Abstract
The phosphatase and tensin homologue on chromosome 10 (PTEN) suppresses the activity of the phosphoinositide-3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway, a signaling cascade critically involved in the regulation of cell proliferation and growth. Human patients carrying germline PTEN mutations have an increased predisposition to tumors, and also display a variety of neurological symptoms and increased risk of epilepsy and autism, implicating PTEN in neuronal development and function. Consistently, loss of Pten in mouse neural cells results in ataxia, seizures, cognitive abnormalities, increased soma size and synaptic abnormalities. To better understand how Pten regulates the excitability of principal forebrain neurons, a factor that is likely to be altered in cognitive disorders, epilepsy and autism, we generated a novel conditional knockout mouse line (NEX-Pten) in which Cre, under the control of the NEX promoter, drives the deletion of Pten specifically in early postmitotic, excitatory neurons of the developing forebrain. Homozygous mutant mice exhibited a massive enlargement of the forebrain, and died shortly after birth due to excessive mTOR activation. Analysis of the neonatal cerebral cortex further identified molecular defects resulting from Pten deletion that likely affect several aspects of neuronal development and excitability.
Keywords: Pten, mTOR, glutamate, autism, seizure, NMDA receptor, epilepsy
Introduction
The PTEN (phosphatase and tensin homolog located on chromosome 10) gene product is a lipid and protein phosphatase that negatively regulates the phosphoinositide-3-kinase (PI3K)/Akt signaling pathway [1]. PTEN opposes the function of PI3K by shifting the balance from phosphatidylinositol (3, 4, 5)-triphosphate (PIP3) to phosphatidylinositol (4,5)-biphosphate (PIP2) [2]. Since PIP3 is required for the activation of 3-phosphoinositide-dependent protein kinase-1 (PDK1), and this kinase in turn phosphorylates and activates Akt, PTEN essentially suppresses Akt activity. Phosphorylated Akt is a crucial regulator of cell survival, growth and differentiation that functions by phosphorylating many downstream targets. One major Akt target is the tuberous sclerosis complex protein, tuberin (TSC2), which is phosphorylated and inhibited by Akt [3,4]. Since TSC inhibits the mammalian target of rapamycin (mTOR) through inactivation of the small GTPase Rheb [5,6], Akt normally promotes mTOR activity. Thus PTEN, by suppressing PI3K/Akt signaling, also suppresses the downstream mTOR kinase activity. mTOR is critically involved in cellular growth and development. This kinase is a core component of distinct protein complexes, mTORC1 and mTORC2 [7]. mTORC1 is primarily involved in the control of protein translation and cellular growth by phosphorylating translation initiation complexes and the ribosomal S6 kinase (p70S6K). mTORC2, on the other hand, participates in a positive feedback loop by phosphorylating Akt at serine 473 [8], a site distinct from that affected by PI3K signaling (threonine 389). Since mTOR promotes protein translation [9,10], unmitigated activity of this kinase in neurons may lead to deregulated synthesis of dendritic and synaptic proteins.
In addition to its well-known association with cancer, PTEN has been implicated in several neurological disorders [11]. Interestingly, mutations in either PTEN or downstream TSC1/TSC2 genes are associated with autism spectrum disorder and epilepsy. These findings suggest that deregulation of the PI3K/Akt/mTOR signaling cascade contributes to the etiology of these diseases. However, the molecular mechanisms underlying altered behavior, cognition and neuronal excitability in autism spectrum disorders and associated epilepsy are not understood. In order to study the role of PTEN deficiency in brain development, several conditional Pten knockout mouse models have been generated. Deletion of Pten in neural progenitor cells, accomplished by the use of a nestin promoter-driven Cre transgene, resulted in increased proliferation, brain enlargement and perinatal lethality [11,12]. Pten deletion driven by a GFAP promoter active in a subset of neuronal progenitor cells (NS-Pten) leads to ataxia, macrocephaly, neuronal hypertrophy and epileptic seizures [13–15]. Treatment with mTOR inhibitors, such as rapamycin or its analogs, rescued the phenotype of these mice and dramatically suppressed seizures, demonstrating the key role of mTORC1 in Pten-dependent epilepsy [15,16]. Interestingly, loss of Pten from a subset of forebrain neurons, achieved by the use of the neural-specific enolase (Nse)-Cre transgene, produced mice that displayed behavioral defects reminiscent of autism [17]. Using a knock down approach, a recent study indicated that Pten reduction in mature neurons enhances the excitatory drive, thus altering the excitatory/inhibitory ratio [18]. In this study, we sought to further investigate the function of Pten in the formation of forebrain cortical structures and in the regulation of neuronal excitability. We generated a novel conditional knockout line (NEX-Pten) by crossing PtenloxP/loxP mice with NEX-Cre transgenic mice, in which virtually all excitatory neurons of the forebrain express Cre [19]. The phenotype of homozygous NEX-Pten conditional knockouts includes premature death, macrocephaly, alterations in forebrain development, and expression of proteins involved in migration, dendrite maturation and neuronal activity in this region.
Materials and Methods
Mice
NEX-Pten mice were generated by crossing homozygous NEX-Cre knockin mice [19] with Cre-negative conditional neuron subset-specific Pten (NS-Pten) knockout mice (PtenloxP/loxP), which have been described previously [15]. NEX-Cre mice (backcrossed for 10 generations to C57Bl/6 mice) were received from Dr. Klaus Nave. In these studies, NEX-Cre+;PtenloxP/+ (heterozygous) mice were used for breeding to generate NEX-Cre+;Pten+/+ (wild type), heterozygous and homozygous (NEX-Cre+;PtenloxP/loxP) knockout mice. The weight of NEX-Pten mouse pups from several litters was recorded daily (Monday-Friday), beginning after birth at postnatal day 0 (P0). For rapamycin treatment, a small cohort of homozygous mutant mice (n=3) received 0.5 mg/kg subcutaneous injections of rapamycin every other day for two weeks starting at postnatal day 1. Survival and weight comparisons were made with untreated mutants of the same age whenever possible. All experiments and animal housing were in accordance with procedures approved by the Animal Protocol Committees at Rutgers University and Baylor College of Medicine, according to the National and Institutional Guidelines for Animal Care established by the National Institute of Health.
Tissue Immunofluorescence
Brains were isolated at P0 immediately after birth, post-fixed in 4% formaldehyde dissolved in phosphate buffered saline (PBS) for 24 hrs, and then transferred to a 30% sucrose solution in PBS for cryoprotection. After sinking in the sucrose solution, brains were frozen in a 30% sucrose/Cryo-OCT compound solution (30:70 mixture) (Fisher) and serially sectioned at 30 µm on a cryostat. To detect potential migration deficits caused by Pten deficiency during development, pregnant dams were treated with 100 mg of 5-bromo-2’-deoxyuridine (BrdU) (Sigma) per kg of body weight by intraperitoneal (IP) injection at embryonic day 15.5 (E15.5) to label proliferating neurons. For immunofluorescence and BrdU labeling, sections were incubated in 2N hydrochloric acid for 30 min at 37°C, followed by neutralization with 100 mM sodium borate (pH 8.5) for 10 min at room temperature. Sections were then permeabilized in 0.1% Triton X-100 (in PBS) for 10 min and blocked with 10% normal goat serum in 0.1% Triton X-100 (in PBS) (blocking solution) for 1 hr at room temperature. Sections were incubated with primary antibodies in blocking solution overnight at 4°C. Primary antibodies were as follows: ratanti-BrdU (1:100; Abcam), mouse anti-Cre (1:100; Covance), rabbit anti-Cux1 (1:100; Santa Cruz Biotechnology), mouse anti-Reelin (CR-50) (1:500; purified from hybridoma cell culture supernatants using Hi-Trap protein G columns (Amersham Biosciences), and rabbit anti-Tbr1 (1:100; Millipore). Sections were then washed and incubated with AlexaFluor 488-, AlexaFluor Cy5- or AlexaFluor 647-conjugated secondary antibody (1:500; Invitrogen) for 1 hr at room temperature. After washing with PBS, sections were mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories). Multiple sections from 2–3 mice per genotype were examined. Representative images were acquired using a Yokogawa CSU-10 spinning disk confocal head attached to an inverted fluorescence microscope (Olympus IX50).
