Abstract
Objectives
This study aimed to investigate the prevalence of antibiotic resistance in fecal Escherichia coli isolates from healthy persons and patients with diarrhea.
Methods
E. coli isolates (n = 428) were obtained from fecal samples of apparently healthy volunteers and hospitalized patients with diarrhea. Susceptibility patterns of isolates to 16 antimicrobial agents were determined by agar disc diffusion.
Results
Most E. coli isolates exhibited less than 10% resistance against imipenem, cefotetan, aztreonam, cefepime, cefoxitin, amikacin and netilamicin, although greater than 65% were resistant to ampicillin and tetracycline. No significant difference in resistance rates for all tested antibiotics was found between isolates from the healthy-and diarrheal-patient groups, including for multi-drug resistance (p = 0.22). The highest number of resistant antibiotics was 12 antibiotics. No significant differences in antibiotic resistance were found among the sex and age strata for isolates from healthy individuals. However, antibiotic resistance rates to cefoxitin, cefotaxime, amikacin, and netilamicin were significantly higher in the isolates of men than those of women (p < 0.05) in isolates from patients with diarrhea. Furthermore, isolates from patients with diarrhea older than 40-years of age showed higher resistance to cefepime and aztreonam (p < 0.05).
Conclusion
High resistance to the antibiotics most frequently prescribed for diarrhea was found in isolates from patients with diarrhea and apparently healthy individuals without any significant difference.
Keywords: Antibiotic resistance, diarrheal patients, Escherichia coli, healthy persons, multi-drug resistance
1. Introduction
Diarrheal diseases continue to be a health problem worldwide. This is especially the case in developing countries, where they are estimated to be responsible for 2.5 million infant deaths per year, with an annual mortality rate of 4.9 per 1000 children and an incidence of 3.2 episodes per child per year among children younger than 5-years of age [1,2]. Antibiotic therapy in hospitals is possibly the most important factor that increases antibiotic-resistant microorganisms [3]. The emergence, propagation, accumulation, and maintenance of antimicrobial resistant pathogenic bacteria have become significant health concerns, and lead to increased morbidity, mortality, and health-care costs as a result of treatment failures and longer hospital stays [4–6].
A recent surveillance study in Korea demonstrated the positive relationship between antibiotic use and antibiotic resistance in several nosocomial pathogens [7]. We therefore aimed to investigate the prevalence of antibiotic resistance in fecal Escherichia coli isolates obtained from hospitalized patients with diarrhea in Korea compared with isolates from apparently-healthy persons who had not visited a health clinic for at least a year.
2. Materials and Methods
2.1. Sampling of feces for surveillance study
The surveillance study was planned by the Laboratory of Enteric Infections of the Korean Center for Disease Control and Prevention. Sampling was carried out from 2004 to 2006 with the help of several public health centers in Guri, Seongnam, and Yoeju in Korea. Specimens were collected from 95 patients with diarrhea who visited clinics because of diarrheal symptoms. The control group comprised 110 apparently-healthy persons living Guri, Seongnam, and Yoeju in Korea who had not visited a health clinic for at least a year (Table 1). Fecal samples were placed in sterile plastic specimen tubes on ice and transported to our laboratory for bacterial isolation within 3 days.
Table 1.
Specimens collected for this study
| Age groups | Control (n = 110) |
Patients with diarrhea (n = 95) |
||
|---|---|---|---|---|
| Male | Female | Male | Female | |
| 1–10 | 0 | 0 | 7 | 15 |
| 11–20 | 2 | 2 | 3 | 5 |
| 21–30 | 9 | 18 | 5 | 7 |
| 31–40 | 3 | 18 | 5 | 6 |
| 41–50 | 6 | 33 | 9 | 3 |
| 51–60 | 0 | 7 | 5 | 4 |
| 61+ | 3 | 9 | 10 | 11 |
| Total | 23 | 87 | 44 | 51 |
2.2. Culture procedures for isolating E. coli
Feces were plated directly on to Mac Conkey agar, or occasionally after enrichment in trypticase soy broth containing vancomycin (Sigma Chemical Co., St. Louis, MO, USA). Candidate colonies were then plated onto trypticase soy agar medium and biochemically characterized using the API20E system (Biomerieux, Marcy l’Etoile, France). For individual samples, one or two E. coli isolates were selected randomly to determine susceptibility.
