Abstract
The nuclear adaptor Ldb1 functions as a core component of multiprotein transcription complexes that regulate differentiation in diverse cell types. In the hematopoietic lineage, Ldb1 forms a complex with the non–DNA-binding adaptor Lmo2 and the transcription factors E2A, Scl and GATA-1 (or GATA-2). Here we demonstrate a critical and continuous requirement for Ldb1 in the maintenance of both fetal and adult mouse hematopoietic stem cells (HSCs). Deletion of Ldb1 in hematopoietic progenitors resulted in the downregulation of many transcripts required for HSC maintenance. Genome-wide profiling by chromatin immunoprecipitation followed by sequencing (ChIP-Seq) identified Ldb1 complex–binding sites at highly conserved regions in the promoters of genes involved in HSC maintenance. Our results identify a central role for Ldb1 in regulating the transcriptional program responsible for the maintenance of HSCs.
In embryonic stem cells (ESCs), the transcription factors Oct-4, Nanog and Sox2 coregulate the expression of genes encoding key molecules involved in self-renewal and differentiation, functioning as core elements in a transcriptional hierarchy essential for the maintenance of pluripotency1,2. Somatic stem cells such as hematopoietic stem cells (HSCs) share several fundamental properties with ESCs, including a dependence on molecules that regulate self-renewal, differentiation, proliferation and cell survival and a requirement for polycomb complex–mediated repression of developmental transcription factors3–5. However, unlike ESCs, HSCs do not have a defined set of core maintenance transcription factors or ‘master regulators’. Oct-4, Nanog and Sox2 are downregulated during early embryogenesis and, with few exceptions, the genes encoding molecules essential for the maintenance of ESCs and HSCs are not identical, which suggests that distinct transcriptional mechanisms regulate the maintenance of ESCs and HSCs.
Ldb1 is a broadly expressed self-dimerizing nuclear factor that functions as a subunit of DNA-binding complexes in diverse cell types6. In hematopoietic cells, Ldb1 forms a multimeric protein complex with the non–DNA-binding adaptor Lmo2 and transcription factors of the zinc-finger family (GATA-1 and GATA-2) and/or the basic helix-loop-helix family (Scl; also known as Tal1, Lyl1 and E2A)7,8. Lmo2, GATA-2, E2A and either Scl or Lyl1 are required for HSC specification and/or HSC maintenance, which suggests that these factors function together in higher-order Ldb1-nucleated multimeric complexes to regulate gene transcription in HSCs9–14.
In this report we demonstrate that Ldb1 is required for HSC specification and for the maintenance of both fetal and adult HSCs in mice but is not essential for ESC maintenance. Induced deletion of Ldb1 in hematopoietic progenitor cells resulted in the rapid depletion of HSCs and the downregulation of many genes encoding molecules known to be required for the specification and/or maintenance of HSCs, which suggests a role for Ldb1 complexes in regulating the transcriptional program necessary for HSC maintenance. Consistent with that hypothesis, genome-wide analysis by chromatin immunoprecipitation coupled with sequencing (ChIP-Seq) identified conserved Ldb1 complex–binding sites in or near 20 of 28 HSC genes analyzed and in 11 of 12 known enhancer elements in the vicinity of these genes. Together our results support a model in which Ldb1 complexes control a core transcriptional program required for HSC maintenance by restricting HSC differentiation.
RESULTS
Ldb1 is required for HSC specification
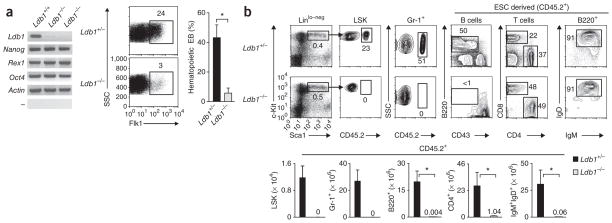
We verified expression of Ldb1 in ESCs by RT-PCR analysis of Ldb1 transcripts and by intracellular staining for Ldb1 protein (Fig. 1a and Supplementary Fig. 1). To investigate whether Ldb1 is required for ESC maintenance, we generated Ldb1−/− ESC lines from a single germline-transmitting Ldb1+/− clone15 by targeting the remaining wild-type Ldb1 allele16. Inactivation of Ldb1 did not affect ESC growth or morphology or the expression of genes encoding molecules essential for the regulation of ESC maintenance, including Nanog, Zfp42 (encoding Rex1) and Pou5f1 (encoding Oct-4; (Fig. 1a and Supplementary Fig. 1)). In addition, the formation of embryoid bodies and the expression of mesodermal genes, including T (encoding brachyury), Bmp4, Smad1 and Kit, were not impaired in the absence of Ldb1 (Supplementary Fig. 1 and data not shown). However, Ldb1−/− embryoid bodies generated very few cells of the hematopoietic lineage in culture, as assessed by expression of the cell surface markers Flk1 and CD45 (Fig. 1a and data not shown). We observed few hematopoietic colonies at day 9 in Ldb1−/− embryoid body methylcellulose cultures17, and the frequency of embryoid bodies that produced hematopoietic colonies was much lower for Ldb1−/− embryoid bodies than for the Ldb1+/− or Ldb1+/+ controls (which showed similar differentiation potential; Fig. 1a). These results suggest that there is normal ESC maintenance but defective hematopoietic specification in the absence of Ldb1.
Figure 1.
Ldb1 is required for hematopoietic specification but is not essential for ESC maintenance. (a) RT-PCR analysis (left) of the expression of various genes (left margin) in Ldb1+/+ ESCs and Ldb1−/− ESCs (two independently generated clones); flow cytometry of cells in embryoid bodies at day 5 derived from Ldb1+/− or Ldb1−/− ESCs (middle); and frequency of Ldb1+/− or Ldb1−/− ESC-derived embryoid bodies (EB) at day 9 with hematopoietic satellite cells (right; identified by Giemsa staining of cytospin preparations). Numbers above outlined areas (middle) indicate percent Flk1+ cells. SSC, side scatter. *P < 0.01 (Student’s t-test). Data represent one of two experiments (error bars, s.d.). (b) Frequency (above) and absolute number (below) of CD45.2+, LSK, granulocyte (Gr-1+) and B cells in the bone marrow and mature T cells in the lymph nodes and mature B cells (B220+) in the spleen of 8-week-old adult Ldb1+/− ESC and Ldb1−/− ESC chimeric mice. Numbers adjacent to outlined areas (above) indicate percent cells in gate; numbers above bars (below) indicate number of cells for bars not visible. Ig, immunoglobulin. *P < 0.01 (Student’s t-test). Data are representative of three experiments with six Ldb1+/− mice and three Ldb1−/− mice.