Immunohistochemistry
For immunohistochemistry, postnatal brains were fixed in 10% (v/v) neutral buffered formalin (NBF, VWR) and embedded in paraffin and 6 µm sagittal sections were cut on a microtome. Prior to antibody incubation, antigen retrieval was performed in 1mM citric acid (pH 6) in a steamer for 30 minutes, plus 20 minutes cooling time. Endogenous peroxidase activity was blocked with 1.8% (v/v) H2O2 (in PBS with 0.1% (v/v) triton X-100) and the sections were incubated with 10% (v/v) normal goat or donkey serum (Jackson ImmunoResearch Laboratories) in PBS with 0.1% (v/v) triton X-100. Sections were incubated with Pten antibodies (mouse monoclonal, 1:1600, clone 6H2.1, Cascade Bioscience) diluted in PBS with 0.1% (v/v) triton X-100 and antibody binding was performed overnight at 4°C. Biotin-labeled secondary antibodies raised in goat (1:200, Jackson ImmunoResearch Laboratories) were then used, followed by peroxidase-conjugated avidin (Vectastain Elite ABC kit, Vector Laboratories). Immunoreactivity was detected by 3,3-diaminobenzidine (DAB) substrate (Vector Laboratories). Slides were counterstained with Mayer’s hematoxylin (VWR) and mounted in Cytoseal 60 (VWR). For histology, thionin staining (FD Neurotechnologies) was performed on 30 µm brain sections fixed in formaldehyde (as described above). Briefly, sections were placed in xylene for 3 min, followed by two incubations in 100% ethanol (3 min each). Next, slides were immersed in 95% and 75% ethanol (3 min each), and then in distilled water 3 times (3 min each). Brain sections were stained in thionin solution for 10 min, rinsed briefly in distilled water, and then immersed in 95% ethanol with 0.1% glacial acetic acid for 2 min. Finally, sections were dehydrated in 100% ethanol with 4 changes (2 min each), cleared in xylene for 3 changes (3 min each) and mounted with Permount (Fisher).
Dissociated Cortical Cultures Preparation and Analysis
Cortical brain tissue was dissected from wild type, heterozygous and homozygous NEX-Pten knockout littermates at P0 (immediately after birth) in cold Hanks Balance Salt Solution (HBSS; Invitrogen), cut into small pieces, and pooled. Neuronal cultures were prepared using a papain dissociation kit (Worthington). Cells were then strained with a 70 µm filter, and centrifuged for 5 min at 300×g. The pellet was re-suspended in a solution of Earle’s Balanced Salt Solution (EBSS)/deoxyribonuclease I (DNase)/ovomucoid protease inhibitor. The re-suspension was layered on top of 5 ml ovomucoid protease inhibitor solution, centrifuged for 6 min at 100×g, and then centrifuged for an additional 6 min at 200×g to evenly coat the cells with the ovomucoid solution. Cells were re-suspended in a mixture of 98% Neurobasal medium, 2% B-27 supplement, 0.5mM glutamine and 0.5 mM PenStrep (Invitrogen), plated onto poly-L-lysine coated glass coverslips at a density of 55,000 cells/cm2, and maintained at 37°C in 5% CO2 in a water-jacked incubator for 15 days in vitro (DIV). Half the culture medium was replaced with freshly prepared medium every 4–5 days for maintenance. Cultures were washed with PBS and then immediately fixed with 4% formaldehyde (in PBS) for 15 min at room temperature. Subsequently, cells were washed 3 times for 5 min with PBS, permeabilized with 0.1% Triton-X (in PBS) for 10 min and blocked with 10% bovine serum albumin (BSA) in 0.1% Triton-X/PBS for 30 min. In order to label actin, rhodamine phalloidin conjugate (10 µl; Invitrogen) was added to the blocking solution for 15 min followed by 3 PBS washes. For Pten immunolabeling, primary antibody (1:250; Cell Signaling) was diluted in 10% goat serum/0.1% Triton-X/PBS and incubated overnight at 4°C. Cells were then washed with PBS and incubated with AlexaFluor 488-conjugated secondary antibody (1:500; Invitrogen) in 0.1% Triton-X/PBS for 1 hr at room temperature. After additional PBS washing, coverslips were mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories). Cortical neurons were imaged at random with a Leica DM5000B epifluorescence microscope using a 20× objective (HC PLAN APO; N.A. 0.7). The soma of Pten-positive and Pten-negative neurons was traced from merged images using the Freehand Selection tool in ImageJ to measure the size. The data were averaged and analyzed using a Students t-test.
Western Blot Analysis
Brains of NEX-Pten mouse pups were isolated immediately after birth at P0 in chilled Hank’s Balanced Salt Solution (HBSS; Invitrogen). Cortex and hippocampus were then dissected and homogenized together in RIPA lysis buffer (50 mM Tris (pH 7.4), 1% NP40, 0.25% sodium deoxycholate, 150 mM NaCl, 1nM EDTA) with protease (cOmplete Mini, Roche) and phosphatase inhibitors (PhosSTOP, Roche). Protein concentrations were determined by the Bradford method. Samples were supplemented with Laemmli sample buffer, boiled for 3 minutes and then subjected to SDS-PAGE. Proteins were electro-transferred by a wet blotting method to a 0.2 µm nitrocellulose membrane. The membranes were washed with TBS-T solution (0.05% Tween-20, 0.8% NaCl, 20 mM Tris (pH 7.5)), and blocked in a 3% nonfat dry milk solution (in TBS-T) for 1 hr, followed by additional washing. Membranes were incubated in 0.3% nonfat dry milk/TBS-T with the appropriate primary antibodies overnight at 4° C. Primary antibodies were as follows: mouse anti-actin (1:10,000; Millipore), rabbit anti-phospho-Akt (Serine 473) (1:1000; Cell Signaling), mouse anti-phospho-Akt (Threonine 389) (1:1000; Cell Signaling), rabbit anti-phospho ribosomal protein S6 (Serine 240/244) (1:1000; Cell Signaling), rabbit anti-Dab1 (1:1000; Rockland), rabbit anti-GluR1 (1:1000; Millipore), rabbit anti-MAP2 (1:1000; Millipore), mouse anti-NeuN (1:1000; Millipore), rabbit anti-NR2A (1:1000; Millipore), rabbit anti-NR2B (1:1000; Millipore), and rabbit anti-PTEN (1:1000; Cell Signaling). After primary antibody incubation, membranes were washed with TBS-T and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. Membranes were incubated with ECL-Plus Western Blotting Detection System (Pierce/Thermo Fisher) to develop antibody signal, and then exposed to autoradiographic films. The levels of each protein analyzed were normalized to the intensity values of actin (loading control). Samples obtained from n=3 different mice of each genotype were run on the same membrane for direct comparison and statistical analysis. Wild type values were averaged and the mean converted to 100%. Data from heterozygous and knockout mice are represented as fold-change over wild type intensity levels.
