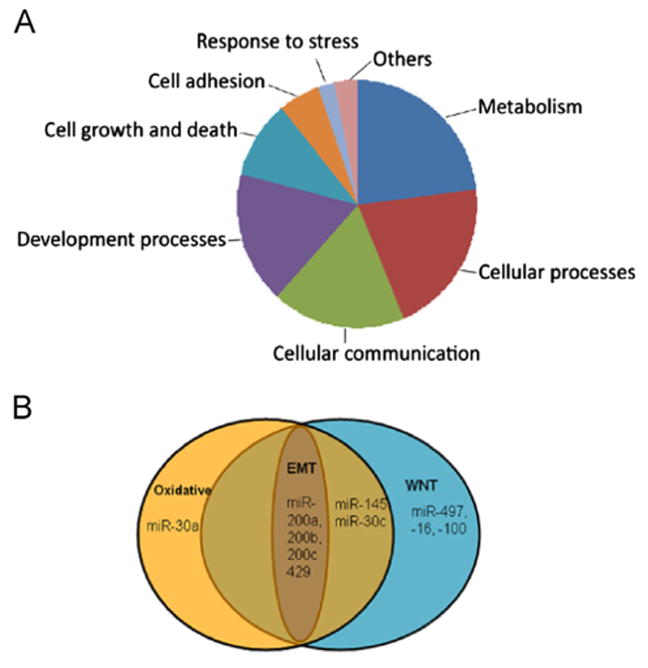
Overrepresented functional categories of the differential expressed miRNAs in HIVAN. (A) The pie-chart shows 8 functional categories and each pie represents a functional category with an overrepresentation of regulatory pathways of miRNA targets. (B) Overlapping of miRNAs in Wnt pathway, oxidative pathway, and EMT pathway.
To identify miRNA-target interaction, we mapped the possible targets of these miRNAs and the molecular pathways they are involved in. We demonstrated 57 genes which could be identified as miRNA targets. These are associated with 29 pathways (Table 1). Of these, major signal transduction pathways were previously described as being involved in various aspects of kidney diseases, such as the mTOR pathway, Wnt pathway, TGFβ pathway, cadherin signaling pathway, oxidative pathway, P53, and JAK/STAT signaling pathway, [3,6,13,14,38,21,25,30].
Table 1.
miRNAs and their targets.
| miRNA | Target genes | Pathways | Log2Tg-rap/Tg |
|---|---|---|---|
| miR-466i | Gli3, Srgap2, Csrnp3, Lmx1a, Nrf1, Strbp, Csnk1e, Csnk1d | Hedgehog signaling pathway, PDGF signaling pathway, Apoptosis, Cell cycle, Cell communication, Response to stress, Wnt pathway | −0.57 |
| miR-467f | Ppp2r2a, Foxj3, Casp2, Cxcr4 | FGF signaling pathway-PI3 kinase pathway, TGF-beta signaling pathway, Apoptosis signaling pathway cytokine signaling pathway | −0.33 |
| miR-669a | Ptpre, Cdc14a, Ank1, Egr1, Rab6, Ankrd45 | Cell cycle, cell communication, Angiotensin II-stimulated signaling through G proteins and beta-arrestin, Response response to stress | −0.40 |
| miR-497/16 | Atp7a, Zbtb34Prkar2a, Akt3Tlk1Traf3 | Heterotrimeric G-protein signaling pathway, Ras Pathway, Hypoxia response via HIF activation, FAS pathway, p53 pathway feedback loops 2, Cell cycle, cytokine-mediated signaling pathway | 1.79 0.84 |
| miR-140 | Pdgfra, Ppp2r3a, Nlk, Wnt9a, Foxp2, Bcl2l1 | FGF pathway, Angiogenesis, Wnt pathway, Cadherin pathway, Insulin/IGF pathway, TGF-beta pathway, Apoptosis pathway | 1.28 |
| miR-145 | Epb4.1l5, Add3, Yes1, Rasa2, Nras, PPP3CA, PPP3R2, DUSP | PDGF pathway, B cell activation, Cadherin pathway, PDGF pathway, FGF pathway, PI3 kinase pathway, VEGF pathway, Ras Pathway, Wnt pathway, Oxidative pathway | 0.71 |
| miR-331 | Cdh9, ING5 | Wnt signaling pathway, Cadherin signaling pathway | 0.86 |
| miR-30a/c | Cyp24a1, Mkrn3, Celsr3, Klhl20, ppp2r5e, APC | Vitamin D metabolism, EGF, FGF receptor pathway, p53 pathway, PI3 kinase pathway, Cadherin pathway, Apoptosis pathway, TGF-beta pathway, Wnt pathway, Oxidative pathway | 0.45 0.33 |
| miR-22 | Arrb1, Esr1, Clic4, Prkar2a, Styx | Wnt pathway, Angiotensin II-stimulated signaling, cytokine pathway, p53 pathway, Dopamine receptor mediated signaling pathway, Beta1 adrenergic receptor pathway, Oxidative stress response | 0.58 |
| miR-378 | Efna5, Arf2, Dcbld2, Frk, Mapk1 | Integrin pathway, Insulin/IGF pathway, TGF-beta pathway, Interleukin pathway, PI3 kinase pathway, Apoptosis pathway, B cell activation, Angiotensin II-stimulated signaling, Ras Pathway | 0.11 |