2.3. Antimicrobial susceptibility testing
Susceptibility testing was conducted using disc diffusion according to the guidelines of the Clinical and Laboratory Standards Institute (formerly National Committee For Clinical Laboratory Standards) [8]. Antimicrobial susceptibility was determined by agar disc diffusion (Kirby-Bauer method) using Mueller–Hinton agar (Difco, MI, USA). The following 16 antibiotics were tested: SAM (ampicillin-sulbactam), AM (ampicillin), TE (tetracycline), ATM (aztreonam), cefotetan, FEP (cefepime), FOX (cefoxitin), CTX (cefotaxime), NN (tobramycin), SXT/TM (trimethoprim-sulfamethoxazole), CF (cephalothin), imipenem, GM (gentamicin), AN (amikacin), TZP (piperacillin/tazobactam), and NET (netilamicin). E. coli ATCC 25922 and E. coli ATCC 35218 were used as control strains.
2.4. Statistic analysis
Antimicrobial susceptibility data were expressed as percentages or the frequency of human isolates. A one-way analysis of variance or χ2 statistics was used to estimate the overall difference between the percentages or frequencies of resistance of E. coli isolates. In all cases, p < 0.05 was regarded as statistically significant.
3. Results
3.1. Bacterial isolate specimens and population characteristics
A total of 428 E. coli isolates were obtained from the collected fecal samples, of which 216 isolates were derived from healthy persons and 212 pathogenic E. coli isolates from patients with diarrhea.
3.2. Isolate antibiotic susceptibility
All isolates were analyzed by agar disc diffusion to determine their susceptibility patterns to the 16 tested antimicrobial agents. A greater percentage of isolates were resistant to AM (76.5% of patients with diarrhea; 77.3% of healthy persons) and TE (66% of patients with diarrhea; 66.5% of healthy persons) in the isolates of both groups, although no isolates showed resistance to imipenem and cefotetan. Isolates showed higher resistance to CF than other antibiotics among the cephems. Among the aminoglycosides, the resistance to GM and NN occurred at higher frequencies in comparison with resistance observed for AN and NET. SXT/TM resistance was also relatively higher than that for other antibiotics. However, most E. coli isolates exhibited a 10% resistance rate against ATM, FEP, FOX, AN, and NET. Resistance rates were compared between the isolates from healthy persons and patients with diarrhea and no significant difference was found between the groups (p < 0.05) (Table 2).
Table 2.
Antibiotic resistance patterns of E. coli strains
| Antimicrobial agents | Antibiotic resistances (%) of isolates |
|
|---|---|---|
| People without diarrhea | Patients with diarrhea | |
| ß-lactams | ||
| AM | 77.3 | 76.5 |
| ß-lactam/ß-lactamase inhibitor combinations | ||
| SAM | 6.5 | 22.6 |
| TZP | 12.5 | 0.9 |
| Cephems | ||
| CF | 30 | 42.9 |
| FEP | 0.9 | 8.9 |
| CTT | 0 | 0 |
| CTX | 3.2 | 13.6 |
| FOX | 3.7 | 2.3 |
| Carbapenems | ||
| IPM | 0 | 0 |
| Aminoglycosides | ||
| AN | 9.6 | 2.1 |
| GM | 39.4 | 29.2 |
| NN | 31 | 17.9 |
| NET | 4.6 | 3.3 |
| Tetracyclines | ||
| TE | 66.5 | 66 |
| Monobactams | ||
| ATM | 1.4 | 9.4 |
| Folate pathway inhibitors | ||
| SXT | 58.3 | 44.8 |
AM = ampicillin; AN = amikacin; ATM = aztreonam; CF = cephalothin; CTT = cefotetan; CTX = cefotaxime; FEP = cefepime; FOX = cefoxitin; GM = gentamicin; IPM = imipenem; NET = netilamicin; NN = tobramycin; SAM = ampicillin-sulbactam; SXT = trimethoprim-sulfamethoxazole; TE = tetracycline; TZP = piperacillin/tazobactam.