Statistical Analyses
Survival curves for NEX-Pten mice were analyzed using a Log-rank (Mantel-Cox) test. The Students t-test was used to compare the body weight of homozygous to wild type mice. Data regarding the survival and the body weight of NEX-Pten homozygous knockout mice with or without the rapamycin treatment were analyzed by Students t-test. For analysis of in vitro soma size, data from Pten-positive and negative neurons were compared using a Students t-test. For Western blot analysis, NEX-Pten wild type and heterozygous or homozygous data were compared using a Students t-test or a one sample t-test. Statistical significance was reported as t(degrees of freedom) = t-score, when p<0.05. Since no significant difference was found between heterozygous and wild type mice, statistical comparisons are only reported between homozygous mutant and wild type mice.
Results
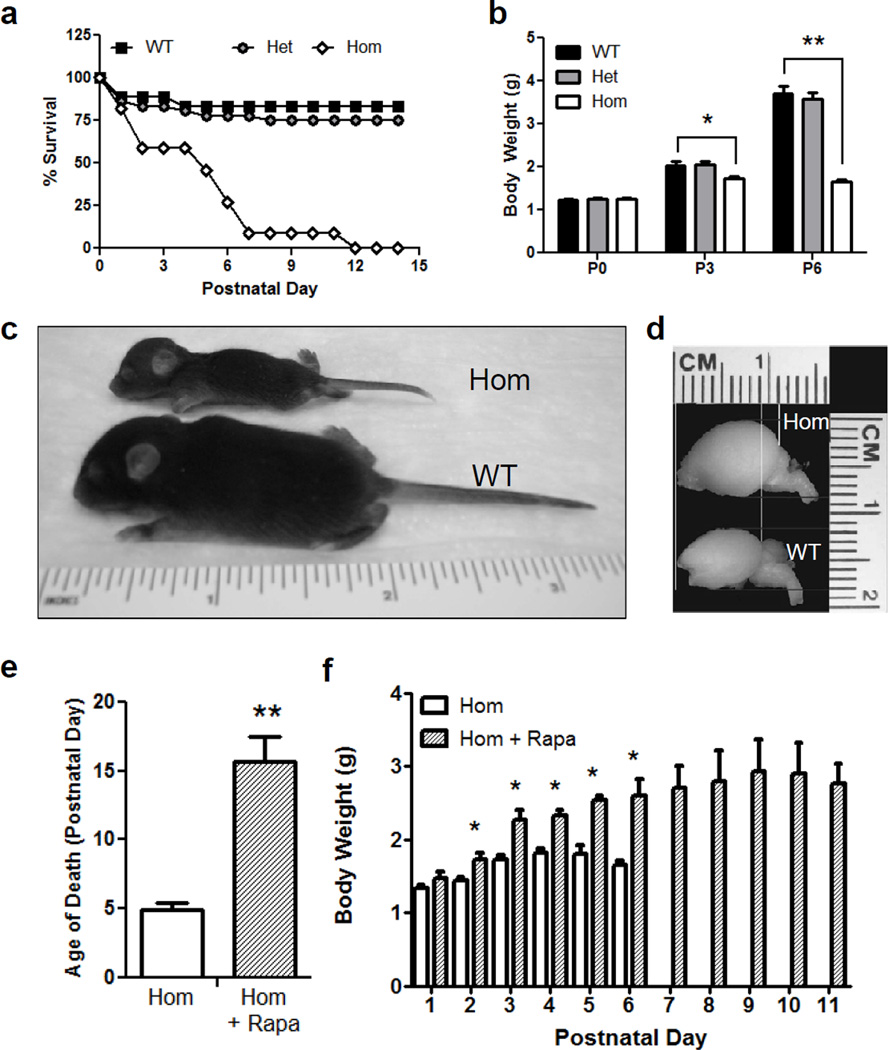
In order to evaluate the effects of Pten deficiency on brain development and neuronal differentiation, we generated a novel conditional Pten knockout line, using the NEX promoter to induce Cre expression and Pten deletion in all forebrain excitatory neurons immediately after they become postmitotic [19]. To generate this NEX-Pten mouse line, homozygous NEX-Cre knockin mice were initially bred with homozygous PtenloxP/loxP mice, producing double heterozygous NEX-Cre+;PtenloxP/wtmice, which appeared healthy and fertile. From the intercrossing of these mice, control and mutant mice of all expected genotypes were obtained. However, while heterozygous Cre+;PtenloxP/wtmice appeared normal, the majority of homozygous Cre+;PtenloxP/loxP knockout mice (n=37) died within one week after birth (fig. 1a). The survival rates between all genotypes were significantly different [X2(2)= 28.09, p<0.0001], such that homozygous Pten knockout mice died prematurely compared to their wild type littermates [X2(1)= 20.79, p<0.0001]. In addition to a reduced lifespan, homozygous Pten knockout mice weighed significantly less than wild type mice by postnatal day (P) 3 [t(25)=2.121, p<0.04], even though no such difference was observed at birth (P0) (p>0.05). Homozygous mutants failed to thrive, continuing to weigh significantly less than wild type at P6 [t(17)=5.796, p<0.0001] (fig. 1b). When a rare homozygous mutant survived past P6, it appeared severely runted (fig. 1c) and was therefore sacrificed at P8. At this age, the brain of the homozygous Pten knockout mouse could be easily distinguished from that of a wild type littermate due to its noticeable increase in size (fig. 1d). To determine whether the premature lethality of homozygous Pten knockout mice was due to excessive activation of mTOR, we treated a cohort of mutant pups (n=3) with the mTORC1 inhibitor rapamycin every two days starting on P1. As compared to untreated mutants (n=37) rapamycin treatment dramatically and significantly increased the survival of homozygous Pten knockout mice, from approximately 5 to 15 postnatal days [t(38)=6.044, p<0.001] (fig. 1e). In addition, the body weight of mutant mice treated with rapamycin (n=3) was significantly increased compared with untreated knockout mice (n=11) starting at postnatal day 2 [t(12)=2.824, and 4.284, for P2 and P3, respectively; t(11)=4.09 for P4; t(4)=5.5 for P5; t(5)= 4.572 for P6; p<0.05] (fig. 1f).
Fig. 1.
Overt phenotype of homozygous NEX-Pten mice. a Survival curve in a cohort of homozygous (Hom), heterozygous (Het) and wild type (WT) NEX-Pten littermates. Homozygous NEX-Pten knockout mice die prematurely, mostly within the first postnatal week. The survival curve for homozygous NEX-Pten mutant mice is significantly different than that of wild type or heterozygous mice (p<0.0001). b Body weight plot from a NEX-Pten littermate cohort. The average body weight of homozygous NEX-Pten mice is significantly less than wild type littermates at postnatal day (P) 3 (p<0.04) and P6 (p<0.001). c A rare homozygous NEX-Pten mouse that survived to P8 appears runted compared to a wild type littermate. d Images of the whole brain from P8 littermates reveal megalencephaly in the homozygous mutant. e Treatment of homozygous NEX-Pten mice with rapamycin (Rapa) significantly prolongs their survival compared untreated mutants (p<0.001). f Rapamycin treatment also significantly increases body weight at several postnatal days(p<0.05).