| miR-100/99 | MTOR, KBTBD8, TARDBP | PDGF signaling pathway, EGF receptor signaling pathway, Hypoxia response via HIF activation, p53 pathway by glucose deprivation | 0.38 0.56 |
| miR-199 | BCAR3, MAP3K4, CDK17, MTOR | PDGF signaling pathway, EGF receptor signaling pathway, Hypoxia response via HIF activation, p53 pathway by glucose deprivation | 0.37 |
| miR-200a/b/c/429 | ZEB2, ZEB1, DUSP Wnt 16, ABL2, MAP2K4 | Insulin/IGF pathway, FAS signaling pathway, FGF signaling pathway, Oxidative stress response, Integrin signalling pathway, p38 MAPK pathway, EGF receptor signaling pathway, Apoptosis signaling pathway | 0.32 0.73 0.1 0.71 |
Interestingly, we found a number of miRNAs in both oxidative and Wnt pathways. For example, miR-30, miR-145 and miR-200 families were involved in oxidative pathway by targeting STYX and DUSP6 genes; at the same time, these miRNAs were involved in Wnt pathways by targeting APC, PPP3CA and Wnt16 genes (Fig. 4B and Table 2). Among these miRNAs, miR-200 families, including miR-200a, miR-200b, miR-200c, miR-429, and miR-141 were demonstrated to regulate cellular phenotype transition by targeting Zeb1 and Zeb2 (Fig. 4B and Table 2). This indicates that miRNAs played a role in amelioration of renal lesions in rapamycin-receiving Tg26 mice.
Table 2.
MicroRNAs overlapping and Wnt pathways and their predicted targets.
| miRNA | Oxidative pathway | Wnt pathway | ||
|---|---|---|---|---|
| mmu-miR-30a | STYX | Serine/threonine/tyrosine interacting protein | APC, PPP2R5E | Adenomatous polyposis coli, protein phosphatase 2, regulatory subunit B′, epsilon isoform |
| mmu-miR-145 | DUSP6 | Dual specificity phosphatase 6 | PPP3CA, PPP3R2 | Protein phosphatase 3, catalytic subunit, alpha isoform, protein phosphatase 3, regulatory subunit B, alpha isoform |
| mmu-miR-200a | DUSP3 | Dual specificity phosphatase 3 | Wnt16 | Wingless-type MMTV integration site family, member 16 |
| mmu-miR-200b | DUSP1 | Dual specificity phosphatase 1 | Wnt16 | Wingless-type MMTV integration site family, member 16 |
| mmu-miR-200c | DUSP1 | Dual specificity phosphatase 1 | Wnt16 | Wingless-type MMTV integration site family, member 16 |
| mmu-miR-429 | DUSP1 | Dual specificity phosphatase 1 | Wnt16 | Wingless-type MMTV integration site family, member 16 |
MiRNA expression in human podocytes (HPs)
To examine whether rapamycin modulated miRNA expression in HIV-1 infected renal cells, we transduced HPs with a recombinant retrovirus encoding HIV-1 genes. We examined expression of miR-99a, miR-100a, miR-199a, and miR-200, which potential target mTOR or regulate EMT by targeting Zeb1 [25,8,11] by real time PCR. Our results showed that the pattern of miRNA expressions in HIV-transduced HPs (HIV/HPs) was similar to that in renal tissues of Tg26 mice. For example, HIV down regulated miR-99a, miR-100a, miR-199a, miR-200a, miR-200b, miR-200c, miR-429 and miR-141 expression, whereas, rapamycin exerted the opposite effect (Fig. 5). The fold changes were in a wide range from 0.5 to 4 fold, however they displayed identical trends (Fig. 5). These findings suggest that rapamycin regulated HIV-induced miRNA expression and provided protection against HIV-induced kidney cell injury in HIVAN mice.