The sex- and age-specific patterns of antibiotic resistance were further analyzed for both groups. In the isolates from healthy persons, no significant differences in antibiotic resistance were found among the sex and age strata. However, in the isolates from patients with diarrhea, sex- and age-specific patterns were observed. As shown in Table 3, the isolates from men showed more resistance to several antibiotics than the isolates from women. Antibiotic resistance rates to CTX (23% vs. 8%), FOX (9% vs. 0%), AN (9% vs. 0%), and NET (9% vs. 0%) were also significantly higher in the isolates from men than those from women (p < 0.05).
Table 3.
Comparison of sex-based antibiotic resistance patterns of the E. coli strains isolated from patients with diarrhea
| Antimicrobial agents | Male |
Female |
||||
|---|---|---|---|---|---|---|
| No. of resistance | No. of non-resistance | Rate of resistance (%) | No. of resistance | No. of non-resistance | Rate of resistance (%) | |
| AM | 36 | 8 | 82 | 40 | 11 | 78 |
| SAM | 11 | 33 | 25 | 16 | 35 | 31 |
| TZP | 1 | 43 | 2 | 1 | 50 | 2 |
| CF | 26 | 18 | 59 | 29 | 22 | 57 |
| FEP | 5 | 39 | 11 | 2 | 49 | 4 |
| CTT | 0 | 44 | 0 | 0 | 51 | 0 |
| CTX∗ | 10 | 34 | 23 | 4 | 47 | 8 |
| FOX∗ | 4 | 40 | 9 | 0 | 51 | 0 |
| IPM | 0 | 44 | 0 | 0 | 51 | 0 |
| AN∗ | 4 | 40 | 9 | 0 | 51 | 0 |
| GM | 18 | 26 | 41 | 18 | 33 | 35 |
| NN | 12 | 32 | 27 | 6 | 45 | 12 |
| NET∗ | 4 | 40 | 9 | 0 | 51 | 0 |
| TE | 32 | 12 | 73 | 33 | 18 | 65 |
| ATM | 5 | 39 | 11 | 2 | 49 | 4 |
| SXT | 24 | 20 | 55 | 25 | 26 | 49 |
∗ A p value <0.05.
AM = ampicillin; AN = amikacin; ATM = aztreonam; CF = cephalothin; CTT = cefotetan; CTX = cefotaxime; FEP = cefepime; FOX = cefoxitin; GM = gentamicin; IPM = imipenem; NET = netilamicin; NN = tobramycin; No. = number; SAM = ampicillin-sulbactam; SXT = trimethoprim-sulfamethoxazole; TE = tetracycline; TZP = piperacillin/tazobactam.
Isolates from patients with diarrhea in the younger than40-years and older than40-years of age groups showed significantly different resistances to SAM (36% vs. 15%), FEP (2% vs. 15%), ATM (2% vs. 15%), and SXT/TM (60% vs. 38%). Isolates from patients with diarrhea older than 40-years of age showed significantly higher resistance to these antibiotics (p < 0.05) (Table 4).
Table 4.