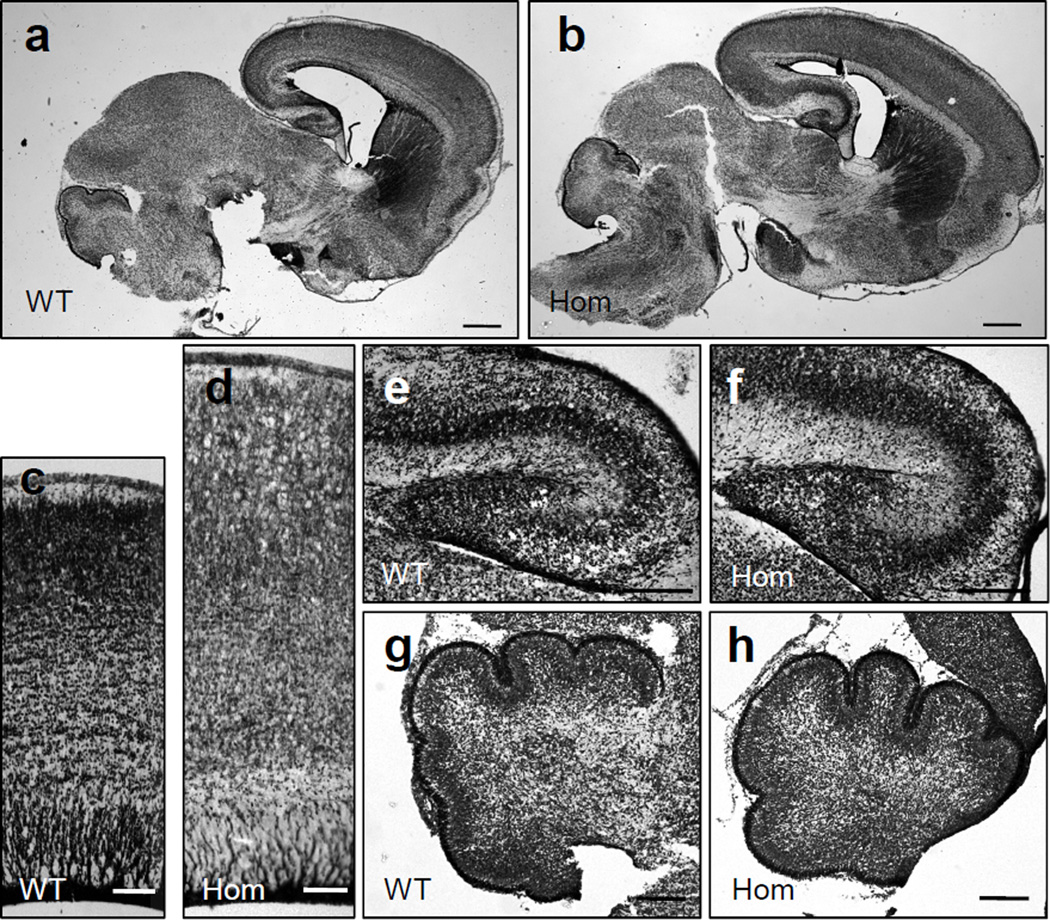
The enlargement of the forebrain, resulting from Pten loss in homozygous mutants, was confirmed by the analysis of brain anatomy in newborn mice. Comparable brain sections obtained from homozygous and wild type littermates were processed by histological staining. Low power images reveal a clear enlargement of forebrain, but not midbrain or hindbrain structures, in homozygous mutants (fig. 2a, b), consistent with the predominantly restricted expression in forebrain regions (fig. 3a) [19]. In addition to the expansion of forebrain structures such as the cerebral cortex (fig. 2c, d) and the hippocampus (fig. 2e, f), distorted and less compact cellular layers were consistently observed in homozygous mutant structures. Hindbrain structures such as the cerebellum, did not differ between genotypes (fig. 2g, h). All brain structures were unaffected in heterozygous Pten knockout mice (data not shown).
Fig. 2.
Histological examination of brain structures in newborn NEX-Pten mice. Sagittal sections obtained from P0 wild type (WT) and homozygous (Hom) NEX-Pten littermates were stained with thionin. Low power images (a, b) show an enlarged forebrain in homozygous mutant mice. Higher magnification images of comparable regions of the lateral cerebral cortex (c, d), hippocampus (e, f), and cerebellum (g, h) reveal a specific enlargement of forebrain structures in homozygous mutant mice. Scale bars = 500 µm (a, b), 100 µm (c, d), 200 µm (e–h).
Fig. 3.
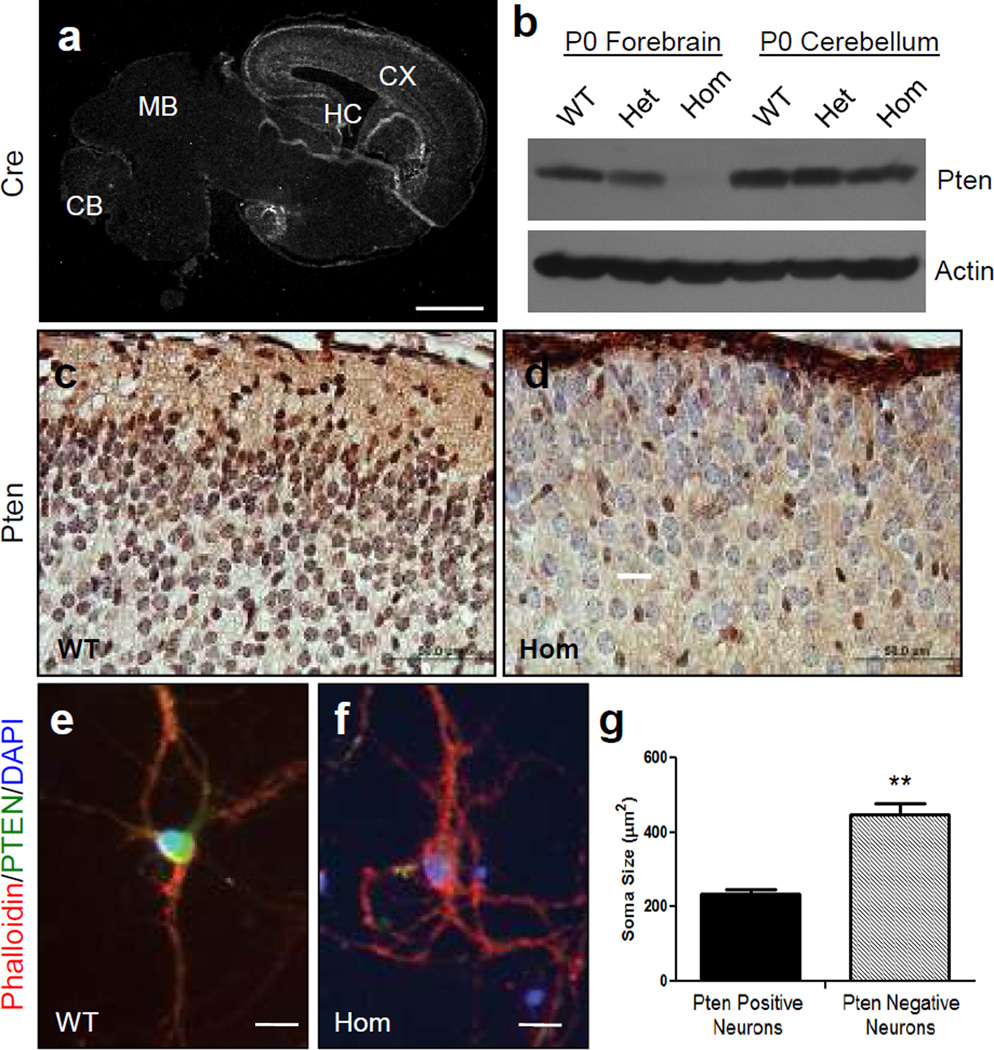
Pten deletion in excitatory cortical neurons results in increased soma size. a Brain sections obtained from P0 wild type (WT) mice in the NEX- Pten colony were processed for immunofluorescence using Cre antibodies. A low magnification confocal image shows widespread expression in the cerebral cortex (CX) and hippocampus (HC), but not in the midbrain (MB) or cerebellum (CB). b Western blot analysis of the forebrain and cerebellum of newborn wild type (WT), heterozygous (Het), and homozygous (Hom) NEX-Pten littermates. The blot, which is representative of data obtained from 3 sets of littermates, was probed with Pten antibodies and then reprobed with actin antibodies to ensure equal protein loading. Pten expression is progressively reduced in the forebrain of heterozygous and homozygous mutants. c, d Pten immunohistochemistry staining (brown) of brain sections obtained from WT and Hom NEX-Pten littermates. Sections were counterstained with cresyl violet. Brightfield images of the upper cortical layers show that the majority of cells are Pten-positive in wild type and Pten-negative in homozygous mutants. Pten-negative cells appeared enlarged, and some protrude in the marginal zone near the pial surface. e, f Confocal images of representative dissociated cortical neurons cultured for 15 DIV and triple-labeled with rhodamine-phalloidin (red), Pten antibodies (green) and DAPI (blue). g Soma size comparison between Pten-positive (n= 35) and Pten-negative cultured neurons (n=22). The difference is statistically significant (p<0.05). Scale bars = 1 mm (a), 50 µm (c and d), 20 µm(e and f).