Hematopoietic progenitor generation in the absence of Ldb1
Ldb1 is required for anterior-posterior patterning and organogenesis during early embryonic development; however, its role in hematopoiesis has remained unclear because of the early death in utero of Ldb1−/− embryos15. To evaluate the developmental potential of Ldb1−/− ESCs in vivo, we generated chimeric mice by injecting Ldb1−/− or Ldb1+/− ESCs (both CD45.2+) into blastocysts from CD45.1 congenic mice deficient in recombination-activating gene 2 (Rag2−/−). All Ldb1−/− ESC chimeras with a high percentage of tissues derived from Ldb1−/− ESCs died during gestation; therefore, we intentionally generated chimeras with a low percentage of cells derived from Ldb1−/− ESCs by injecting fewer Ldb1−/− ESCs (or control ESCs) into Rag2−/− blastocysts. In addition, as the in vitro differentiation potential of Ldb1+/− and Ldb1+/+ ESCs was equivalent and as Ldb1+/− mice showed no hematological defects (Fig. 1a and data not shown), we used Ldb1+/− and Ldb1+/+ ESCs and mice interchangeably as controls in these and subsequent experiments. All adult Ldb1−/− ESC chimeras had few mature CD4+ or CD8+ T lymphocytes and mature B lymphocytes positive for immunoglobulin M and immunoglobulin D (Fig. 1b and Supplementary Fig. 2b). As lymphocyte development is blocked at an immature stage in Rag2−/− mice, mature lymphocytes in chimeric mice were derived from Ldb1−/− ESCs, as confirmed by the finding that all lymphocytes were CD45.2+ (originated from Ldb1−/− ESCs; Fig. 1b). However, Ldb1−/− ESC–derived cells with low to negative expression of lineage markers and positive for expression of the markers Sca-1 and c-Kit (Linlo–negSca-1+c-Kit+ (LSK cells)), which include HSCs18, were undetectable in the bone marrow of all Ldb1−/− ESC adult chimeras regardless of their age (Fig. 1b and data not shown). CD45.2+ B lymphocyte progenitors (B220+CD43+) and Gr-1+ granulocytes (which are short-lived and require constant replenishment from bone marrow progenitors)19 were also absent from the bone marrow of all Ldb1−/− ESC adult chimeric mice (Fig. 1b). Ldb1−/− ESC–derived immature CD4+CD8+ thymocytes were present in young adult Ldb1−/− ESC chimeras (<8 weeks old), but these cells were absent from older mice (Supplementary Fig. 2a), which indicated that although they were fewer in number, hematopoietic progenitors were generated from Ldb1−/− ESCs but eventually underwent depletion in adult mice.
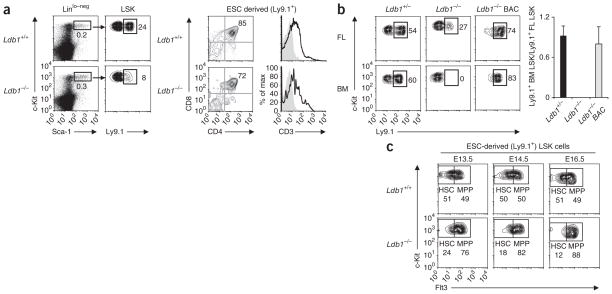
To determine if Ldb1−/− hematopoietic progenitors could be detected at earlier stages of development, we examined fetal liver cells from chimeras generated by injection of Ldb1−/− ESCs into C57BL/6 (B6) Ldb1+/+ blastocysts (Fig. 2). We identified ESC-derived cells of the hematopoietic lineage through the use of the Ly9.1 marker, as hematopoietic cells derived from ESCs of the 129 strain express Ly9.1, but blastocysts of the B6 strain do not. Low numbers of Ldb1−/− LSK cells were present in fetal livers of all Ldb1−/− ESC chimeras at all stages of gestation analyzed (embryonic days 13.5–16.5 (E13.5–E16.5); Fig. 2a and data not shown), which demonstrated that LSK cells were generated from Ldb1−/− ESCs and that the absence of detectable LSK cells in adult mice was not caused by lower expression of c-Kit and/or Sca-1. In addition, Ldb1−/− fetal liver LSK cells were able to give rise to CD4+CD8+ cells in the fetus (Fig. 2a and data not shown), which demonstrated that they retained normal lymphoid differentiation potential.
Figure 2.
Ldb1−/− hematopoietic progenitors are present in chimeric fetal livers but are unable to reconstitute hematopoiesis in irradiated recipients. (a) LSK cells in chimeric embryos at E13.5, generated by injection of Ldb1+/+ or Ldb1−/− ESCs into B6 (Ldb1+/+) blastocysts (left); ESC-derived (Ly9.1+) LSK cells were detected by staining for Ly9.1. Right, staining of gated Ly9.1+ (ESC-derived) thymocytes from chimeras at E18.5 with anti-CD4 plus anti-CD8 or anti-CD3. Numbers adjacent to outlined areas (left) and numbers in top right quadrants (right) indicate percent cells in gate. Data are representative of five experiments. (b) Flow cytometry of donor fetal liver (FL) cells at E15.5 and recipient bone marrow (BM) cells: Ldb1+/− or Ldb1−/− ESCs or Ldb1−/− ESCs reconstituted with a bacterial artificial chromosome containing Ldb1 (Ldb1−/− BAC) (Ly9.1+) were injected into Rag2−/− (Ly9.1−) blastocysts to generate chimeric embryos, followed by collection of fetal liver cells at E15.5 and injection into irradiated Rag2−/− (Ly9.1−) mice; 16 weeks later, bone marrow from recipient mice was analyzed for the presence of ESC-derived (Ly9.1+) LSK cells. Numbers adjacent to outlined areas (left) indicate percent Ly9.1+ LSK cells. Right, summary of data at left. Data are representative of four independent experiments (error bars, s.d.). (c) Flow cytometry of fetal liver cells from chimeric embryos at E13.5–E16.5, generated by injection of Ldb1+/+ or Ldb1−/− (Ly9.1+) ESCs into B6 (Ldb1+/+, Ly9.1−) blastocysts, assessing c-Kit versus Flt3 profiles on gated ESC-derived (Ly9.1+) LSK cells. Numbers adjacent to outlined areas indicate percent Flt3− LSK cells (HSC) or Flt3+ LSK cells (MPP). Data are representative of seven experiments with a total of two to three fetal livers per each time point.
LSK populations include a small number of long-term repopulating HSCs (LTR-HSCs) with the capacity for indefinite self-renewal and multilineage differentiation18. The presence of LTR-HSCs can be established experimentally by testing if fetal liver cells can reconstitute hematopoiesis in lethally irradiated adult mice18. Control Ldb1+/− ESC–derived fetal liver cell populations included LTR-HSCs as donor (Ly9.1+) LSK cells, and immature B220+CD43+ B cells and immature CD4+CD8+ T cells were present in recipient mice 16 weeks after transfer (Fig. 2b and Supplementary Fig. 3b). In contrast, although low numbers of mature T and B lymphocytes were present in the spleens of Rag2−/− mice given injection of Ldb1−/− fetal liver cells, we detected no donor LSK cells, B cell progenitors or immature T cells (Fig. 2b and Supplementary Fig. 3b). We confirmed the absence of Ldb1−/− LTR-HSCs in secondary (bone marrow) transfer experiments (Supplementary Fig. 3c). These results demonstrate that Ldb1−/− fetal liver LSK cell populations included progenitors that could transiently support hematopoiesis but did not include LTR-HSCs. In addition, they show that the defect in Ldb1−/− ESC–derived hematopoietic progenitors was cell autonomous.