Comparison of age-based antibiotic resistance patterns of the E. coli strains isolated from patients with diarrhea
| Antimicrobial agents | Younger than 40-years-old age |
Older than 40-years-old age |
||||
|---|---|---|---|---|---|---|
| No. of resistance | No. of non-resistance | Rate of resistance | No. of resistance | No. of non-resistance | Rate of resistance | |
| AM | 45 | 8 | 85 | 23 | 11 | 68 |
| SAM∗ | 19 | 34 | 36 | 5 | 29 | 15 |
| TZP | 0 | 53 | 0 | 2 | 32 | 6 |
| CF | 31 | 22 | 58 | 18 | 16 | 53 |
| FEP∗ | 1 | 52 | 2 | 5 | 29 | 15 |
| CTT | 0 | 53 | 0 | 0 | 34 | 0 |
| CTX | 5 | 48 | 9 | 6 | 28 | 18 |
| FOX | 2 | 51 | 4 | 2 | 32 | 6 |
| IPM | 0 | 53 | 0 | 0 | 34 | 0 |
| AN | 2 | 51 | 4 | 2 | 32 | 6 |
| GM | 19 | 34 | 36 | 15 | 19 | 44 |
| NN | 8 | 45 | 15 | 8 | 26 | 24 |
| NET | 2 | 51 | 4 | 2 | 32 | 6 |
| TE | 36 | 17 | 68 | 25 | 9 | 74 |
| ATM∗ | 1 | 52 | 2 | 5 | 29 | 15 |
| SXT∗ | 32 | 21 | 60 | 13 | 21 | 38 |
∗A p value <0.05.
AM = ampicillin; AN = amikacin; ATM = aztreonam; CF = cephalothin; CTT = cefotetan; CTX = cefotaxime; FEP = cefepime; FOX = cefoxitin; GM = gentamicin; IPM = imipenem; NET = netilamicin; NN = tobramycin; No. = number; SAM = ampicillin-sulbactam; SXT = trimethoprim-sulfamethoxazole; TE = tetracycline; TZP = piperacillin/tazobactam.
3.3. Multi-drug resistance patterns
Percentages of multiple drug resistance in E. coli isolates for each group are given in Figure. Among the isolates from healthy persons, 84% (181/216 isolates) exhibited resistance to two or more antimicrobials. Moreover, the resistance to four or more antibiotics occurred at a frequency of 46%. Seven of the isolates were resistant to eight antibiotics (SAM/AM/TE/NN/SXT/GM/AN/TZP or AM/TE/FOX/NN/SXT/GM/TZP/NET).
Figure.
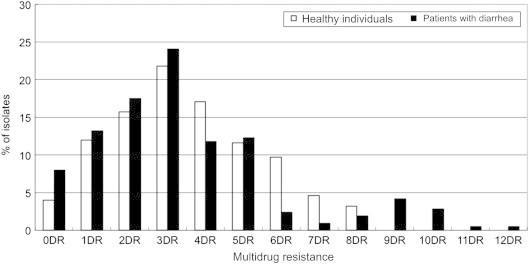
Antibiotic multi-resistance patterns of E. coli strains isolated from patients with diarrhea and healthy individuals. DR = drug resistance.
Among the isolates from patients with diarrhea, 78.8% (167/212) exhibited resistance to two or more antimicrobials. There was no significant difference in multi-drug resistance between isolates from healthy persons and those from patients with diarrhea (p = 0.22). The rates of antibiotic resistance to four or more antibiotics (37%) were similar to the rates of healthy persons. However, the number of resistant antibiotics was higher in the E. coli isolates from the patients with diarrhea than those from healthy persons. Resistance to over nine antibiotics was detected only in the patients with diarrhea. The highest rate of resistance was to 12 antibiotics (SAM/AM/TE/ATM/FEP/FOX/CTX/NN/CF/GM/AN/NET). Resistance to AM in combination with TE was the most frequently observed in isolates (data not shown).
4. Discussion
Judicious use of antimicrobials may be beneficial in preserving antimicrobial efficacy and substantially reducing diarrheal illness. However, antibiotic therapy can further increase drug resistance in microorganisms [3]. In this study, we examined antimicrobial resistance of E. coli isolates from hospitalized patients due to diarrhea and compared them to E. coli isolates from healthy persons. The highest levels of resistance were observed against AM and TE for both commensal and pathogenic E. coli, which may be caused by the frequent use of these antibiotics and the transfer of plasmids between bacteria [9,10].