To further verify the spatial and cellular specificity of our NEX-Pten knockout line, we examined Cre and Pten protein expression in newborn mice. Immunofluorescence data confirmed the previously reported predominant expression of NEX-Cre in the forebrain (fig. 3a). This expression pattern correlated with the loss of Pten, as Western blot analysis demonstrated a gradual reduction of Pten expression in neonatal forebrain structures of heterozygous and homozygous NEX-Pten knockout mice, but not in hindbrain structures such as the cerebellum (fig. 3b). Pten immunohistochemistry further revealed that the majority of neurons in the neocortex of homozygous knockout mice were indeed Pten-negative, whereas most neurons present in similar areas of the neocortex of wild type mice were Pten-positive (fig. 3c, d). Thus, the pattern of Pten deletion in the neocortex of homozygous mutants is consistent with the previously reported specific expression of NEX-Cre in excitatory, glutamatergic principal neurons [19]. The few Pten-positive cells seen in the neocortex of homozygous knockout mice could be interneurons or glial cells, which do not express NEX-Cre [19].
Pten-negative cortical neurons in homozygous mutants were obviously enlarged compared to Pten-positive neurons in wild type controls, and sometimes protruded into the marginal zone, a region that is normally cell-poor (fig. 3c, d). To examine their size in more detail, we co-cultured cortical neurons from embryos obtained from the mating of two NEX-Pten heterozygotes. Pten-negative and Pten-positive neurons were identified by Pten immunofluorescence, and were double labeled by rhodamine-conjugated phalloidin to stain the cell bodies, and by DAPI to visualize the nuclei. The cell bodies were then traced to measure soma size. The data indicate that Pten-negative neurons were indeed significantly larger than Pten-positive neurons and exhibited thicker neuritic processes (fig. 3e, f, g). These findings are entirely consistent with previous reports, describing neuronal hypertrophy of mutant neurons in different Pten conditional mutants [13–15].
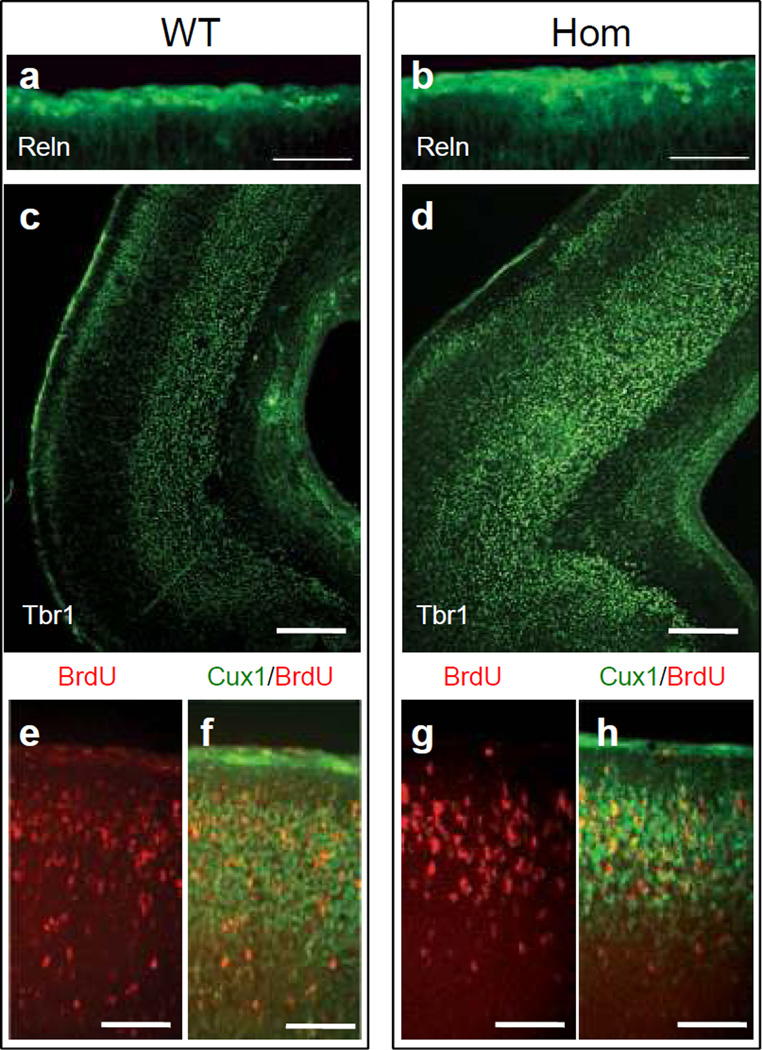
To better examine the effect of Pten loss on the migration and positioning of excitatory neurons into cortical layers, we conducted immunofluorescence experiments in the newborn brain. Since the migration of principal cortical neurons is largely controlled by Reelin (Reln) [20], we first examined the expression of this protein. The Reln signal was predominantly observed in the marginal zone of both wild type and homozygous NEX-Pten mutants (fig. 4a, b). The size and layer-localization of Reln-positive Cajal-Retzius cells appeared similar in both genotypes, consistent with the lack of NEX-Cre expression in these cells [19], although their distribution appeared slightly uneven in the mutant cortex. Principal cortical neurons, which lack Pten expression, were visualized by Tbr1 and Cux1 immunofluorescence and BrdU incorporation. Tbr1 immunofluorescence readily identifies early-born neurons residing in deep cortical layers of wild type mice (fig. 4c). Many Tbr1-positive neurons were also correctly positioned in deep cortical layers in the homozygous mutant neocortex. However, these layers appeared less compact than in the wild type, and some Tbr1-labeled neurons were also present at the surface of the cortex, near or within the marginal zone (fig. 4d). Late-born cortical neurons labeled with BrdU at embryonic day 15.5 (E15.5) migrated properly to the upper layers of the newborn cortex in Pten homozygous mutants as well as wild type littermates (fig. 4e, f, g, h). Additionally, expression of Cux1, an upper layer marker, did not differ between mutant and control mice (fig. 4f, h), suggesting that cell type specification and radial migration are largely unaltered by the cell autonomous loss of Pten in principal neurons of the neocortex.
Fig. 4.