To verify that the defects of Ldb1−/− hematopoietic progenitors were due to loss of Ldb1 expression and not to artifacts caused by ESC manipulation, we reconstituted Ldb1 expression in Ldb1−/− ESCs by transfection of a bacterial artificial chromosome containing the entire Ldb1 gene. Fetal liver cells derived from those ESCs at E15.5 were able to establish long-term colonization of recipient bone marrows (Fig. 2b and Supplementary Fig. 3b). This demonstrated that the defect in Ldb1−/− ESC–derived hematopoietic progenitors can be attributed entirely to the absence of Ldb1.
Requirement for Ldb1 in fetal HSC maintenance
The absence of Ldb1−/− LTR-HSCs in fetal and adult chimeric mice suggested the possibility that Ldb1 is required for HSC maintenance. However, as hematopoietic specification was impaired in the absence of Ldb1 (Fig. 1a), it was possible that LTR-HSCs were not generated or that Ldb1−/− LTR-HSCs were defective in their ability to efficiently home to and colonize the fetal liver and adult bone marrow. To address these issues, we made use of a loxP-flanked allele of Ldb1 (Ldb1fl) generated by gene targeting20. Inactivation of Ldb1fl was mediated by Cre recombinase expressed transgenically from the promoter of the gene encoding the tyrosine kinase Tie2 (Tie2-Cre)21, as published experiments with mice with conditional knockout of loxP-flanked alleles of the gene encoding Scl (Tal1; called ‘Scl’ here) mediated by Tie2-Cre have demonstrated that Tie2-Cre results in efficient deletion of this gene in fetal liver HSCs; however, gene deletion does not take place until after HSC specification22. To increase the efficiency of Ldb1 inactivation, we deleted one Ldb1 allele in the germline to generate Ldb1fl/Δ mice. Similar to Ldb1+/− mice, Ldb1fl/Δ mice were viable and fertile and showed no hematopoietic defects (data not shown).
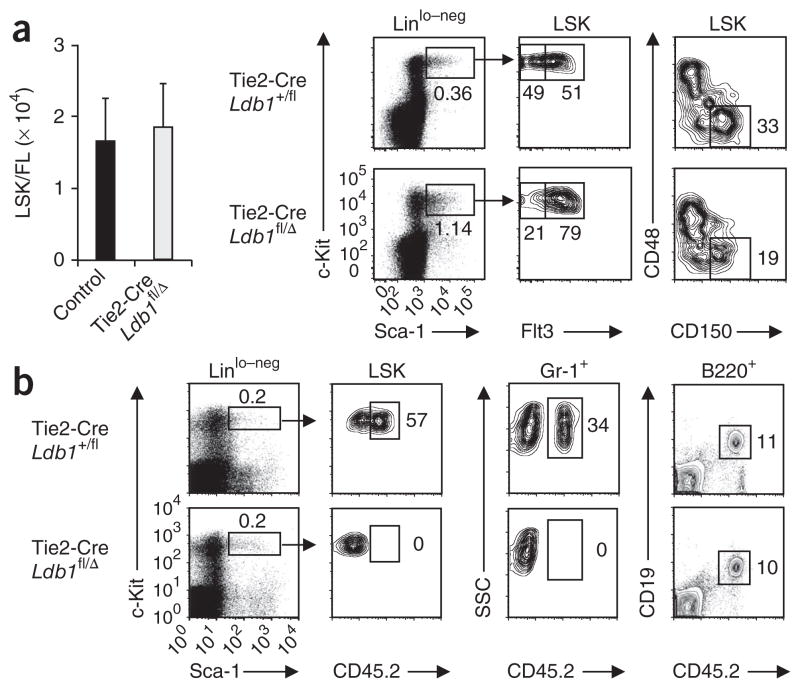
Tie2-Cre Ldb1fl/Δ embryos developed normally through E12.5, but by E13.5, all embryos showed widespread hemorrhage and edema, and we detected no live embryos after E14.5 (ref. 17). We confirmed Tie2-Cre-mediated deletion of Ldb1 by PCR analysis of genomic DNA from total fetal liver cells (Supplementary Fig. 4a). Tie2-Cre Ldb1fl/Δ fetal livers and fetal livers from littermate control mice (Ldb1fl/Δ, Ldb1+/fl and Tie2-Cre Ldb1+/fl) at E12.5 contained similar numbers of LSK cells with equivalent expression of the chemokine receptor CXCR4 (Fig. 3a and Supplementary Fig. 4b), which indicated that HSC population expansion and homing of HSCs to fetal liver were not impaired in Tie2-Cre Ldb1fl/Δ mice. However, LSK cells from Tie2-Cre Ldb1fl/Δ fetal livers at E12.5 did not show long-term repopulating potential, as donor-derived CD45.2+ LSK cells were undetectable in recipient bone marrows 16 weeks after adoptive transfer (Fig. 3b). Thus, continuous expression of Ldb1 is essential for the maintenance of fetal LTR-HSCs.
Figure 3.
Ldb1−/− fetal hematopoietic progenitor (LSK) populations do not contain LTR-HSCs. (a) Fetal liver LSK cells (left) in embryos at E12.5: Tie2-Cre Ldb1fl/Δ, 1.87 × 104 ± 0.61 × 104; control (Ldb1fl/Δ, Ldb1+/fl and Tie2-Cre Ldb1+/fl littermates), 1.68 × 104 ± 0.57 × 104 (mean ± s.d.). P = 0.52 (Student’s t-test). Middle, flow cytometry of Linlo–neg fetal liver cells from Tie2-Cre Ldb1+/fl and Tie2-Cre Ldb1fl/Δ littermates at E12.5, stained for c-Kit and Sca-1 (left), followed by analysis of Flt3 expression (right) in the gate outlined at left (LSK cells; arrows). Numbers below outlined areas indicate percent LSK cells (left plots) or percent HSCs (left area; Flt3−) and MPPs (right area; Flt3+) among LSK cells (right plots). Far right, flow cytometry of fetal liver LSK cells at E12.5; numbers adjacent to outlined areas indicate percent CD48−CD150+ LSK cells. Data are from one representative of two experiments. (b) Flow cytometry of donor-derived (CD45.2+) cells in irradiated Rag2−/− (CD45.1) hosts 16 weeks after adoptive transfer of total fetal liver cells from Tie2-Cre Ldb1+/fl or Tie2-Cre Ldb1fl/Δ mice at E12. Numbers adjacent to outlined areas indicate percent in each gate. Data are from one representative of two experiments.