Fig. 5.
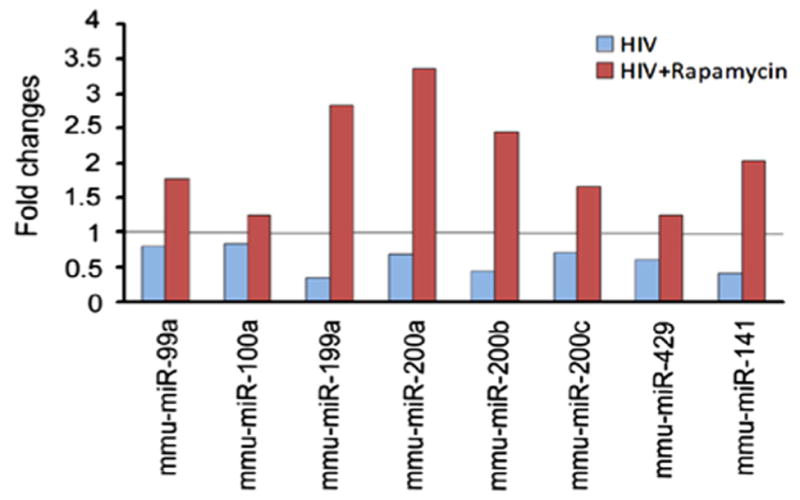
MiRNAs expression in HIV-transduced human podocytes (HIVHPs). HPs were infected with HIV-1 virus for 72 h. Real-time PCR analysis showed that HIV induced up regulation of miR-99a, miR-100a, miR-199a, miR-200a, miR-200b, miR-200c, miR-429 and miR-141; whereas rapamycin led to inverse expression of these miRNAs in HIV/HPs. U6 snRNA was used as an internal control. The relative expression level of specific miRNA was calculated using modified 2−ΔΔCt method.
Discussion
The present study provides insights into the modulatory effects of miRNA expression in the pathogenesis of HIVAN. Rapamycin reversed HIV-induced down regulation of miRNA expression in renal tissues of HIVAN mice. In vitro studies confirmed the modulatory effects of rapamycin on miRNA expression in HIV/HPs. These studies support the notion that rapamycin has potential to attenuate the progression of HIVAN phenotype through modulation of miRNA expression.
Rapamycin is currently used alone or in combination with other immunosuppressive drugs to prevent renal graft rejection. Among the potential positive clinically relevant effects is rapamycin’s capacity to interfere with the virus replication. It has been reported that rapamycin exhibits anti-HIV properties in multiple ways, e.g., inhibition of LTR-mediated HIV gene transcription, increasing the anti-HIV activity of HIV-entry inhibitors, and down regulation of CCR5 ([7,12] and [24]). On that account, the potential benefits of rapamycin as a therapeutic drug for the treatment of HIVAN has been studied [35]. A recent report demonstrated that rapamycin attenuated transcription of HIV LTR both in HIVAN mice and HIV-infected podocytes [28]. These properties point out that rapamycin is not only an immunosuppressant but has a potential to emerge as an antiviral drug for the treatment of HIV infected patients who are susceptible for developing HIVAN.