Analysis of cortical layer formation in newborn NEX-Pten mice. Brain sections obtained from P0 wild type (WT) and homozygous (Hom) NEX-Pten littermates were processed for immunofluorescence using Reln (a and b) or Tbr1 (c and d) antibodies. A pregnant dam was injected with BrdU to label newly generated neurons at embryonic day 15.5. Brain sections were processed for immunofluorescence using BrdU and Cux1 antibodies (e–h). Confocal images show predominant expression of Reln in the marginal zone, and Tbr1 in deep layers of the caudal cerebral cortex in both genotypes. BrdU (red) and Cux1 (green) labeling was predominant in upper layers of the cerebral cortex in both genotypes. Scale bars = 100 µm (a and b), 200 µm(c and d), and 50 µm (e–h).
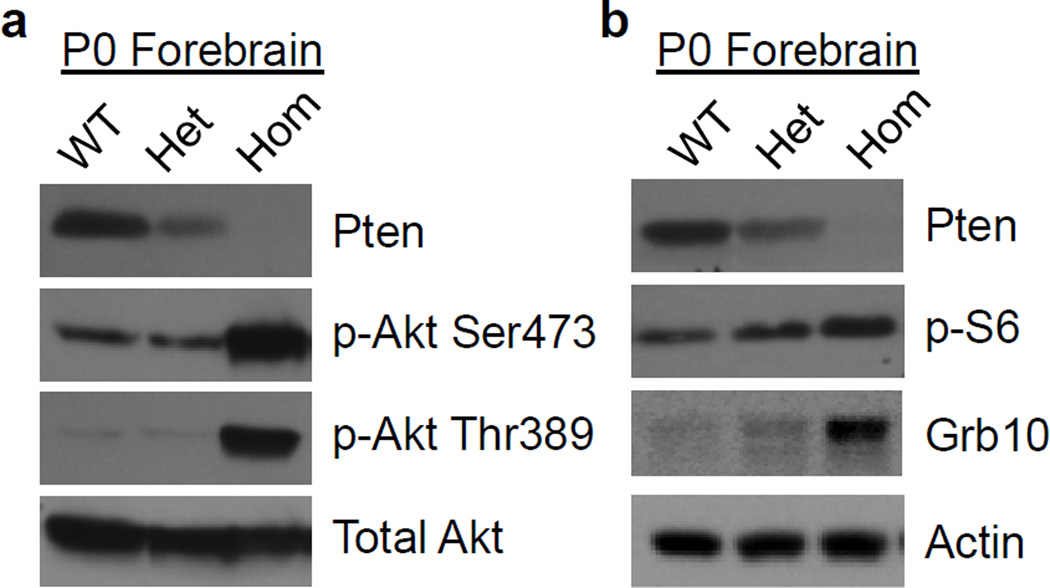
Since Pten normally acts to suppress the PI3K/Akt/mTOR pathway, we hypothesized that the abnormal forebrain development observed in Nex-Pten homozygous mutants would correlate with altered signaling. Using Western blots, components of this signaling cascade were analyzed from multiple mice (n>3 per genotype) and the values were normalized to that of actin to take into account potential increases in general translation mechanisms or loading errors. In lysates of the forebrain dissected from newborn pups, a progressive reduction of Pten expression was observed in heterozygous and homozygous mutant mice (fig. 3b, 5a, b). Statistical analysis confirmed a significant reduction in Pten expression in homozygous knockout mice compared to wild type littermate samples [t(4)=3.851, p<0.05]. Homozygous Pten knockout mice also exhibited dramatic increases in the levels of Akt phosphorylation at two different sites, serine 473 (the target of mTORC2) and threonine 389 (the target of the PI3K–dependent Pdk1 kinase) (fig. 5a). We found that both phosphorylation events were significantly increased in homozygous Pten knockout compared to wild type mice [t(4)=5.484 and 5.044, for serine 473 and threonine 389, respectively, p<0.05], whereas total levels of Akt were unaffected. These data indicate elevated activity of PI3K and mTORC2 in homozygous mutants. Next, we examined the activity of mTORC1 by analyzing targets such as ribosomal protein S6 and Grb10. Levels of phospho-S6 were noticeably increased in homozygous NEX-Pten mice (fig. 5b). Total levels of Grb10, a newly identified substrate of the mTORC1 that is stabilized by phosphorylation [21,22], were also significantly elevated in samples from homozygous mutants [t(8)=4.084, p<0.004] (fig. 5b). The data indicate elevated mTORC1 activity in homozygous NEX-Pten mutant mice. No changes in PI3K or mTOR signaling were observed in the cerebellum of homozygous NEX-Pten mice, as expected (data not shown). Together, our data demonstrate that the loss of Pten in excitatory neurons of the forebrain leads to elevated activity of the PI3K/Akt/mTOR signaling cascade.
Fig. 5.
Upregulation of the PI3K/Akt/mTOR signaling pathway in the forebrain of newborn homozygous NEX-Pten mice. a Representative Western blots of Pten, phospho- Akt serine 473 (p-Akt Ser473), phospho- Akt threonine 389 (p-Akt Thr389) and total Akt in NEX-Pten littermates of the indicated genotypes. b Representative Western blots of Pten, phospho-ribosomal protein S6 (Serine 240/244) (p-S6), Grb10 and actin in NEX-Pten littermates of the indicated genotypes. Loss of Pten in homozygous mutant mice correlates with the increased phosphorylation of Aktand mTOR targets. Images are representative of data obtained from 3–6 animals per genotype.