LSK cells can be fractionated into two populations on the basis of expression of the tyrosine kinase receptor Flt3. Flt3− LSK cell populations include LTR-HSCs, as well as HSCs with limited self-renewal capability (short-term repopulating HSCs)23,24. Flt3+ LSK cells (multipotent progenitor populations (MPPs)) are generated from Flt3− LSK cells and are multipotent but lack self-renewal potential23,24. Fetal liver LSK cells from control embryos at E12.5 included similar numbers of Flt3− HSCs and Flt3+ MPPs (Fig. 3a). However, in Tie2-Cre Ldb1fl/Δ embryos at E12.5, the number of Flt3− LSK cells was consistently lower than the number of Flt3+ LSK cells (Fig. 3a). We confirmed the lower number of HSCs in Tie2-Cre Ldb1fl/Δ fetal livers by an alternative staining method that identifies a cell population (Lin−Sca-1+c-Kit+CD48−CD150+) highly enriched for HSCs25 (Fig. 3a). Notably, the percentage of cycling and apoptotic (annexin V–positive) LSK cells was similar in Tie2-Cre Ldb1fl/Δ and control fetal livers (Supplementary Fig. 4c, d). We also detected no defects in cell cycle or survival in Ldb1−/− fetal liver LSK cells from Ldb1−/− ESC chimeras (Supplementary Fig. 5a). Analysis of Ldb1−/− fetal liver LSK cells during progressive days of gestation demonstrated that the HSC/MPP ratio decreased between E13.5 and E16.5 (Fig. 2c and Supplementary Fig. 5b, c), which indicated that HSCs underwent gradual depletion. These findings suggested that the loss of HSCs after inactivation of Ldb1 was caused by their differentiation rather than by defects in proliferation or survival. To determine if Ldb1 is also required at other stages of hematopoiesis, we evaluated the colony-forming potential of Tie2-Cre Ldb1fl/Δ fetal liver cells at E12.5 in methylcellulose cultures. Tie2-Cre Ldb1fl/Δ fetal liver cells generated considerably fewer colonies than did control fetal liver cells; this mainly reflected a lower number of definitive erythroid colony-forming progenitors (Supplementary Fig. 4e, f). Together with published studies17, these results identify a selective requirement for Ldb1 downstream of the HSC in cells of the erythroid lineage.
Requirement for Ldb1 in the maintenance of adult HSCs
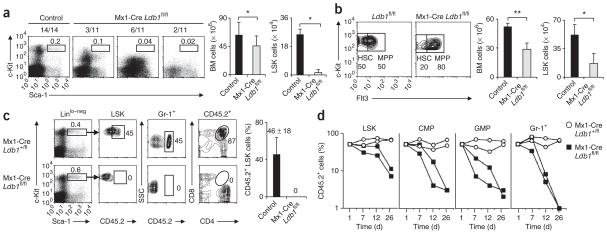
To determine if Ldb1 is required for the maintenance of adult HSCs, we used adult Ldb1fl/fl mice with expression of Cre driven by the interferon-inducible promoter of the gene encoding Mx1 (Mx1-Cre) and inactivated Ldb1 by injecting polyinosinic-polycytidylic acid (poly(I:C))26. We confirmed the induction of Cre recombinase by poly(I:C) in adult bone marrow LSK cells in Mx1-Cre Rosa26–loxP–stop–loxP–green fluorescent protein reporter mice and verified deletion of Ldb1 in bone marrow cells by PCR amplification of genomic DNA (Supplementary Fig. 6). We administered three injections of poly(I:C) to adult Mx1-Cre Ldb1fl/fl mice and control mice (Ldb1fl/fl, Ldb1+/fl and Mx1-Cre Ldb1+/fl). Analysis of bone marrow cells on day 12 after the first poly(I:C) treatment showed that Mx1-Cre Ldb1fl/fl mice had a much lower number and percentage of LSK cells even at this early time point (Fig. 4a). LSK cells were absent or much lower in number in all Mx1-Cre Ldb1fl/fl mice that had received poly(I:C) injections (n = 11). The degree to which Linlo–negSca-1lo–negc-Kit+ cell populations, which are composed mainly of non–self-renewing lineage-committed progenitors, were diminished varied between individual mice; this probably reflected differences in the kinetics and efficiency of progenitor cell depletion. To evaluate the effect of Ldb1 deletion on the LSK phenotype, we analyzed Mx1-Cre Ldb1fl/fl mice at earlier time points after poly(I:C) injection. Similar to results obtained with Tie2-Cre Ldb1fl/Δ fetal liver LSK cells, deletion of Ldb1 in adult LSK cells resulted in a lower ratio of Flt3− cells to Flt3+ cells (HSCs/MPPs; Fig. 4b).
Figure 4.
Ldb1 is continuously required for the maintenance of adult HSCs. (a) LSK profile (left) of bone marrow cells from mice injected with poly(I:C) on days 1, 3 and 5, assessed on day 12 by staining of gated Linlo–neg bone marrow cells for c-Kit and Sca-1. Control, Ldb1fl/fl, Ldb1+/fl and Mx1-Cre Ldb1+/fl mice. Numbers above outlined areas indicate percent LSK cells; numbers above plots indicate mice with a phenotype similar to that shown in the plot/total mice. Right, total bone marrow and LSK cells from control mice (n = 14) and Mx1-Cre Ldb1fl/fl mice (n = 11). *P < 0.05 (Student’s t-test). Data are from one representative of five experiments (error bars, s.d.). (b) Flt3 expression (left) by LSK cells from adult Ldb1fl/fl or Mx1-Cre Ldb1fl/fl littermates injected with poly(I:C) on days 1, 3 and 5, assessed on day 6. Numbers adjacent to outlined areas indicate percent Flt3− LSK cells (HSC) or Flt3+ LSK cells (MPP). Right, total bone marrow and LSK cells from control mice (n = 3) and Mx1-Cre Ldb1fl/fl mice (n = 3). *P < 0.05 and **P < 0.01 (Student’s t-test). Data are representative of two experiments (error bars, s.d.). (c) Flow cytometry (left) of bone marrow cells and thymocytes from irradiated Rag2−/− (CD45.1) recipient mice 6 months after injection of 50:50 mixtures of bone marrow cells from Mx1-Cre Ldb1fl/fl (CD45.2) mice or littermate control (CD45.2) mice and B6 (CD45.1) mice that had been injected with poly(I:C) as described in a. Numbers adjacent to outlined areas indicate percent cells in each gate. Right, summary of data at left. Data are from one representative of three independent experiments (error bars, s.d.). (d) CD45.2+ cells among bone marrow cells from premade bone marrow chimeras generated with 50:50 mixtures of bone marrow cells from Mx1-Cre Ldb1fl/fl (CD45.2) mice or control Mx1-Cre Ldb1+/fl (CD45.2) mice and bone marrow cells from B6 (CD45.1) mice after injection of poly(I:C) (three times, every other day). CMP, common myeloid progenitor, GMP, granulocyte macrophage progenitor. Data are from one representative of two experiments.