In the Enterobacteriaceae, resistance to AM is mainly because of ß-lactamases, such as the TEM-1 and SHV-1 enzymes, that hydrolytically cleave the ß-lactam ring. Plasmid-encoded derivatives of the ß-lactamases that show an enhanced spectrum of catalytic activity have been known since the early 1980s [11]. Furthermore to the large number of ESBL-TEM and ESBL-SHV variants, other plasmid-encoded ESBL, such as CTX-M enzymes, are now frequently reported [12].
TE resistance in bacteria is mediated by four mechanisms: efflux, ribosomal protection, enzymatic inactivation, and target modification [13]. Widespread resistance to broad-spectrum TE has been caused, in part, by heavy clinical use and misuse in the human population. In the United States and other parts of the world, TEs, alone or in combination with other antibiotics, are used for the treatment of infectious diseases, as well as for prophylaxis, both orally and topically, because of their excellent safety profile and low cost [14]. Aminoglycoside resistance in E. coli most often occurs by aminoglycoside-modifying enzymes encoded on transmissible plasmids [14,15]. We did not determine the drug therapy for participants with diarrheal illness or the clinical impact of such treatment.
We found that higher resistance to the antibiotics most frequently prescribed for diarrhea was found in the isolates of not only the patients with diarrhea, but also apparently-healthy persons. The higher resistance in the greater than 40-year-old group of patients with diarrhea may be explained by the longer exposure of these individuals to antibiotics. The reason antibiotic resistance was different between the two sexes remains unknown, and therefore requires further investigation.
Acknowledgement
This study was supported by a grant from the Korean National Institute of Health, Republic of Korea (2004–2006).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Passariello A., Terrin G., Baldassarre M.E. Diarrhea in neonatal intensive care unit. World J Gastroenterol. 2010 Jun 7;16(21):2664–2668. doi: 10.3748/wjg.v16.i21.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosek M., Bern C., Guerrant R.L. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81(3):197–204. [PMC free article] [PubMed] [Google Scholar]
- 3.Tacconelli E., De Angelis G., Cataldo M.A. Antibiotic usage and risk of colonization and infection with antibiotic-resistant bacteria: a hospital population-based study. Antimicrob Agents Chemother. 2009 Oct;53(10):4264–4269. doi: 10.1128/AAC.00431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy S.B., Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004 Dec;10(Suppl. 12):122–129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 5.WHO . 2001. Global strategy for containment of antimicrobial resistance.http://www.who.int/drugresistance/guidance/en/index.html [Date accessed: 25 March 2011] [Google Scholar]
- 6.Salma T.G. Gram-negative antibiotic resistance: there is a price to pay. Crit Care. 2008;12(Suppl. 4):S4. doi: 10.1186/cc6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Y.A., Jung S.I., Chang M.O. Relationship between antibiotic use and antibiotic resistance in major nosocomial pathogens at a university hospital. Chonnam Med J. 2008 Dec;44(3):137–143. [in Korean] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute (CLSI) CLSI; Wayne, PA: 2009. Performance standards for antimicrobial susceptibility testing. 19th Informational Supplement. CLSI Document M100–S19. [Google Scholar]
- 9.Roberts M.C. Tetracycline therapy: update. Clin Infect Dis. 2003 Feb 15;36(4):462–467. doi: 10.1086/367622. [DOI] [PubMed] [Google Scholar]
- 10.Uma B., Prabhakar K., Rajendran S. Antibiotic sensitivity and plasmid profiles of Escherichia coli isolated from pediatric diarrhea. J Glob Infect Dis. 2009 Jul;1(2):107–110. doi: 10.4103/0974-777X.56255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kliebe C., Nies B.A., Meyer J.F. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985 Aug;28(2):302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livermore D.M., Woodford N. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas, and Acinetobacter. Trends Microbiol. 2006 Sep;14(9):413–420. doi: 10.1016/j.tim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Chopra I., Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001 Jun;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies J., Wright G.D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997 Jun;5(6):234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 15.Galimand M., Courvalin P., Lambert T. Plasmid-mediated high level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003 Aug;47(8):2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