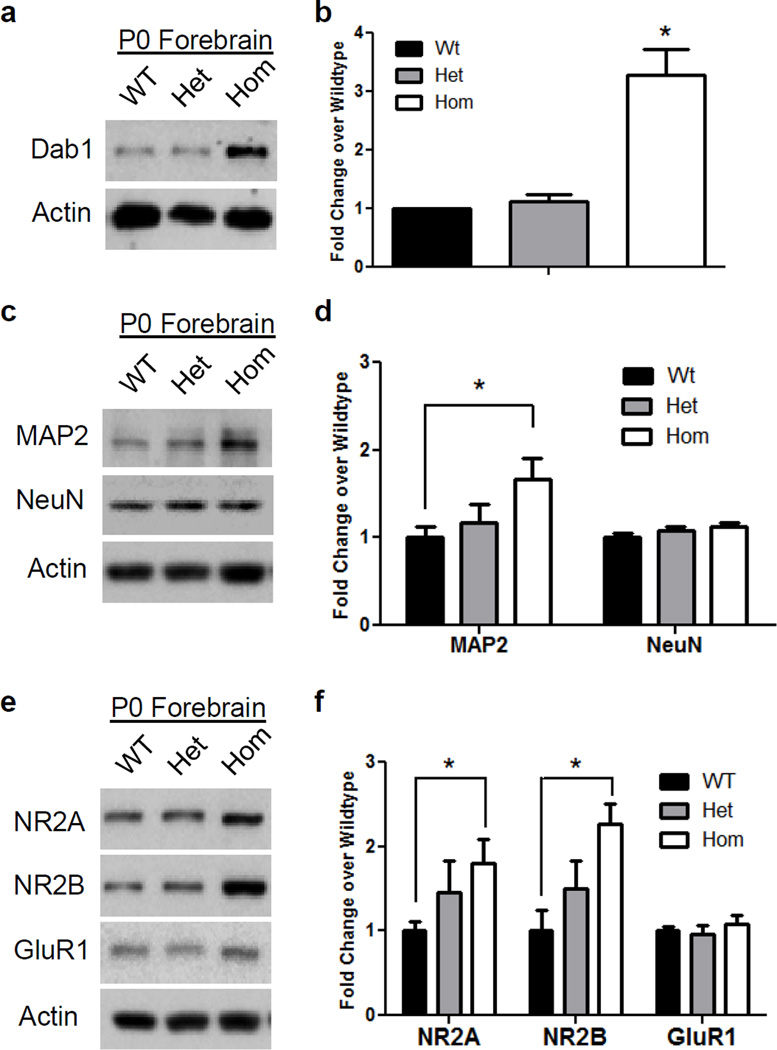
To investigate the consequence of the excessive activation of the PI3K/Akt/mTOR signaling pathway, we examined several key proteins that are involved in different aspects of brain development, such as migration, differentiation and synaptic function. All proteins examined were normalized to the levels of actin to take into account potential changes in protein levels or loading errors. Given the distortion of cellular layers observed in homozygous mutant mice, we examined the levels of Dab1, an adapter protein that is a critical mediator of Reelin in the control of layer formation [20]. We found that Dab1 levels were significantly increased approximately 3 fold in the forebrain of homozygous, but not heterozygous, NEX-Pten mice [one sample t-test: t(2)=5.267, p<0.03] (fig. 6a, b). To examine the consequences of Pten deletion on neuronal differentiation, we analyzed the expression levels of mature neuron markers, such as MAP2 and NeuN. We found that homozygous Pten knockout mice displayed significantly increased levels of MAP2 in the cortex [t(10)=2.473, p<0.05], but had similar levels of NeuN, compared to wild type littermates (fig. 6c, d). Heterozygous mutants, on the other hand, displayed levels of MAP2 and NeuN that were similar to those of wild type samples. To investigate the effects of Pten deficiency in excitatory neurons on synaptic function, we focused on the expression of glutamate receptors in the forebrain of newborn pups. We found that levels of the NMDA receptor subunits NR2A and NR2B were significantly increased in homozygous Pten knockout [t(10)=2.627 for NR2A and t(8)=3.659 for NR2B, p<0.05] compared to wild type mice (fig. 6e, f). Levels of NR2A and NR2B in heterozygous mice appeared slightly increased compared to wild type samples, however, the difference did not reach statistical significance (p>0.05). Levels of the AMPA receptor GluR1 were unaltered in heterozygous as well as homozygous mutant mice (p>0.05) (fig. 6e, f), suggesting that Pten deletion in excitatory forebrain neurons specifically affects the expression of NMDA receptor subunits.
Fig. 6.
Abnormal expression of proteins involved in brain development and synaptic plasticity in the forebrain of newborn homozygous NEX-Pten mice. Representative Western blots show upregulation of Dab1 (a), MAP2 and NeuN (c), NR2A, NR2B and GluR1 (e) in homozygous (Hom) compared to heterozygous (Het) and wild type (WT) mice. Quantitative analysis of data obtained from 5–6 mice per genotype confirms that Dab1 (b), MAP2 (d), NR2A and NR2B levels (f) normalized to actin are significantly higher in homozygous than in wild type samples (p<0.05).
Discussion
The Pten phosphatase has been previously implicated in several aspects of neuronal development, including soma size determination, axon specification, dendrite branching and synapse formation (reviewed by [23]). These neuroanatomical changes correlate with altered behavior and neuronal physiology in Pten mutant mice, which are reminiscent of autistic-like traits and epilepsy in human patients carrying PTEN mutations. However, the molecular mechanisms underlying the etiology of these neurological abnormalities have not been elucidated. Here we generated a novel conditional knockout mouse in which Pten was deleted in all excitatory neurons of the forebrain by the selective expression of Cre driven by the NEX promoter [19]. Our initial characterization of this line revealed that homozygous mutants are severely affected by macrocephaly and neuronal hypertrophy, and die prematurely shortly after birth. Postnatal treatment with the mTORC1 inhibitor rapamycin prolonged survival, demonstrating the role of this signaling complex in the overt phenotype of the mutant mice. Premature death, however, did not prevent us from analyzing the development of the neonatal cerebral cortex at the anatomical and molecular level. Notably, we found that loss of Pten in excitatory forebrain neurons did not affect their ability to migrate into the cortical plate and positioning into appropriate cellular layers, consistent with our previous observations in NS-Pten mice [15]. However, overmigration into the marginal zone and a distortion of cortical cellular layers was observed in homozygous mutants. A similar overmigration phenotype was previously reported in Cullin5 knockout mice, which express high levels of the Reelin signaling protein Dab1 [24]. The Reelin-Dab1 signaling pathway is required for radial migration and layer formation. Levels of Dab1 are regulated by ubiquitination and Cullin-5 proteasome degradation, and these processes are believed to ensure that neurons terminate their upward migration once the marginal zone is reached [25]. We found a striking increase in the levels of this protein in the homozygous mutant forebrain, which could account for the overmigration phenotype. Our data are thus consistent with the model that Dab1 downregulation is necessary to arrest radial migration at the top of the cortical plate. Preliminary data indicate that the Dab1 overexpression is not due to increase in mRNA levels (G.H.L., unpublished results), suggesting that upregulation of one or more components of the PI3K/Akt/mTOR pathway alters protein translation or stability.
Pten loss in homozygous NEX-Pten mice also caused specific increases in the levels of the dendritic marker MAP2, consistent with previous reports of dendrite hypertrophy in upper cortical layers [26–28]. It is not presently clear whether MAP2 protein upregulation causes dendrite hypertrophy or simply reflects the overgrowth of these processes. We also found that forebrain regions of NEX-Pten homozygous mutants express higher levels of the NMDA receptor subunits 2A and 2B, whereas levels of other glutamate receptor subunits, such as GluR1, were unaffected. Given the role of the NMDA receptor in synaptic plasticity, these findings reveal a candidate molecular mechanism by which Pten controls this process in excitatory forebrain neurons.
Our present findings that Pten loss alters the expression of NMDA receptor subunits are consistent with previous studies indicating that Pten is present in synaptic fractions, where it physically associates with this receptor and modulates synaptic plasticity [29,30]. Further studies are required to determine whether the elevated levels of NR2A and NR2B found in NEX-Pten homozygous mutant mice result from mTOR-mediated increases in local protein translation, or by increased protein stability. In a recent study, we found that levels of NR2A and NR2B were decreased in NS-Pten knockout mice, in which gene deletion occurred in selected neuronal progenitors [30]. This apparent discrepancy could be due to the different cell specificity of the knockout lines. Alternatively, it could be due to the different age of the mice at the time of analysis: adult mice were analyzed in our previous study, whereas newborn mice are analyzed here. In light of the present findings, we hypothesize that a loss of Pten leads to an initial increase in NR2A and NR2B levels in developing neurons, which may trigger compensatory, homeostatic mechanisms leading to reduced expression of these synaptic proteins in the adult brain. Given the early lethality of our NEX-Pten mutant mice, we unfortunately are not able to directly test this hypothesis in vivo. Future studies with dissociated neuronal cultures derived from homozygous NEX-Pten mutant embryos will be required to address this issue.
Germline mutations in PTEN in humans have been associated with a variety of neurological disorders, including autism and epilepsy [31–35]. Since these are heterozygous mutations, it will be interesting to determine whether our newly generated NEX-Pten heterozygous mice, which appear normal at first glance, exhibit behavioral, physiological or molecular synaptic deficits upon closer examination at later ages. In this study, we noted a modest increase in NR2A and NR2B levels in newborn NEX-Pten heterozygous mice. However, the values did not reach statistical significance. Additional analysis of animals at different ages and more quantitative methods of analysis may be required to obtain conclusive data on the expression of these receptor subunits in heterozygous mice. Furthermore, it will be worthwhile to investigate whether these mice are more susceptible to seizure in response to insults, such as traumatic brain injury or chemical convulsants. Since heterozygous NEX-Pten mice appear healthy and have a normal life span, these experiments are feasible and are likely to be informative.