We assessed the effect of Ldb1 deletion on adult LTR-HSCs by adoptive transfer of a 50:50 mixture of bone marrow cells from poly(I:C)- injected Mx1-Cre Ldb1fl/fl (CD45.2) mice and B6-CD45.1 mice into irradiated Rag2−/− (CD45.1) recipient mice. At 6 months after bone marrow transfer, LSK cells from Mx1-Cre Ldb1fl/fl donor mice (CD45.2+ cells) were undetectable in recipient mice (Fig. 4c). We also confirmed the absence of HSCs in recipient mice by the absence of CD45.2+ granulocytes, immature CD4+CD8+ thymocytes and immature B cells (Fig. 4c and data not shown). To evaluate the consequences of Ldb1 deletion under conditions that do not require homing or population expansion of HSCs, we first generated bone marrow chimeras with a 50:50 mixture of bone marrow from Mx1-Cre Ldb1fl/fl (CD45.2) mice and B6 (CD45.1) mice. After confirming stable bone marrow chimerism, we injected mice with poly(I:C) and analyzed hematopoietic cells at various time points after Ldb1 deletion. In these experiments, Mx1-Cre Ldb1fl/fl LSK cells also underwent rapid depletion after inactivation of Ldb1 (Fig. 4d), which demonstrated that the defect in adult HSCs was cell autonomous.
Regulation of HSC-maintenance genes by Ldb1
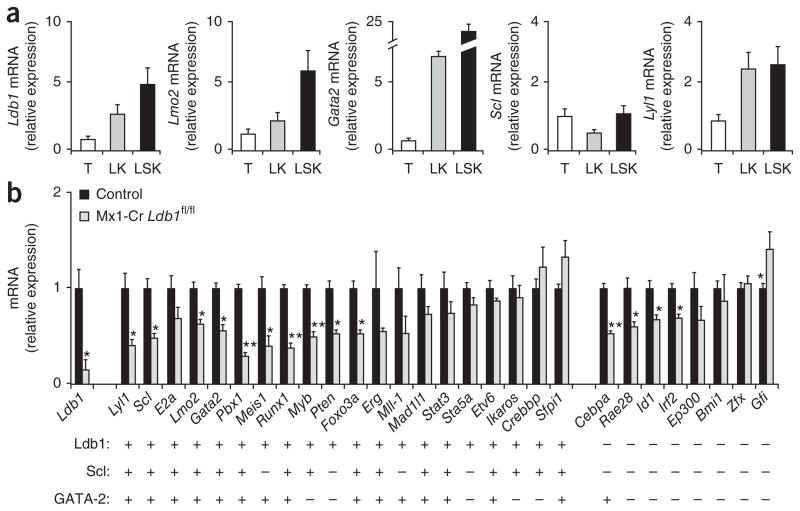
The continuous requirement for Ldb1 in both fetal and adult HSCs suggested that multimeric Ldb1-nucleated transcription complexes may regulate the expression of genes essential for HSC maintenance. Analysis of sorted LSK cells by real-time PCR showed that hematopoietic progenitor cells had high expression of Ldb1, as well as genes encoding putative Ldb1 complex subunits, including Lmo2, Lyl1, Scl and Gata2 (but not Gata1; Fig. 5a and data not shown). We next evaluated the effect of Ldb1 downregulation on the expression of a set of 28 genes shown by published gene-deletion studies to be essential for the specification and/or maintenance and self-renewal of HSCs3,10,27–29. We enriched cell populations for bone marrow hematopoietic progenitor cells by depletion of Lin+ cells on day 3 after injection of poly(I:C), when the percentage and number of CD34− (and Flt3−) LSK cell progenitors were similar in control and Mx1-Cre Ldb1fl/fl mice (Supplementary Fig. 7). Ldb1 transcripts in progenitor-enriched bone marrow cell populations from poly(I:C)-treated Mx1-Cre Ldb1fl/fl mice at this time point were diminished to <25% of the abundance in control mice (Fig. 5b). Notably, transcripts of 14 of the 28 HSC-maintenance genes assayed, including Pbx1, Meis1, Runx1, Myb, Pten and Foxo3a were also significantly lower in abundance after deletion of Ldb1 (Fig. 5b). Among the downregulated transcripts were those encoding subunits of the Ldb1 complex (Lmo2, Scl, Lyl1 and Gata2), which suggested a positive autoregulatory function for Ldb1 complexes in the expression of these genes. In contrast, transcripts encoding the transcription factor Gfi1 were greater in abundance after deletion of Ldb1 (Fig. 5b), which suggested an inhibitory function for Ldb1 complexes in the regulation of this gene.
Figure 5.
Ldb1 regulates the expression of many transcription factors required for HSC maintenance. (a) Expression of transcripts encoding Ldb1 and subunits of the Ldb1 complex in hematopoietic progenitors. T, total adult bone marrow; LK, sorted Linlo–neg c-Kit+ bone marrow; LSK, sorted LSK bone marrow. Data are from one representative of two independent experiments. (b) Real-time RT-PCR analysis of various transcripts (horizontal axis) in bone marrow cells obtained from adult Mx1-Cre Ldb1fl/fl mice or control Mx1-Cre Ldb1+/+ mice 2 d after the first of two injections of poly(I:C) (day 3), then enriched for hematopoietic progenitors (lineage depletion). Results in a and b are presented relative to expression of mRNA for the endogenous reference gene Actb. Below, genes with (+) or without (−) Ldb1-, Scl- and/or GATA-2-binding sites in the gene body or within 5 kb of the transcription start site, assessed by ChIP-Seq. *P < 0.05 and **P < 0.01 (Student’s t-test). Data are representative of three independent experiments (error bars, s.d.).
To determine if Ldb1 complexes bind to sites at or near genes involved in HSC maintenance, we next analyzed bone marrow enriched for progenitor cells by ChIP-Seq screens30,31 with antibody to Ldb1 (anti-Ldb1) as well as with anti-Scl and anti-GATA-2, as these represent likely DNA-binding subunits of Ldb1 complexes expressed in HSCs. Notably, we detected Ldb1 complex–binding sites, which were localized to 200–base pair genomic fragments by ChIP-Seq analysis, in the promoter (<5 kb from the transcription start site) and/or in the gene body of 20 of the 28 HSC-maintenance genes analyzed (Fig. 5b, Supplementary Fig. 8 and Supplementary Tables 1 and 2). The enrichment for Ldb1 complex–binding sites in the HSC gene set was significant relative to the abundance of binding sites in randomly selected gene sets (P < 0.00000007; hypergeometric test). Of the 14 genes downregulated after deletion of Ldb1, 10 (71%) contained Ldb1 complex–binding sites in their promoter or gene body, and half of all the genes involved in HSC maintenance with Ldb1 complex–binding sites contained a binding site in the promoter (Supplementary Table 1).