In addition to the inhibition of HIV gene transcription, rapamycin has also been reported to ameliorate renal lesions in HIVAN through different mechanisms. Recently, the role of mTOR pathway has been demonstrated in the development of HIVAN proliferative phenotype in cystic tubules of HIVAN mice [16]. By targeting mTORC1 in kidney cells, rapamycin inhibits HIV-induced activation of mTORC1 which manifested in the form of attenuated renal lesions, proteinuria and uremia. Besides inhibiting the mTOR pathway, rapamycin has also been reported to attenuate HIVAN phenotype by modulating HIV-mediated renal cell EMT [35]. HIV-1 promotes enhanced expression of Snail/ZEB transcription factors, which reversed the repression of the transcription of E-cadherin. Rapamycin treated HIVAN mice not only showed an attenuated renal cell expression of ZEB1/2 but also displayed restoration of renal cell expression of E-cadherin [35]. Basd upon these findings, we evaluated the alterations in miRNA expressions in rapamycin-receiving HIVAN mice. Our microarray analysis revealed that rapamycin modulated renal tissue miRNA expression in HIVAN mice. Interestingly, rapamycin displayed a reverse trend in renal tissues of miRNA expression in HIV transgenic mice. Comparison of these differentially regulated miRNAs and their potentially targeted genes, suggested that these genes were involved in critical pathological processes of HIVAN, such as Wnt signaling [13], cadherin signaling [14], and oxidative stress [20]. These results point out that miRNAs also contributed to rapamycin-induced amelioration of HIVAN.
Oxidative stress and endoplasmic reticulum stress have been demonstrated to play a crucial role in renal tubular damage in acute kidney injury as well as chronic kidney disease [22]. Our recent studies also showed that HIV gene expression stimulates ROS generation and induces renal tubular cells apoptosis both in vivo and in vitro [33], indicating that oxidative stress plays a role in the development of HIVAN. There have been many reports recently suggesting the involvement of miRNA in oxidative stress response [20,22]. It has been indicated that miR-205 exhibits a protective role against both oxidative and endoplasmic reticulum stress [22]. MiR-200a and miR-141 could modulate the oxidative stress response by targeting p38α in cancer cells [20]. Our current data demonstrates that HIV induced down regulation of miR-200 expression in both renal tissues of HIVAN mice and HIV-transduced podocytes. On the contrary, rapamycin showed an opposite effect by reversing miR-200 expression, indicating that miR-200 is involved in the oxidative stress response in HIV-mediated renal cell injury. Further functional roles of the specific miRNAs in HIVAN is worth exploring in future studies.
Interestingly, miR-200 has been demonstrated to play a critical role in the cellular phenotype transition (EMT) in cancer cells as well as in kidney cells by targeting Zeb1 and Zeb2 [11,13,27,34]. Rapamycin modulates HIVAN phenotype through inhibition of EMT in renal cells [35]. In those studies, rapamycin preserved renal epithelial cell expression of E-cadherin in HIVAN mice by inhibiting renal cell ZEB expression; whereas, HIVAN mice showed enhanced proliferation of glomerular and tubular cells [35]. Our present data suggest that rapamycin not only modulates miRNA expression, especially reversed miR-200 expression but also attenuates the progression of HIVAN. Since EMT has been implicated in the development of HIVAN phenotype, it appears that rapamycin could have attenuated renal cell EMT through modulation of miR-200 expression; these findings provide a new insight into the mechanism of rapamycin-induced amelioration of HIVAN.
The role of miRNAs in mTOR regulation in cancer cell is being investigated extensively. These studies demonstrated that miR-99, miR-100, and miR-199 could suppress tumor growth by targeting mTOR [8,23]. However, the role of miRNA in mTOR regulation in HIVAN needs further analysis. We showed that miR-99a, miR-100a and miR-199a were decreased in renal tissues of HIVAN mice. The expression of these miRNAs was also suppressed in HIV-transduced podocytes. Since rapamycin reversed the expression of these miRNAs both in HIVAN mice and HIV-transduced podocytes, it would indicate that miRNAs played a role in rapamycin-mediated activation of mTOR in HIVAN. Thus, it would be critical to further investigate the functional role of the miRNAs in the regulation of mTOR pathway in the progression of HIVAN.
In summary, we have demonstrated miRNA expression profile in rapamycin treated HIV-1 transgenic mice (Tg26) and involvement of these miRNAs in HIV-1 transduced human podocytes. Our results suggest that miRNAs play a role in the pathogenesis of HIVAN. Further functional investigation of these miRNAs in HIVAN model will not only enhance our understanding of the pathogenesis but would also lead to development of novel therapeutic strategies for HIVAN patients.