In conclusion, we generated a novel conditional knockout mouse line that will facilitate further analysis of the role of Pten in the maturation of excitatory neurons of the forebrain. These studies will lead to a better understanding of the mechanisms underlying cognitive dysfunction and epilepsy associated with PTEN mutations in human patients.
Acknowledgements
The authors would like to thank Drs. Klaus-Armin Nave and Sandra Goebbels for the gift of NEX-Cre knockin mice, and Prescott Leach for his assistance with statistical analysis and critical reading of the manuscript. This work was supported in part by a Research Grant from the New Jersey Governors Council for Medical Research and Treatment of Autism and a Challenge Award from the Citizens United for Research in Epilepsy (G.D.), a predoctoral Fellowship from the Epilepsy Foundation (C.N.S.), NIH/NINDS RO1 NS039943 and NS049427 (A.E.A.), the Foundation of UMDNJ Society of Research Scholars and a Leathem Steinetz Stauber McCallum 2011 Summer Research Award (T.M.K.).
References
- 1.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of pkb/akt-dependent cell survival by the tumor suppressor pten. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 2.Maehama T, Dixon JE. The tumor suppressor, pten/mmac1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 3.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 4.Inoki K, Li Y, Zhu T, Wu J, Guan KL. Tsc2 is phosphorylated and inhibited by akt and suppresses mtor signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 5.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, tuberin and hamartin, control mtor signaling by acting as a gtpase-activating protein complex toward rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 7.Laplante M, Sabatini DM. Mtor signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of akt/pkb by the rictor-mtor complex. Science (New York, NY. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 9.Gong R, Park CS, Abbassi NR, Tang SJ. Roles of glutamate receptors and the mammalian target of rapamycin (mtor) signaling pathway in activity-dependent dendritic protein synthesis in hippocampal neurons. J Biol Chem. 2006;281:18802–18815. doi: 10.1074/jbc.M512524200. [DOI] [PubMed] [Google Scholar]
- 10.Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang N, El-Hayek YH, Gomez E, Wan Q. Phosphatase pten in neuronal injury and brain disorders. Trends Neurosci. 2007;30:581–586. doi: 10.1016/j.tins.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the pten tumor suppressor gene in vivo. Science (New York, NY. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 13.Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J, Tsao MS, Shannon P, Bolon B, Ivy GO, Mak TW. Deletion of pten in mouse brain causes seizures, ataxia and defects in soma size resembling lhermitte-duclos disease. Nat Genet. 2001;29:396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- 14.Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ. Pten regulates neuronal soma size: A mouse model of lhermitte-duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- 15.Ljungberg MC, Sunnen CN, Lugo JN, Anderson AE, DArcangelo G. Rapamycin suppresses seizures and neuronal hypertrophy in a mouse model of cortical dysplasia. Dis Model Mech. 2009;2:389–398. doi: 10.1242/dmm.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunnen CN, Brewster AL, Lugo JN, Vanegas F, Turcios E, Mukhi S, Parghi D, DArcangelo G, Anderson AE. Inhibition of the mammalian target of rapamycin blocks epilepsy progression in ns-pten conditional knockout mice. Epilepsia. 2011;52:2065–2075. doi: 10.1111/j.1528-1167.2011.03280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luikart BW, Schnell E, Washburn EK, Bensen AL, Tovar KR, Westbrook GL. Pten knockdown in vivo increases excitatory drive onto dentate granule cells. J Neurosci. 2011;31:4345–4354. doi: 10.1523/JNEUROSCI.0061-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA. Genetic targeting of principal neurons in neocortex and hippocampus of nex-cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- 20.D'Arcangelo G. Reelin mouse mutants as models of cortical development disorders. Epilepsy Behav. 2006;8:81–90. doi: 10.1016/j.yebeh.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mtor-regulated phosphoproteome reveals a mechanism of mtorc1-mediated inhibition of growth factor signaling. Science (New York, NY. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies grb10 as an mtorc1 substrate that negatively regulates insulin signaling. Science (New York, NY. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Diepen MT. Eickholt BJ: Function of pten during the formation and maintenance of neuronal circuits in the brain. Dev Neurosci. 2008;30:59–64. doi: 10.1159/000109852. [DOI] [PubMed] [Google Scholar]
- 24.Feng L, Allen NS, Simo S, Cooper JA. Cullin 5 regulates dab1 protein levels and neuron positioning during cortical development. Genes Dev. 2007;21:2717–2730. doi: 10.1101/gad.1604207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerjan G. Gleeson JG: A missed exit: Reelin sets in motion dab1 polyubiquitination to put the break on neuronal migration. Genes Dev. 2007;21:2850–2854. doi: 10.1101/gad.1622907. [DOI] [PubMed] [Google Scholar]
- 26.Chow DK, Groszer M, Pribadi M, Machniki M, Carmichael ST, Liu X, Trachtenberg JT. Laminar and compartmental regulation of dendritic growth in mature cortex. Nat Neurosci. 2009;12:116–118. doi: 10.1038/nn.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser MM, Bayazitov IT, Zakharenko SS, Baker SJ. Phosphatase and tensin homolog, deleted on chromosome 10 deficiency in brain causes defects in synaptic structure, transmission and plasticity, and myelination abnormalities. Neuroscience. 2008;151:476–488. doi: 10.1016/j.neuroscience.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Blundell J, Ogawa S, Kwon CH, Zhang W, Sinton C, Powell CM, Parada LF. Pharmacological inhibitionof mtorc1 suppresses anatomical, cellular and behavioral abnormalities in neural-specific pten knock-out mice. J Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurado S, Benoist M, Lario A, Knafo S, Petrok CN, Esteban JA. Pten is recruited to the postsynaptic terminal for nmda receptor-dependent long-term depression. EMBO J. 2010;29:2827–2840. doi: 10.1038/emboj.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventruti A, Kazdoba TM, Niu S, D'Arcangelo G. Reelin deficiency causes specific defects in the molecular composition of the synapses in the adult brain. Neuroscience. 2011;189:32–42. doi: 10.1016/j.neuroscience.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 31.Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline pten tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buxbaum JD, Cai G, Chaste P, Nygren G, Goldsmith J, Reichert J, Anckarsater H, Rastam M, Smith CJ, Silverman JM, Hollander E, Leboyer M, Gillberg C, Verloes A, Betancur C. Mutation screening of the pten gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conti S, Condo M, Posar A, Mari F, Resta N, Renieri A, Neri I, Patrizi A, Parmeggiani A. Phosphatase and tensin homolog (pten) gene mutations and autism: Literature review and a case report of a patient with cowden syndrome, autistic disorder and epilepsy. J Child Neurol. 2011 doi: 10.1177/0883073811420296. [DOI] [PubMed] [Google Scholar]
- 34.McBride KL, Varga EA, Pastore MT, Prior TW, Manickam K, Atkin JF, Herman GE. Confirmation study of pten mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010;3:137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- 35.Orrico A, Galli L, Buoni S, Orsi A, Vonella G, Sorrentino V. Novel pten mutations in neurodevelopmental disorders and macrocephaly. Clin Genet. 2009;75:195–198. doi: 10.1111/j.1399-0004.2008.01074.x. [DOI] [PubMed] [Google Scholar]