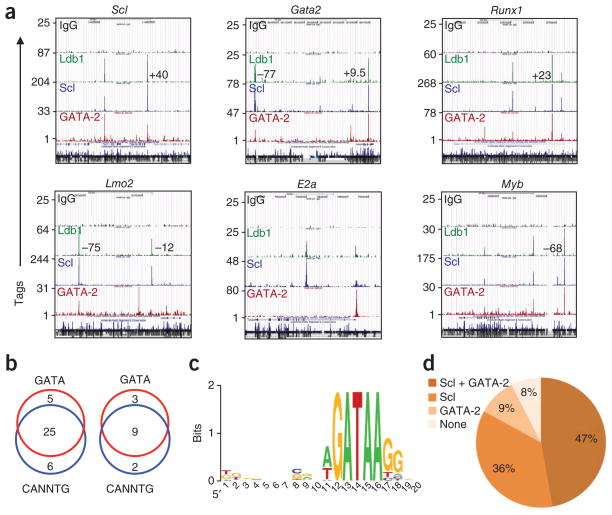
The ChIP-Seq results for Ldb1, Scl and GATA-2 were highly correlated at sites in or near HSC genes (Figs. 5b and 6a, d and Supplementary Fig. 8), which indicated that most Ldb1 complexes that bound at these sites contained Scl and/or GATA-2. Of the 20 genes in which we detected Ldb1 complex–binding sites in the promoter or gene body with anti-Ldb1, 90% contained an Scl- or GATA-2-binding site, and 65% contained binding sites for both Scl and GATA-2 (Fig. 5b). Moreover, of the 39 DNA fragments in the HSC gene set identified as Ldb1 complex–binding sites by ChIP-Seq with anti-Ldb1, 90% were also identified as Scl- and/or GATA-2 binding sites (Supplementary Fig. 9). Consistent with those results, 30 of 39 of the Ldb1 complex–binding sites contained one or more conserved GATA motifs; 31 of 39 sites contained one or more conserved CANNTG (E-box) motifs (where ‘N’ is any nucleotide); and 25 of 39 sites contained both motifs (Fig. 6b and Supplementary Table 2). The GATA sites closely matched the revised consensus GATA transcription factor–binding motif (WGATAA, where W represents A or T)32 (Fig. 6c). We found that 70% of all GATA motifs were paired with complete (CANNTG) or partial (TG) E-box motifs located seven to ten base pairs upstream of the GATA sequence (CANNTG-N7–10-GATA or TG-N7–10-GATA), and one third of the DNA fragments contained partial or complete E-box–E-box paired motifs (CANNTG-N7–10-CANNTG or TG-N7–10-CANNTG; Supplementary Table 2 and data not shown). Overall, 69% (27 of 39) of the DNA fragments contained at least one paired motif matching a known Ldb1 complex–binding site7,33 (Supplementary Table 2).
Figure 6.
Ldb1 complex–binding sites are present in a high percentage of genes critical for HSCs. (a) Selected genes involved in HSC maintenance with Ldb1 complex–binding sites in the promoter and/or gene body, as determined by ChIP-Seq analysis with control antibody to immunoglobulin G (IgG), anti-Ldb1, anti-Scl and anti-GATA-2. Numbers in plots indicate positions of binding sites at known distal regulatory elements. Sequence conservation track is shown at the bottom of each browser shot. Data are representative of two experiments. (b) Ldb1 complex–binding site fragments in the HSC-maintenance gene set (left) or in known regulatory elements near these genes (right) with conserved GATA motifs and/or E-box (CANNTG) motifs. Data are representative of two experiments. (c) Consensus sequence motif of sites containing a GATA-binding sequence in Ldb1 complex–binding sites at the promoter, gene body and/or known enhancers of HSC maintenance genes. Letter size indicates nucleotide frequency, scaled to the information content (measure of conservation) at each position; colors distinguish the nucleotides. Data are representative of two experiments. (d) Prevalence of Scl or GATA-2 binding at HSC gene sites identified by ChIP-Seq with anti-Ldb1. Data set includes the 53 DNA fragments in Supplementary Tables 1 and 3. Data are representative of two experiments.
We detected one or more highly conserved binding consensus sequences for other transcription factors, including Runx1, ETS (PU.1), Gfi1, Myb, C/EBP, Meis1, Nkx, Foxo and Smad (Supplementary Table 2), in each of the DNA fragments identified as Ldb1-binding sites, which indicated that the Ldb1-binding sites were in important regulatory domains. In addition, we detected Ldb1 complex–binding sites in 11 of 12 known regulatory elements in the vicinity of the genes involved in HSC maintenance (Fig. 6a, Supplementary Tables 3 and 4 and Supplementary Fig. 8). Binding of Scl and GATA-2 correlated with binding of Ldb1 at these sites (Fig. 6a, d and Supplementary Figs. 8 and 9) and the distribution of E-box and GATA motifs, as well as E-box–GATA paired motifs, in these binding sites was similar to that observed for sites in the promoter or gene body of genes involved in HSC maintenance (Fig. 6b and Supplementary Table 4). We also identified Ldb1 complex–binding sites near Cebpa and Gfi1, genes that did not contain such binding sites in their promoter or gene body but were significantly downregulated (Cebpa) or upregulated (Gfi1) after downregulation of Ldb1 (Fig. 5b and Supplementary Fig. 8). These binding sites contained conserved GATA and E-box sequences as well as conserved binding motifs for Ets, Gfi1, Runx and Meis1 (data not shown), which suggested they were in previously unknown regulatory elements.
DISCUSSION
Our results have demonstrated an essential and continuous requirement for Ldb1 for the maintenance of both fetal and adult HSCs. Most proteins identified as having an important role in HSC maintenance are required in fetal or adult HSCs but not both3,29,34–36. The phenotype elicited by Ldb1 inactivation also differed from that of other genes critical for HSC maintenance in the rapidity with which HSCs were lost and the finding of no overt defects in the proliferation or survival of HSC cells. A possible explanation for our findings is that Ldb1 complexes coordinate the expression of genes that function to prevent HSC differentiation. Consistent with that idea, deletion of Ldb1 resulted in skewing of the ratio Flt3− LSK cells to Flt3+ LSK cells (HSCs/MPPs), which indicated the loss of HSCs was accompanied by a transient increase in the number of non–self-renewing MPPs.