Acknowledgments
This work was supported by grant RO1DK084910 (PCS), RO1DK083931 (PCS) and 1R01DK098074 (PCS) from National Institutes of Health, Bethesda, MD.
References
- 1.Atta MG, et al. Diagnosis and natural history of HIV-associated nephropathy. Adv Chronic Kidney Dis. 2010;17:52–58. doi: 10.1053/j.ackd.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Baskerville S, Bartel DP, et al. Microarray profiling of micro-RNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt K, Zhou L, Mi QS, Huang S, She JX, Dong Z, et al. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cells survival. Mol Med. 2010;16:409–416. doi: 10.2119/molmed.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, Bertram JF, Jeyaseelan K, et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81:617–627. doi: 10.1038/ki.2011.448. [DOI] [PubMed] [Google Scholar]
- 5.Cheng K, Rai P, Plagov A, Lan X, Subrati A, Husain M, Malhotra A, Singhal PC, et al. MicroRNAs in HIV-associated nephropathy (HIVAN) Exp Mol Pathol. 2012;94:65–72. doi: 10.1016/j.yexmp.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung AC, Huang XR, Meng X, Lan HY, et al. miR-192 mediates TGF-beta/Smad3-driven. J Am Soc Nephrol. 2010;21:1317–1325. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Benedetto F, Di Sandro S, De Ruvo N, Montalti R, Ballarin R, et al. First report on a series of HIV patients undergoing rapamycin monotherapy after liver transplantation. Transplantation. 2010;89:733–738. doi: 10.1097/TP.0b013e3181c7dcc0. [DOI] [PubMed] [Google Scholar]
- 8.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L, et al. MiR-199a3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 9.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J, et al. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Nat Acad Sci USA. 2010;107:14339–14344. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Pagano F Negrini, Gomella LG, Croce CM, Baffa R, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 12.Heredia A, Latinovic O, Gallo RC, Melikyan G, Reitz M, et al. Reduction of CCR5 with low-dose rapamycin enhances the antiviral activity of vicriviroc against both sensitive and drug-resistant HIV-1. Proc Nat Acad Sci USA. 2008;105:20476–20481. doi: 10.1073/pnas.0810843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Y, Ratnam K, Chuang PY, Fan Y, Zhong Y, Dai Y, Mazloom AR, Chen EY, D′Agati V, Xiong H, Ross MJ, Chen N, Ma′ayan A, He JC, et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat Med. 2012;18:580–588. doi: 10.1038/nm.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R, et al. A microRNA circuit mediates transforming growth factor-β1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011;80:358–368. doi: 10.1038/ki.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopp JB, Klotman ME, Adler SH, Bruggeman LA, Dickie P, Marinos NJ, Eckhaus M, Bryant JL, Notkins AL, Klotman PE, et al. Progressive glomerulosclerosis and enhanced renal accumulation of basement membrane components in mice transgenic for human immunodeficiency virus type 1 genes. Proc Nat Acad Sci USA. 1992;89:1577–1581. doi: 10.1073/pnas.89.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar D, Konkimalla S, Yadav A, Sataranatarajan K, Kasinath BS, Chander PN, Singhal PC, et al. HIV-associated nephropathy: role of mammalian target of rapamycin pathway. Am J Pathol. 2010;177:813–821. doi: 10.2353/ajpath.2010.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y, Ridzon D, Wong L, Chen C, et al. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenzen JM, Haller H, Thum T, et al. MicroRNAs as mediators and therapeutic targets in. Nat Rev Nephrol. 2011;7:286–294. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 19.Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, Ross MD, Gusella GL, Benson G, D′Agati VD, Hahn BH, Klotman ME, Klotman PE, et al. Replication and com-partmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med. 2002;8:522–526. doi: 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- 20.Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F, et al. MiR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 21.Mori H, Inoki K, Masutani K, Wakabayashi Y, Komai K, Nakagawa R, Guan KL, Yoshimura A, et al. The mTOR pathway is highly activated in diabetic nephropathy and rapamycin has a strong therapeutic potential. Biochem Biophys Res Commun. 2009;384:471–475. doi: 10.1016/j.bbrc.2009.04.136. [DOI] [PubMed] [Google Scholar]