Although the precise subunit composition of Ldb1-nucleated complexes expressed in HSCs remains to be defined, several lines of evidence indicate that they include Lmo2, GATA-2, E2A and Scl (or Lyl1). Each of these subunits has been detected in multimeric Ldb1 complexes7,8,33,37–39, and each has high expression in hematopoietic progenitors. Lmo2, GATA-2, E2A, Scl or Lyl1, and now Ldb1, have been shown to be required for HSC specification and/or HSC maintenance9–14. The high correlation of the binding of Ldb1, Scl and GATA-2 at sites containing conserved E-box–GATA sequence motifs further supports the idea that these proteins function cooperatively in the same complex. Although each of the putative subunits of the Ldb1 complex in HSCs is necessary for HSC maintenance, the phenotype elicited by deletion of Ldb1 was particularly severe in that HSCs were lost far more rapidly after deletion of Ldb1 than after deletion of any other single subunit, which suggested that whereas Ldb1 is indispensable, some functional redundancy exists among the other subunits. Indeed, it has been shown that either Scl or Lyl1 is required for HSC maintenance, which demonstrates that these closely related basic helix-loop-helix proteins have partially redundant functions in hematopoietic progenitor cells11.
Our ChIP-Seq screening confirmed published data obtained by conventional ChIP demonstrating that the Lmo2 enhancers located −75 kilobases (kb) and −25 kb relative to the transcription start site40 and the Runx1 HSC enhancer located +23 kb relative to the transcription start site38 bind Ldb1, Scl, Lmo2 and GATA-2. In addition, we identified Ldb1 complex–binding sites in known regulatory elements in or near Gata2, Scl, Lyl1, c-Myb and Erg, which have been shown before to bind Scl41. Notably, the single enhancer element not bound by Ldb1 complexes (Scl; +19 kb relative to the transcription start site) is known to bind a different multimeric complex composed of GATA-2 and the Ets proteins Fli-1 and Elf-1 in HSC lines42.
Our data suggest that Ldb1 complexes accomplish an important function in regulating the transcription of a large number of genes critical to HSC maintenance. Although in some cases the effect of Ldb1 downregulation on gene expression seemed to be indirect, ChIP-Seq screening identified Ldb1 complex–binding sites in or near a large percentage of the 28 HSC genes surveyed. Nearly all of these Ldb1 complex–binding sites contained consensus partial or complete paired E-box–GATA or E-box–E-box motifs shown before to be Ldb1 complex–binding sites7,33, as well as conserved binding sites for other hematopoietic transcription factors. In addition, we identified Ldb1 complex–binding sites at 11 of 12 known hematopoietic enhancer elements. Together these findings support a model whereby Ldb1 complexes regulate a core transcriptional program necessary for HSC maintenance. Ldb1 complexes show several parallels to Oct-4, Nanog and Sox2, the core transcriptional mediators of ESC maintenance. Similar to Ldb1 complex subunits, Oct-4, Nanog and Sox2 function cooperatively, perhaps through the formation of a multimeric complex, bind to the same genomic sites together with additional transcription factors, show autoregulatory activity, and coregulate the expression of many genes essential for the maintenance of stem cells1,2. Notably, although it was expressed in ESCs, Ldb1 was not required for ESC maintenance, which suggests that it may instead function to ‘prime’ ESCs for differentiation after the induction of lineage specific transcription factors that require Ldb1 for their activity.
Ldb1 has high expression in HSCs and erythroblasts but is considerably downregulated in all other hematopoietic lineages17. Commitment of HSCs to the erythroid lineage is triggered by induction of GATA-1 and the (GATA-1-mediated) repression of Gata2, which results in a switch in the subunit composition of Ldb1 complexes43. Accordingly, the triggering event for HSC differentiation might involve either downregulation of Ldb1 or the formation of Ldb1 complexes with different subunit composition (GATA-1 versus GATA-2). Overexpression of Lmo2 in immature thymocytes reinstates a self-renewal program closely resembling that in HSCs, which suggests that sustained expression of partial or complete Ldb1 complexes in T cell progenitors promotes self-renewal44. However, Ldb1 complexes that contain GATA-1 induce erythroid gene expression and therefore function to promote the terminal differentiation of erythroid cells17,37,45. In summary, our results have established a critical role for Ldb1 in the maintenance of fetal and adult HSCs and have provided evidence that Ldb1-nucleated complexes function by regulating a core transcriptional program required for HSC ‘stemness’ and by restricting HSC differentiation.
ONLINE METHODS
Mice
B6, congenic B6-CD45.1, Rag2−/− and congenic Rag2−/−-CD45.1 mice were from Taconic Farms. Tie2-Cre-transgenic, Mx1-Cre–transgenic and Rosa26–green fluorescent protein Cre reporter mice (B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J) were from Jackson Laboratories. All mice were bred and maintained in a National Institutes of Health Research Animal Facility in accordance with the specifications of the Association for Assessment and Accreditation of Laboratory Animal Care. Mouse protocols were approved by the Animal Care and Use Committee of the National Institutes of Health.
ESC lines
The generation of Ldb1+/− ESCs15, Ldb1−/− ESCs16 and Ldb1fl ESCs20 has been described. Reconstitution of Ldb1 expression in Ldb1−/− ESCs was achieved by the introduction of a bacterial artificial chromosome (RP23-209B17; Invitrogen) containing Ldb1 into Ldb1−/− ESCs by electroporation. Reconstitution of Ldb1 expression was confirmed by RT-PCR analysis of Ldb1 transcripts and by intracellular staining. LacZ+ Ldb1−/− ESCs were generated as shown in the Supplementary Methods.
Flow cytometry
Conjugated antibodies, including isotype-matched control mouse and hamster immunoglobulin G antibodies were from BD Bioscience or eBioscience. Antibodies used for flow cytometry and the composition of the lineage mixture used for the detection of LSK cells are described in the Supplementary Methods. Cells were incubated with blocking antibody (2.4G2), then stained with fluorchrome-conjugated antibodies, then analyzed with a FACSCalibur or LSR II. Cells were sorted with a FACSAria.
ESC culture and embryoid body formation
Undifferentiated ESCs were cultured in Knockout DMEM (Invitrogen) containing 15% (vol/vol) FCS and leukemia-inhibitory factor on mitomycin-treated embryonic fibroblast feeder cells. For the generation of embryoid bodies, ESC cultures were treated with trypsin until clusters of five to ten cells were formed. Trypsin treatment was stopped by the addition of 10 ml Iscove’s modified Dulbecco’s medium containing 10% (vol/vol) FCS and cells were incubated for 45 min at 37 °C to allow reattachment of embryonic fibroblasts. ESCs were transferred to 10-cm bacterial Petri dishes and were incubated for 3–6 d in Iscove’s modified Dulbecco’s medium plus 15% (vol/vol) FCS in the presence of stem cell factor (40 ng/ml) and were observed for the formation of embryoid bodies. Reagents from Stem Cell Technologies were used according to the manufacturer’s protocols for methylcellulose cultures.
RT-PCR and real-time quantitative RT-PCR
For gene-expression studies, total cell RNA was isolated with a PicoPure RNA isolation kit (Arcturus), then 100 ng of each RNA sample was reverse-transcribed with SuperScript First-Strand Synthesis system (Invitrogen) and assayed by RT-PCR (primers, Supplementary Table 5). Transcripts were quantified with a Roche LightCycler 480. Duplicates were run for each sample in a 96-well plate; Actb (encoding β-actin) was used as the endogenous reference gene. The relative quantification method was used. Gene expression was normalized to that of Actb, and the expression of the mRNA of interest was then presented as a ratio relative to expression of the same gene in control bone marrow cells. The specificity of the products was confirmed on the basis of melting curves and electrophoresis.
Generation of chimeric mice via blastocysts
Chimeric mice were generated by injection of four to six ESCs into B6, B6 (CD45.1+) Rag2−/− or Rag2−/− (CD45.1+) blastocysts. Two Ldb1−/− ESC clones and two Ldb1−/− ESC clones reconstituted with a bacterial artificial chromosome containing Ldb1 were used in these experiments. As the individual clones yielded identical phenotypes, a single clone was used for most experiments.
Generation of fetal liver and adult bone marrow chimeras
For experiments involving adoptive transfer of fetal liver cells, 5 × 106 chimeric fetal liver cells at E15.5 or total fetal liver cells from Tie2-Cre Ldb1fl/Δ mice and littermate controls at E12 were injected intravenously into irradiated (650 rads) Rag2−/− (CD45.1) recipients. For experiments involving adoptive transfer of adult bone marrow, a total of 5 × 106 cells (50:50 mixture of Mx1-Cre Ldb1fl/fl (CD45.2+) bone marrow and B6 (CD45.1+) bone marrow) was transferred into irradiated Rag2−/− (CD45.1) recipients.
Conditional inactivation of Ldb1 in adult mice
For inactivation of Ldb1 in adult hematopoietic cells, 250 μl poly(I:C) (1 mg/ml; Amersham Biosciences) was injected intraperitoneally into each mouse every other day for a total of three doses unless noted otherwise. The first injection day was designated day 1. Bone marrow cells were either analyzed directly for the presence of LSK cells or were mixed 50:50 with bone marrow cells from identically treated B6 (CD45.1) mice, then adoptively transferred into irradiated Rag2−/− (CD45.1) mice for analysis of activity of LTR-HSCs.
ChIP-Seq
ChIP-Seq for Ldb1, Scl, GATA-2 and immunoglobulin G (control) was done as described30. The SISSRs algorithm (site identification from short sequence reads)31 was used for the identification of binding sites (Supplementary Methods).
Supplementary Material
Acknowledgments
We thank D. El-Khoury and A. Grinberg for technical support, and M. Mukhopadhyay for advice about mouse experiments. Supported by the Intramural Research Program of the National Institutes of Health (Eunice Kennedy Shriver National Institute of Child Health and Human Development, L.L., J.Y.L., T.C., M.G., I.T., Y.Z., H.W. and P.E.L.; National Institute of Environmental Health Sciences, R.J.; National Heart, Lung, and Blood Institute, K.C. and K.Z.) and the National Institutes of Health (DK068634 to E.H.B.).
Footnotes
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
L.L. designed and (with assistance from P.E.L.) did all of the experiments; L.L. and P.E.L. designed the study and wrote the manuscript; K.C. and K.Z. assisted in designing and doing the ChIP-Seq experiments; R.J. did the statistical analysis of ChIP-Seq data; E.H.B. provided reagents and input for the ChIP-Seq experiments; J.Y.L., T.C., M.G., I.T., Y.Z. and S.M.H. assisted with specific mouse experiments and provided input on experimental design; and H.W. provided the Ldb1 mouse strains.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Accession codes. GEO: ‘sequence-read’ data for Figures 5 and 6, Supplementary Tables 1–4 and Supplementary Figures 8 and 9, GSE26031.
References
- 1.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 3.Akala OO, Clarke MF. Hematopoietic stem cell self-renewal. Curr Opin Genet Dev. 2006;16:496–501. doi: 10.1016/j.gde.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 5.Majewski IJ, et al. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 2008;6:e93. doi: 10.1371/journal.pbio.0060093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadman IA, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier N, et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues NP, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 10.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci USA. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren AJ, et al. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 13.Ling KW, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhyay M, et al. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- 16.Hwang M, Gorivodsky M, Kim M, Westphal H, Geum D. The neuronal differentiation potential of Ldb1-null mutant embryonic stem cells is dependent on extrinsic influences. Stem Cells. 2008;26:1490–1495. doi: 10.1634/stemcells.2007-1099. [DOI] [PubMed] [Google Scholar]
- 17.Li L, et al. A requirement for Lim domain binding protein 1 in erythropoiesis. J Exp Med. 2010;207:2543–2550. doi: 10.1084/jem.20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison SJ, Uchida N, Weissman IL. The biology of hematopoietic stem cells. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 19.Lord BI, et al. Myeloid cell kinetics in mice treated with recombinant interleukin-3, granulocyte colony-stimulating factor (CSF), or granulocyte-macrophage CSF in vivo. Blood. 1991;77:2154–2159. [PubMed] [Google Scholar]
- 20.Zhao Y, et al. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA. 2007;104:13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kisanuki YY, et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 22.Schlaeger TM, Mikkola HK, Gekas C, Helgadottir HB, Orkin SH. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood. 2005;105:3871–3874. doi: 10.1182/blood-2004-11-4467. [DOI] [PubMed] [Google Scholar]
- 23.Adolfsson J, et al. Upregulation of Flt3 expression within the bone marrow Lin−Sca1+c-kit+ stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, et al. Identification of Lin−Sca1+kit+CD34+Flt3− short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 25.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 27.Wilson A, Laurenti E, Trumpp A. Balancing dormant and self-renewing hematopoietic stem cells. Curr Opin Genet Dev. 2009;19:461–468. doi: 10.1016/j.gde.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453:306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
- 29.Lessard J, Faubert A, Sauvageau G. Genetic programs regulating HSC specification, maintenance and expansion. Oncogene. 2004;23:7199–7209. doi: 10.1038/sj.onc.1207940. [DOI] [PubMed] [Google Scholar]
- 30.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grutz GG, et al. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 1998;17:4594–4605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hock H, Orkin SH. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr Opin Hematol. 2006;13:1–6. doi: 10.1097/01.moh.0000190111.85284.8f. [DOI] [PubMed] [Google Scholar]
- 35.Galan-Caridad JM, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anguita E, et al. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nottingham WT, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landry JR, et al. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood. 2008;111:3005–3014. doi: 10.1182/blood-2007-07-098830. [DOI] [PubMed] [Google Scholar]
- 40.Landry JR, et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood. 2009;113:5783–5792. doi: 10.1182/blood-2008-11-187757. [DOI] [PubMed] [Google Scholar]
- 41.Wilson NK, et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- 42.Gottgens B, et al. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grass JA, et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCormack MP, et al. The lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 45.Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.