- 22.Muratsu-Ikeda S, Nangaku M, Ikeda Y, Tanaka T, Wada T, Inagi R, et al. Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PLoS One. 2012;7:e41462. doi: 10.1371/journal.pone.0041462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG, Olokpa E, Itamochi H, Ueno, Hawkins SM, Anderson ML, Matzuk MM, et al. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 2010;24:447–463. doi: 10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicoletti F, Fagone P, Meroni P, McCubrey J, Bendtzen K, et al. mTOR as a multifunctional therapeutic target in HIV infection. Drug Discov Today. 2011;16:715–721. doi: 10.1016/j.drudis.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Oneyama C, Ikeda J, Okuzaki D, Suzuki K, Kanou T, Shintani Y, Morii E, Okumura M, Aozasa, Okada M, et al. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene. 2011;30:3489–3501. doi: 10.1038/onc.2011.63. [DOI] [PubMed] [Google Scholar]
- 26.Pandey P, Brors B, Srivastava PK, Bott A, Boehn SN, Groene HJ, Gretz N, et al. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics. 2008;9:624. doi: 10.1186/1471-2164-9-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SM, Gaur AB, Lengyel E, Peter ME, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rai P, Plagov A, Kumar D, Pathak S, Ayasolla KR, Chawla A, Mathieson PW, Saleem MA, Husain M, Malhotra A, Singhal PC, et al. Rapamycin-induced modulation of HIV gene transcription attenuates progression of HIVAN. Exp Mol Pathol. 2013;94:255–261. doi: 10.1016/j.yexmp.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren Z, Liang W, Chen C, Yang H, Singhal PC, Ding G, et al. Angiotensin II induces nephrin dephosphorylation and podocyte injury: role of caveolin-1. Cell Signal. 2012;24:443–450. doi: 10.1016/j.cellsig.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy J, Paquette JS, Fortin JF, Tremblay MJ, et al. The immunosuppressant rapamycin represses human immunodeficiency virus type 1 replication. Antimicrob Agents Chemother. 2002;46:3447–3455. doi: 10.1128/AAC.46.11.3447-3455.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saal S, Harvey SJ, et al. MicroRNAs and the kidney: coming of age. Curr Opin Nephrol Hypertens. 2009;18:317–323. doi: 10.1097/MNH.0b013e32832c9da2. [DOI] [PubMed] [Google Scholar]
- 33.Salhan D, Sagar A, Kumar D, Rattanavich R, Rai P, Maheshwari S, Adabala M, Husain M, Ding G, Malhotra A, Chander PN, Singhal PC, et al. HIV-associated nephropathy: role of AT2R. Cell Signal. 2012;24:734–741. doi: 10.1016/j.cellsig.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong M, Jiang L, Zhou Y, Qiu W, Fang L, Tan R, Wen P, Yang J, et al. The miR-200 family regulates TGF-β1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol. 2012;302:F369–379. doi: 10.1152/ajprenal.00268.2011. [DOI] [PubMed] [Google Scholar]
- 35.Yadav A, Kumar D, Salhan D, Rattanavich R, Maheshwari S, Adabala M, Ding G, Singhal PC, et al. Sirolimus modulates HIVAN phenotype through inhibition of epithelial Mesenchymal transition. Exp Mol Pathol. 2012;93:173–181. doi: 10.1016/j.yexmp.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan JS, Reed A, Chen F, Stewart CN, Jr, et al. Statistical analysis of real-time PCR data. BMC Bioinf. 2006;7:1471–2105. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC, et al. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am J Hypertens. 2010;23:78–84. doi: 10.1038/ajh.2009.208. [DOI] [PubMed] [Google Scholar]
- 38.Wang B, Koh P, Winbanks C, Coughlan MT, McClelland A, Watson A, Jandeleit-Dahm K, Burns WC, Thomas MC, Cooper ME, Kantharidis P, et al. miR-200a Prevents renal fibrogenesis through repression of TGF-β2 expression. Diabetes. 2011;60:280–287. doi: 10.2337/db10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]


